Abstract
The first virus-like particle to be tested for use as a vaccine carrier was based on the hepatitis B virus nucleocapsid protein. This viral subunit, while not infectious on its own, is a 36-nm particle that is highly immunogenic during a natural infection. The self-assembly and high degree of immunogenicity is maintained when expressed as a recombinant protein and, moreover, can confer a high degree of immunogenicity on foreign antigens linked to the particle, either chemically or genetically. This review describes the current state of the hepadnaviral core protein as a vaccine carrier.
Keywords: HBcAg, HBV subunit vaccine platform, virus-like particle, WHcAg
The growing body of knowledge on immune epitopes and the resulting increase in the number of neutralizing peptide epitopes identified has further enhanced interest in the use of these subunits for the design of prophylactic and therapeutic vaccines [1]. Indeed, vaccines that target peptide subunits of a pathogen offer a number of advantages, including the ability to precisely target B- and T-cell epitopes known to be neutralizing, rather than using the whole organism or even a complete protein from a given pathogen. Because the target epitopes are small they can easily be mutated to match any naturally occurring sequences in the wild population or escape mutants that arise. Additionally, subunit vaccines have the advantages of being easy to produce and purify, which yields lower costs, greater safety and improved stability compared with live-attenuated or killed pathogens. However, the use of peptide epitopes for vaccines is limited by the fact that these small antigens are poorly immunogenic and must be conjugated to an immunogenic carrier for delivery to the immune system. One method of delivering peptide antigen epitopes that has proven very successful is the virus-like particle (VLP), which is an inert, noninfectious, self-assembling particle [2].
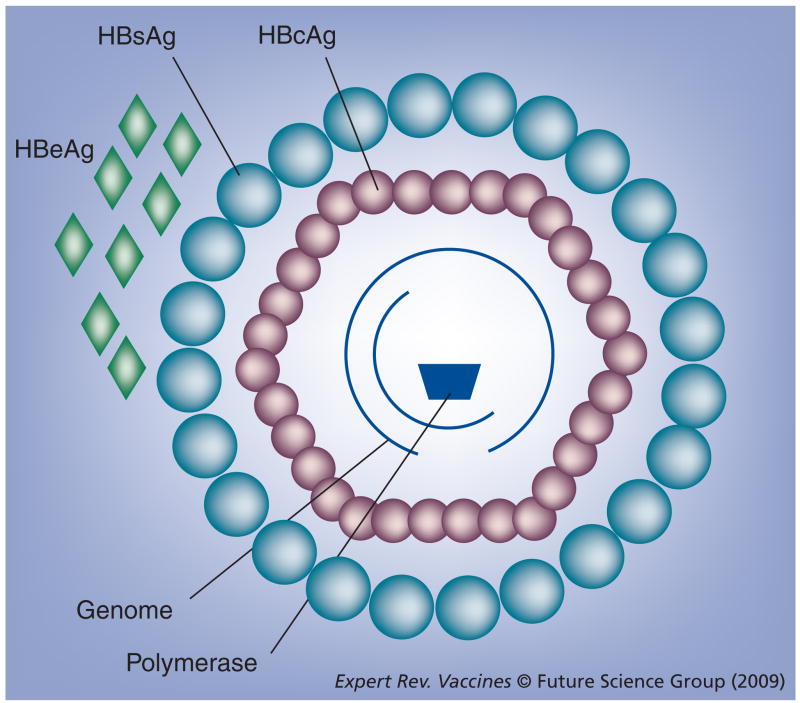
In the 1980s, HBV (Hepadnaviridae) provided the source for one of the prototypical VLPs when the HBV core antigen (HBcAg) was used to display a foreign epitope, namely one from the foot-and-mouth disease virus (Picornaviridae) [3]. HBcAg forms the nucleocapsid of the intact HBV virion, which is a small 42-nm virus surrounded by a lipid envelope with embedded HBV surface antigen (HBsAg) surrounding the HBcAg, which in turn encapsidates the partially double-stranded HBV genome [4,5]. HBV also encodes a splice variant of the core protein, known as the secreted e antigen (HBeAg), and a polymerase associated with the genome (Figure 1). The surface antigen has been used as a vaccine carrier, but HBcAg is significantly more immunogenic than HBsAg both during a natural infection [6] and as a recombinant protein [7]. Moreover, it is increasingly clear that this HBcAg-based VLP possesses a number of characteristics that make it highly favored as an efficient and flexible carrier for foreign immune epitopes.
Figure 1. Diagrammatic representation of the HBV virion.
Large outer circles represent HBsAg embedded in the lipid envelope. Smaller inner circles represent core proteins that form the HBcAg nucleocapsid, while diamonds represent the secreted form of the core protein, HBeAg (expressed during infection). The partial rings represent the partially dsDNA genome and the trapezoid represents the viral polymerase. The X protein is not represented.
HBcAg: HBV core antigen; HBeAg: HBV e antigen; HBsAg: HBV surface antigen.
The HBV core protein is a 21-kDa polypeptide, which spontaneously assembles into an icosahedral particulate structure of approximately 36 nm diameter made up of 120 homodimers, that is, 240 copies of the HBcAg protein. Crystallographic and cryoelectron microscopy studies have elucidated the structure of HBcAg particles and show that dimers align to form spikes on the surface of the particle, each composed of four long bundled α-helices connected by a loop [8–10]. We and others determined that the immunodominant B-cell epitope on HBcAg is localized around amino acids 76–82 [11,12]. This is consistent with the finding that a heterologous sequence inserted into the internal 75–83 loop region of HBcAg is significantly more antigenic and immunogenic than the same sequence positioned at the N- or C-terminus [12], and that the inserted sequence is more immunogenic than it is in the context of its native protein. The arginine-rich nucleic acid-binding domain in the C-terminal tail is not required for assembly of the particles, and as little as the first 140 amino acids of the core gene are sufficient to achieve assembly [13,14].
Researchers have faced two challenges in applying the HBcAg technology to VLP vaccine design for human use. First, this VLP is based on a human pathogen, so it will not be effective in the 450 million chronic HBV carriers worldwide and may be less effective for those already exposed to the virus. Second, many of the foreign epitopes inserted into the core gene destroy the self-assembly property of the HBcAg particle. The first challenge has been addressed by using the core particles of related hepadnaviruses that infect species other than humans, primarily woodchuck and duck hepadnavirus [15–17], although hepadnaviruses have also been reported in several squirrel species, herons, storks, kangaroos and even reptiles [4]. The second challenge has been addressed in two ways: thorough characterization and optimization of the features that limit assembly and by a novel technology that relieves internal stresses imposed by insertion of certain proteins into the loop of the HBcAg gene [18,19]. This review will cover the properties of hepadnavirus core proteins that make them well suited as vaccine carriers, summarized in Box 1, as well as how research on the core particles from different species has enhanced the technology.
Box 1. Characteristics indicating that hepadnaviral core proteins are excellent carrier moieties.
Efficient self-assembly into 36-nm particles
Fully amenable to Escherichia coli expression
Availability of free sulfhydryls on the particle surface
High immunogenicity during HBV infection in humans
No MHC nonresponder phenotypes identified
Noncytotoxic in humans
Highly immunogenic in animal models (i.e., >100 × vs HBsAg)
T-cell independence
Function as T-cell carriers for envelope antigens within the virion
Delivery of internalized TLR ligands directly to B cells
Flexibility of many insertion sites and C-terminal modifications enable greater than 95% efficiency of assembly*
Rodent core proteins circumvent HBcAg-specific immune tolerance in humans*
Use of rodent core proteins will not compromise use of the anti-HBcAg diagnostic assay*
Characteristics common only to nonhuman hepadnaviral core proteins.
Note: due to conservation at the amino acid level (i.e., 67%), many characteristics are shared among the human and rodent hepadnaviral core proteins except, of course, bullet points 4 and 6, which are based on observation in natural human infection. HBcAg: HBV core antigen; HBsAg: HBV surface antigen; TLR: Toll-like receptor.
Structure & flexibility to carry foreign epitopes
The hepadnaviral core protein has proven highly flexible, partly owing to the multiple roles it plays during the natural hepadnaviral infection. Evidence of the multiple roles is seen in the two in-frame initiation codons of the hepadnaviral core gene. Translation from the first codon results in synthesis of the precore protein, which includes 29 N-terminal residues that represent a hydrophobic signaling sequence that directs the nascent precore protein to the endoplasmic reticulum. The precore protein is processed and secreted as HBeAg, which possesses immunologic properties strikingly different from HBcAg [20]. Initiation of translation from the second AUG results in expression of the 21-kDa core protein, which is not proteolytically processed. This protein self-assembles into the 240-subunit icosahedral HBcAg nucleocapsid particle, which in conjunction with the HBV polymerase, is capable of specifically encapsidating the viral pregenomic RNA [8,21]. This assembly process occurs not only during the course of native virion assembly, but also when expressed in heterologous systems, both prokaryotic and eukaryotic, including expression in plants [22]. When expressed in exogenous systems, the C-terminal nucleic acid-binding domain of the full-length core particle will encapsidate host RNA of approximately 100–3000 bp in length during assembly [13].
While the hepadnaviral e protein and core particle are serologically distinct, these antigens are cross-reactive at the T helper (Th) cell level of recognition since most of their primary sequence is shared as the same open reading frame (ORF) [23]. For example, in B10.S (H-2S) mice and B10 (H-2b) mice, the dominant Th recognition sites are at amino acids 120–131 and 129–140, respectively. Both of these sequences are identical between HBeAg and HBcAg as they are contained within the overlapping ORF region of the proteins.
The core antigen protein contains four cysteine residues, all of which are involved to varying extents in forming disulfide bonds between monomers (although not within a single polypeptide chain), but experiments on HBcAg have shown that formation of these disulfide bonds is not required for particle assembly [24]. This feature allows flexibility to genetically insert heterologous epitopes into the core particle without the consideration for preserving the native disulfide bridges. The cysteine at position 48 in HBcAg is known to be exposed and approximately half of these residues are tied up in intermonomer disulfide bonds, leaving the remaining half of the residues available to react with alkylating agents [24]. This means that there are approximately 120 cysteine residues on the surface of each core particle (at position 48 and conserved in the rodent hepadnaviral core proteins) with free sulfhydryl groups as ‘handles’ to chemically couple antigens.
In addition to the native cysteine residues, it is possible to genetically engineer reactive groups onto the core particle and chemically link peptides and larger protein domains. Inserting a lysine residue into the immunodominant loop allowed chemical conjugation of peptides to the HBcAg VLP, resulting in a sufficiently high peptide density to elicit a strong antibody response to the coupled peptide in the absence of exogenous adjuvants [25,26]. This HBcAg VLP conjugation technology has been used to couple a 23-amino acid peptide corresponding to the extracellular domain of the M2 protein for use as a universal influenza A vaccine. However, this chemically conjugated VLP elicited only weak protective efficacy in mice [27]. A recently published report examined genetic fusion of M2 to HBcAg, and chemical coupling of M2 to the carboxyl groups of M2. By coupling to carboxyl groups on the HBcAg particle, 4300 sites are available and the researchers achieved 32% loading for approximately 1400 M2 peptides per particle, compared with the 100% efficiency of genetic fusion yielding 240 M2 peptides per HBcAg particle. Immunizing with an equal mass of M2 peptide revealed that genetic fusion of M2 to HBcAg elicited higher anti-M2 peptide antibody responses and more potent protection in a mouse model of influenza infection [28].
The ability of the hepadnaviral core particle to self-assemble, despite disruption of the native disulfide bridges and insertion of a lysine residue in the loop, underscores the resiliency of the protein’s assembly function, suggesting that the core particle is well suited for genetic insertion of foreign antigens. This theory was born out by the early success in inserting a wide variety of foreign epitopes, as large as 55 amino acids, into HBcAg and achieving assembly of the chimeric particles [3,12,29–37]. However, there are limits to the genetic insertions tolerated by the hepadnaviral core proteins and many epitopes inserted into HBcAg can disrupt assembly, causing some groups to turn to chemical linkage systems [25]. The disruption by insertion of foreign sequences can affect both the expression level and/or the correct assembly of chimeric core particles. This phenomenon has been investigated by analyzing the biochemical and biophysical properties of the amino acid sequence inserted into chimeric particles, revealing that parameters such as length, hydrophobicity, high β-strand index or large volume may impede the proper folding and assembly of chimeric HBcAg particles [38]. Our own work has indicated that insert charge is the most important characteristic in terms of determining chimeric core particle assembly (see later).
Our laboratory has approached the problem of some insertions abrogating assembly by exploiting the inherent flexibility of the hepadnaviral core particle. We have concentrated our efforts on the core particle from one of the rodent hepadnaviruses, namely the woodchuck hepadnaviral core antigen (WHcAg), which is 67% identical and 78% similar to HBcAg at the amino acid level. Multiple epitopes were systematically tested by insertion at numerous sites within and outside the immunodominant region of the WHcAg protein. These epitopes and insertion sites were tested with a panel of modified C-terminal tails and we discovered that the optimal insertion site and C-terminal tail can be different for each epitope sequence [18]. Thus, by determining the correct combination of insertion site and C-terminal tail, WHcAg particles could be successfully assembled for most inserted epitopes. An additional discovery allowed the successful particle assembly for more than 95% of the epitopes chosen for genetic insertion into WHcAg. It was noted that the presence of highly basic amino acids in an insert epitope correlated with poor or negative assembly of chimeric core particles. However, acidifying the insert with amino acids (such as glutamic acid), either flanking the insert or substitution at noncritical sites within the insert, rescued assembly of the core particle that would not assemble with the basic insert [18]. This acidification worked for HBcAg and WHcAg, as well as the core proteins from ground squirrel and arctic ground squirrel hepadnaviruses.
Another successful approach has been developed to insert large domain epitopes and even entire proteins into the immunodominant region of the HBcAg protein. For proteins whose N- and C-termini are sufficiently closely juxtaposed, it is possible to engineer them into the HBcAg loop region and achieve independent folding of the core and fused proteins and also assembly of the chimeric core particles [39,40]. However, for proteins whose N- and C-termini are distant in the tertiary structure, long linkers must be used and chimeric HBcAg particle assembly is perturbed, giving rise to irregular particles and various aggregates [41]. This was addressed by cleaving the HBcAg protein in the immunodominant loop adjacent to the inserted protein. Cleaving the protein and relieving the constraint on the HBcAg monomers makes particle assembly significantly more efficient [19]. This technology allows the genetic fusion of large domains or proteins to the HBcAg particle in addition to the incorporation of small targeted epitopes.
Inherent immunogenicity
The HBcAg particle is known to be highly immunogenic in humans, because during an HBV infection virtually 100% of patients produce high titer antibody to it despite the fact that the core particle is an internal virion structure [42]. In both acute and chronic HBV infection, the antibody titer raised to HBcAg is significantly greater than the antibody raised even to the envelope and may be the only component of the virus to induce antibody production in patients with asymptomatic chronic infections [43]. The core particle itself is apparently not cytotoxic and is well tolerated, as the anti-HBV core protein (HBc) response is observed both in the presence or absence of liver damage [44].
Research on HBcAg VLPs has been aided by the fact that the high immunogenicity observed in humans is similarly seen in mice [23,45]. Moreover, in mice the core particles derived from rodent hepadnaviruses (woodchuck, ground squirrel and arctic ground squirrel) are at least as immunogenic as HBcAg [16]. In testing the HBcAg and three rodent hepadnaviral core particles in a broad panel of mouse strains, a strong IgG anti-core response was measured and no nonresponder strains were identified, consistent with the data on the anti-HBc response in humans. The enhanced immunogenicity of core particles seems to derive from factors intrinsic to the core particle structure.
Classic cognate T-cell help requires direct T-cell–B-cell interaction. Therefore, the antigenic determinants recognized by Th and B cells must be covalently linked on a single molecule. However, particulate antigens, such as viruses, may represent a special case, since B cells expressing an immunoglobulin receptor for any determinant exposed on the particle would internalize all the molecules of a given particle. Consequently, T-cell recognition of an epitope within a particle would be predicted to supply Th-cell function to numerous B-cell specificities, representing intermolecular/intrastructural T-cell help. In view of this phenomenon, we examined the ability of HBcAg-primed T cells to function as Th cells for antibody production to envelope HBsAg epitopes, even though HBsAg and HBcAg are on distinct molecules, albeit within the same virions. Mice primed with HBcAg and subsequently challenged with subimmunogenic amounts of virions (HBV) produced anti-HBs (HBV surface protein). This result indicated that HBcAg-primed T cells could function to help anti-envelope antibody production, and the Th-cell activity required the HBcAg and HBsAg to be present within the same particle, since the mixture was ineffective [7]. To confirm the T-cell nature of this effect, an identical experiment was performed using a peptide containing the HBcAg-derived synthetic T-cell recognition site, p120–131, as the priming antigen. Mice primed with p120–131 and challenged with HBV produced anti-envelope antibodies, whereas mice primed with p120–131 and challenged with an HBcAg/HBsAg mixture did not. These results were the first indication that HBcAg may represent an efficient carrier moiety for heterologous epitopes. The results of these experiments are consistent with serological data suggesting that HBcAg/HBe-specific Th cells may also mediate anti-envelope antibody production during HBV infection [43].
The high immunogenicity of hepadnaviral core particles can also be attributed to functions independent of T cells. We have examined the ability of HBcAg to activate B cells directly by immunizing euthymic and athymic mice with various doses of HBcAg [46]. Euthymic mice produced dose-dependent anti-HBc antibody at 10 days and a four- to 16-fold increase in anti-HBc titer at 24 days. However, athymic mice also produced dose-dependent anti-HBc antibody at 10 days after immunization, but showed no increase in the anti-HBcAg titer at 24 days. To determine if T-cell independence required the HBcAg to be particulate, we also immunized euthymic and athymic mice with the nonparticulate HBeAg. Athymic mice were nonresponsive to HBeAg, indicating that the response to HBeAg was T-cell dependent and that T-cell independence requires the particulate structure. However, the particulate protein structure, while necessary, was not itself sufficient for T-cell independence as illustrated by the fact that the HBsAg (a 22-nm particle) is a T cell-dependent antigen. Rather, the particulate structure more likely acts to display the epitopes in a repetitive array with spacing for optimal crosslinking of the B-cell receptor [47–49].
Since HBcAg can act as a T-cell-independent antigen and activate B cells directly in the absence of T cells, it was of interest to determine if this T-cell independence could be conveyed to an antigen coupled to HBcAg. BALB/c nu/nu (athymic) mice were immunized with dinitrophenol (DNP) conjugates: HBcAg–DNP1 or BCG–DNP50. BALB/c nu/nu mice produced anti-DNP antibodies after HBcAg–DNP1 but not after BCG–DNP50 immunization. This indicates that the T-cell-independent characteristic of HBcAg can be conveyed to a normally T-cell-dependent antigen by coupling to the particle. Other putative T-cell carrier moieties (i.e., BCG, tetanus toxoid or diphtheria toxoid) do not possess this characteristic since these proteins are not T-cell-independent antigens.
A variety of theories have been proposed to explain the robust immunogenicity of core particles, including binding of the C-terminal domain to membrane heparin sulfates, interaction with Toll-like receptor (TLR)2, and contaminating TLR ligands derived from the bacterial purification [50–53]. However, the hepadnaviral core particle characteristic most likely to explain the enhanced immunogenicity, especially in terms of antibody production, is that B cells are the primary antigen-presenting cell (APC) for HBcAg [48]. This was a surprising finding since dendritic cells (DCs) are normally regarded as being the primary professional APC. Not only are B cells the primary APC for core particles, but we further found that specific subsets, splenic B1a and B1b cells, are the most efficient presenters for core particles [54]. Naive B1a cells are capable of binding HBcAg particles, perhaps explaining their high efficiency in presentation of core particles. Conversely, both in vitro cell assays and in vivo immunizations demonstrated that DCs are unable to efficiently present core particles because they do not internalize this particulate antigen at physiologic concentrations [54].
A significant consequence of B cells being the primary APC for core particles is that immune stimulants integral to the particle, for example, the RNA encapsidated by the core particles, can be delivered directly to the antibody-producing cell along with the antigen, which leads to co-ligation of the B-cell receptor and TLR and the resulting immune synergy [54,55]. We have demonstrated that the host RNA (Escherichia coli, yeast and mammals) encapsidated in core particles acts as a TLR7 ligand and enhances immunogenicity of the antigen [54]. For example, the full-length HBcAg containing RNA elicited a 25-fold higher antibody response than the truncated version that lacks encapsidated RNA. However, this increase in immunogenicity is entirely abrogated in TLR7 knockout mice, demonstrating that the improved immune response to full-length core particles is attributable to the encapsidated RNA and that this RNA acts through TLR7.
Although anti-HBc antibody production can occur through a T-cell-independent pathway, the protein nature of HBcAg also allows for T-cell recognition, unlike a number of strictly T-cell-independent antigens. In fact, HBcAg represents a very efficient T-cell immunogen. The order of T-cell responsiveness among a panel of B10, H-2 congenic strains correlates with their ability to produce anti-HBc antibody in vivo [45]. It is noteworthy that HBcAg-primed T cells of high responding strains (i.e., B10.BR, B10.S and B10.D2) demonstrate HBcAg-specific T-cell activation (i.e., IL-2 production) at an in vitro HBcAg concentration as low as 0.03 ng/ml, which is approximately 100-fold more efficient than HBsAg-specific T cells from similarly high responder strains. This characteristic is useful in vaccine design because foreign CD4+ T-cell sites can be fused to the core particle and are functional [56].
Insertion of foreign epitopes into core particles
The immunologic characteristics of HBcAg have long been known to be very favorable for its use as a vaccine carrier. The multiplicity of epitopes fused to the HBcAg gene over the last 20 years has grown past the point of enumeration being informative. However, a relatively recent list has been compiled [57]. The most recent advance in the use of core genes as vaccine carriers has been the development of core particles derived from the rodent hepadnaviruses. The woodchuck-derived WHcAg has proven to be at least as immunogenic as the human-derived HBcAg, but is not burdened by the problem of pre-existing immunity and, more importantly, avoids immune tolerance in the human population since WHcAg is minimally cross-reactive with HBcAg at the T-cell level [15,16].
We have subjected WHcAg chimeric particles to the same detailed analysis applied to HBcAg, testing insertion of a large panel of model epitopes at various positions on the WHcAg particle. Model epitopes were genetically inserted at the N- and C-terminal ends of the core gene as well as at several sites around the immunodominant spike, and both accessibility and immunogenicity were assessed by the ability to bind monoclonal antibodies and raise a polyclonal antibody response in mice, respectively. As was previously found for HBcAg chimeric particles, we found that insert position has a significant influence on the display and immunogenicity of the epitope carried by the WHcAg particle [12,18]. The overall trend is that C-terminally fused epitopes are the least well exposed and least immunogenic, while N-terminally fused eptiopes are better exposed and more immunogenic, and insertion of epitopes into the loop provides for the best accessibility and highest immunogenicity. The precise insertion point within (or adjacent to) the immunodominant spike that provides the best immunogenicity varies by epitope and, similar to the case for optimal assembly, is influenced by the sequence of the C-terminal domain [18]. The deliberate and systematic analysis of the suitability of multiple insertion sites for various inserted sequences has enabled the routine use of the full-length core gene or one that includes the C-terminal domain to bind RNA and encapsidate it into the chimeric particle. This RNA has the advantage of producing a better antibody response against the insert as a result of the integral TLR signaling [54].
In addition to the knowledge derived from the thorough and systematic characterization of the WHcAg particle as a vaccine platform, this rodent hepadnaviral particle has the inherent advantage of being derived from a nonhuman pathogen. The cross-reactivity between HBcAg and WHcAg (as well as the core particles derived from the ground squirrel and arctic ground squirrel hepadnaviruses) have been directly assessed. Antibody against HBcAg does not cross-react with WHcAg and shows very little cross-reactivity with the other rodent hepadnaviral core particles, thus pre-existing antibody to HBcAg in the human population should not interfere with the WHcAg as a vaccine carrier [16]. HBcAg and WHcAg are only partially cross-reactive at the CD4+ T-cell level. Therefore, use of the rodent core particles can circumvent immune tolerance to the HBcAg present in the 450 million chronic HBV carriers worldwide [15].
Application of core particle VLPs as human vaccine carriers
Studies in the 1980s demonstrated that the repeat sequences in circumsporozoite coat (CS) protein are the dominant target of protective antibodies against human and rodent malaria parasites, Plasmodium falciparum and Plasmodium berghei, respectively [58–61]. We have inserted the DNA sequence encoding a P. berghei CS repeat sequence, that is, (DP4NPN)2, into a recombinant HBcAg to produce chimeric vaccine particles. P. berghei is the causative agent of rat malaria and is capable of infecting mice, and therefore can be used to study protection in a very accessible model.
The P. berghei CS repeat sequence inserted into the immunodominant loop of HBcAg was expressed in E. coli and purified chimeric HBcAg–CS particles were found to elicit high titer anti-CS IgG antibodies. In challenge studies, mice immunized with HBcAg–CS showed 100% protection against P. berghei, while control mice immunized with HBcAg or PBS showed no protection [56]. These results were confirmed and extended in a subsequent set of experiments in which the CS repeat epitope from the Plasmodium yoelii rodent malaria was inserted into the HBcAg and used to immunize mice. The HBcAg–CS specific for P. yoelii malaria also raised high anti-CS IgG antibodies and protected 90 and 100% of mice in separate challenge studies [62].
The successful production of a model malaria vaccine based on a hepadnaviral core particle laid the groundwork for the first use of an HBcAg-based vaccine in human clinical trials. Malariavax (also known as ICC-1132) is an experimental human malaria vaccine conceptually based on the HBcAg–CS rodent malaria vaccines described above. Malariavax is a VLP based on HBcAg and contains the P. falciparum (human malaria) CS repeat B-cell epitopes inserted in the immunodominant loop, as well as a universal human T-cell epitope from the P. falciparum CS fused to the C-terminus of the carrier. Based on promising animal data in mice and nonhuman primates [63–65], human clinical trials were initiated in which volunteers were vaccinated with either multiple doses in alum adjuvant or a single dose in the water-in-oil adjuvant, Montanide ISA 720 [66–69]. A single-dose vaccination elicited modest anti-CS antibody production in the majority of volunteers and only weak T-cell responses, so the lack of protection against malarial sporozoite challenge was unsurprising [69]. In animal studies, a booster immunization results in an increase in anti-CS antibody titer, so it is likely that the efficacy of this vaccine could be improved simply by using multiple vaccinations.
The other HBcAg-based VLP vaccine to have been tested in humans is ACAM-FLU-A, which targets the influenza A M2 protein, proposed to contain a universal influenza epitope, M2e, owing to its high sequence conservation [70,71]. While the results of the clinical trial have not yet been published in a peer-reviewed journal, Acambis announced that the trial demonstrated the vaccine to be safe and immunogenic. The vaccine, as described in the scientific literature, was constructed by fusing the M2e epitope to the N-terminus of HBcAg, consistent with this epitope being at the N-terminus of the native M2 protein [72]. In a mouse model, the HBcAg–M2 chimeric VLP provided 90–100% protection in a lethal influenza challenge study. Protection against lethal flu challenge can also be achieved by immunizing mice via the intranasal route [73].
The early promise and later success of the core particle-based VLPs demonstrated the value of self-assembling viral nucleocapsids for use as vaccine carriers and consequently other VLPs are being developed. VLPs based on the human papillomavirus (HPV; Papillomaviridae) and bacteriophage Qβ (Leviviridae) coat proteins have each been extensively developed as carriers of chemically conjugated epitopes [74,75]. Genetic fusion of foreign epitopes has been less widespread, but some success has been reported with the HPV capsid as well as with some novel VLPs derived from plant viruses [76–78].
Summary
The hepadnaviral core particle was among the first of the VLP vaccine carrier moieties, owing to its many advantageous characteristics. These characteristics include the robust self-assembly of a single protein into a 240-subunit particle, the ability to use yeast and bacterial expression systems, high immunogenicity, and the flexibility to genetically fuse foreign epitopes and still preserve the advantages of the native particle. The reasons core particles were originally chosen as promising entities for vaccine carriers are many of the same reasons that they continue to be developed and that, more than 20 years later, they remain one of the best-developed chimeric vaccine carrier moieties. The flexibility of the core gene and robustness of assembly have aided in developing new improvements to the platform, including the encapsidation of TLR ligands and fusion of larger and more difficult immunogens. Studies in vitro and in vivo have elucidated some of the underlying mechanisms of the core particles’ high immunogenicity, which should allow further enhancements to the technology. Two different vaccines based on hepadnaviral core particles have now been tested in clinical trials and were found to be safe, and improved dosing will likely provide more robust demonstration of effectiveness in humans.
Expert commentary
The use of killed and live-attenuated pathogens has provided many effective vaccines that have had a great positive impact on human health through the prevention of diseases. Some pathogens have proven resistant to this vaccine strategy for various reasons and the scientific community has developed new approaches, including the use of pathogen subunits known to be capable of eliciting a neutralizing immune response. A major limitation of this approach is that clean, safe proteins are generally not very immunogenic on their own. The most promising method of addressing this limitation has been the use of highly immunogenic, noninfectious VLPs to deliver the relevant epitopes to the immune system. The hepadnaviral core proteins are one of the most promising of the VLP platforms because they are so immunogenic, eliciting strong B-cell and T-cell responses, and they are sufficiently flexible to allow the recombinant or chemical inclusion of a wide variety of foreign insertions. The vaccines based on HBcAg VLPs have already show protective efficacy in animal models of malaria, influenza and Lyme disease. With the existing knowledge that low doses of these HBcAg-based VLPs are safe, the current focus is on finding the proper formulation for eliciting human immune responses strong enough to neutralize the targeted pathogens.
Five-year view
There is a great deal of interest in developing subunit vaccines and many are already in clinical trials. The indications are that an effective formulation for the HBcAg-based VLPs will soon be established and we are optimistic that this knowledge will lead to the demonstration of protective efficacy for one or more diseases. Such a demonstration will underscore the value of this vaccine platform, and we believe that in 5 years the result of this development will be that the focus will be in the process of shifting from assessments of safety and potency to identifying epitopes and disease indications that are best suited to the particular strengths of the HBcAg-based VLPs.
Key issues.
The use of subunit vaccines offers the advantages of increased safety and precisely targeted immune responses.
The HBV-based core protein was the first recombinant virus-like particle (VLP) shown to convey a high level of immunogenicity to a foreign peptide.
The hepadnaviral core proteins represent a highly immunogenic and very flexible platform for delivering poorly immunogenic subunits.
Rodent hepadnaviral core proteins posses all of the advantages of the human HBV core antigen (HBcAg), but avoid the problem of immune tolerance to a human pathogen.
VLPs based on HBcAg have shown protective efficacy in animal models.
The HBcAg VLP has been shown in multiple clinical trials to be safe, and additional formulations are likely to provide better immune responses in humans.
Footnotes
Financial & competing interests disclosure
David C Whitacre is an employee, and David R Milich is a founder and shareholder in VLP Biotech, Inc., which develops vaccines based on hepadnaviral core proteins. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
David C Whitacre, Email: dwhitacre@vrisd.org, Vaccine Research Institute of San Diego, 10835 Road to the Cure, Suite 150, San Diego, CA 92121, USA, Tel.: +1 858 581 3960, Fax: +1 858 581 3970 and VLP Biotech, Inc., 10835 Road to the Cure, Suite 155, San Diego, CA 92121, USA, Tel.: +1 858 272 8757, Fax: +1 858 272 9361.
Byung O Lee, Email: blee@vrisd.org, Vaccine Research Institute of San Diego, 10835 Road to the Cure, Suite 150, San Diego, CA 92121, USA, Tel.: +1 858 581 3960, Fax: +1 858 581 3970.
David R Milich, Email: dmilich@vrisd.org, Vaccine Research Institute of San Diego, 10835 Road to the Cure, Suite 150, San Diego, CA 92121, USA, Tel.: +1 858 581 3960, Fax: +1 858 581 3970.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1••.Peters B, Sidney J, Bourne P, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3(3):e91. doi: 10.1371/journal.pbio.0030091. The authors describe the conception, establishment, organization and goals of the Immune Epitope Database ( www.iedb.org), which contains a publicly-searchable database of known immune epitopes. The epitopes and the information about them are useful for the design of subunit vaccines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grgacic EV, Anderson DA. Virus-like particles: passport to immune recognition. Methods. 2006;40(1):60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Clarke BE, Newton SE, Carroll AR, et al. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature. 1987;330(6146):381–384. doi: 10.1038/330381a0. Original description of the recombinant virus-like particle (VLP) used to display a foreign peptide epitope. [DOI] [PubMed] [Google Scholar]
- 4.Howard CR. The biology of hepadnaviruses. J Gen Virol. 1986;67(Pt 7):1215–1235. doi: 10.1099/0022-1317-67-7-1215. [DOI] [PubMed] [Google Scholar]
- 5.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64(1):51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoofnagle JH, Gerety RJ, Barker LF. Antibody to hepatitis-B-virus core in man. Lancet. 1973;2(7834):869–873. doi: 10.1016/s0140-6736(73)92004-7. [DOI] [PubMed] [Google Scholar]
- 7•.Milich DR, McLachlan A, Thornton GB, Hughes JL. Antibody production to the nucleocapsid and envelope of the hepatitis B virus primed by a single synthetic T cell site. Nature. 1987;329(6139):547–549. doi: 10.1038/329547a0. First description of intermolecular/intrastructural T-cell help for HBV antigens. [DOI] [PubMed] [Google Scholar]
- 8.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386(6620):88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 9.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386(6620):91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 10.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3(6):771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 11.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63(2):798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Schodel F, Moriarty AM, Peterson DL, et al. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J Virol. 1992;66(1):106–114. doi: 10.1128/jvi.66.1.106-114.1992. Description of how insert position within HBV core antigen (HBcAg) affects the immunogenicity of the inserted foreign epitope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64(7):3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts NR, Conway JF, Cheng N, et al. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 2002;21(5):876–884. doi: 10.1093/emboj/21.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Billaud JN, Peterson D, Lee BO, et al. Advantages to the use of rodent hepadnavirus core proteins as vaccine platforms. Vaccine. 2007;25(9):1593–1606. doi: 10.1016/j.vaccine.2006.11.013. Here the use of the rodent hepadnaviral core proteins is presented as a VLP vaccine carrier to avoid the limitations of the HBcAg-based platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billaud JN, Peterson D, Schodel F, et al. Comparative antigenicity and immunogenicity of hepadnavirus core proteins. J Virol. 2005;79(21):13641–13655. doi: 10.1128/JVI.79.21.13641-13655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paoletti LC, Peterson DL, Legmann R, Collier RJ. Preclinical evaluation of group B streptococcal polysaccharide conjugate vaccines prepared with a modified diphtheria toxin and a recombinant duck hepatitis B core antigen. Vaccine. 2001;20(3–4):370–376. doi: 10.1016/s0264-410x(01)00364-4. [DOI] [PubMed] [Google Scholar]
- 18.Billaud JN, Peterson D, Barr M, et al. Combinatorial approach to hepadnavirus-like particle vaccine design. J Virol. 2005;79(21):13656–13666. doi: 10.1128/JVI.79.21.13656-13666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Walker A, Skamel C, Vorreiter J, Nassal M. Internal core protein cleavage leaves the hepatitis B virus capsid intact and enhances its capacity for surface display of heterologous whole chain proteins. J Biol Chem. 2008;283(48):33508–33515. doi: 10.1074/jbc.M805211200. By splitting the core protein into two halves, the authors show that very large inserts can be accommodated at the site of the immunodominant loop. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou JH, Laub O, Rutter WJ. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci USA. 1986;83(6):1578–1582. doi: 10.1073/pnas.83.6.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziermann R, Ganem D. Homologous and heterologous complementation of HBV and WHV capsid and polymerase functions in RNA encapsidation. Virology. 1996;219(2):350–356. doi: 10.1006/viro.1996.0260. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Santi L, LePore K, Kilbourne J, Arntzen CJ, Mason HS. Rapid, high-level production of hepatitis B core antigen in plant leaf and its immunogenicity in mice. Vaccine. 2006;24(14):2506–2513. doi: 10.1016/j.vaccine.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 23.Milich DR, McLachlan A, Stahl S, et al. Comparative immunogenicity of hepatitis B virus core and E antigens. J Immunol. 1988;141(10):3617–3624. [PubMed] [Google Scholar]
- 24.Zheng J, Schodel F, Peterson DL. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J Biol Chem. 1992;267(13):9422–9429. [PubMed] [Google Scholar]
- 25.Jegerlehner A, Tissot A, Lechner F, et al. A molecular assembly system that renders antigens of choice highly repetitive for induction of protective B cell responses. Vaccine. 2002;20(25–26):3104–3112. doi: 10.1016/s0264-410x(02)00266-9. [DOI] [PubMed] [Google Scholar]
- 26.Jegerlehner A, Storni T, Lipowsky G, Schmid M, Pumpens P, Bachmann MF. Regulation of IgG antibody responses by epitope density and CD21-mediated costimulation. Eur J Immunol. 2002;32(11):3305–3314. doi: 10.1002/1521-4141(200211)32:11<3305::AID-IMMU3305>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 27.Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172(9):5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 28.Fu TM, Grimm KM, Citron MP, et al. Comparative immunogenicity evaluations of influenza A virus M2 peptide as recombinant virus like particle or conjugate vaccines in mice and monkeys. Vaccine. 2009;27(9):1440–1447. doi: 10.1016/j.vaccine.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Beesley KM, Francis MJ, Clarke BE, et al. Expression in yeast of amino-terminal peptide fusions to hepatitis B core antigen and their immunological properties. Biotechnology (NY) 1990;8(7):644–649. doi: 10.1038/nbt0790-644. [DOI] [PubMed] [Google Scholar]
- 30.Borisova GP, Berzins I, Pushko PM, et al. Recombinant core particles of hepatitis B virus exposing foreign antigenic determinants on their surface. FEBS Lett. 1989;259(1):121–124. doi: 10.1016/0014-5793(89)81509-1. [DOI] [PubMed] [Google Scholar]
- 31.Brown AL, Francis MJ, Hastings GZ, et al. Foreign epitopes in immunodominant regions of hepatitis B core particles are highly immunogenic and conformationally restricted. Vaccine. 1991;9(8):595–601. doi: 10.1016/0264-410x(91)90248-5. [DOI] [PubMed] [Google Scholar]
- 32.Clarke BE, Brown AL, Grace KG, et al. Presentation and immunogenicity of viral epitopes on the surface of hybrid hepatitis B virus core particles produced in bacteria. J Gen Virol. 1990;71(Pt 5):1109–1117. doi: 10.1099/0022-1317-71-5-1109. [DOI] [PubMed] [Google Scholar]
- 33.Francis MJ, Hastings GZ, Brown AL, et al. Immunological properties of hepatitis B core antigen fusion proteins. Proc Natl Acad Sci USA. 1990;87(7):2545–2549. doi: 10.1073/pnas.87.7.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriarty A, McGee JS, Winslow B, et al. Vaccines 90: Modern Approaches to New Vaccines Including Prevention of AIDS. Vol. 225. Cold Spring Harbor Press; NY, USA: 1990. Expression of HIV gag and env B-cell epitopes on the surface of HBV core particles and analysis of the immune responses generated to those epitopes. [Google Scholar]
- 35.Schodel F, Milich DR, Will H. Hepatitis B virus nucleocapsid/pre-S2 fusion proteins expressed in attenuated Salmonella for oral vaccination. J Immunol. 1990;145(12):4317–4321. [PubMed] [Google Scholar]
- 36.Stahl SJ, Murray K. Immunogenicity of peptide fusions to hepatitis B virus core antigen. Proc Natl Acad Sci USA. 1989;86(16):6283–6287. doi: 10.1073/pnas.86.16.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Brunn A, Brand M, Reichhuber C, Morys-Wortmann C, Deinhardt F, Schodel F. Principal neutralizing domain of HIV-1 is highly immunogenic when expressed on the surface of hepatitis B core particles. Vaccine. 1993;11(8):817–824. doi: 10.1016/0264-410x(93)90356-3. [DOI] [PubMed] [Google Scholar]
- 38•.Karpenko LI, Ivanisenko VA, Pika IA, et al. Insertion of foreign epitopes in HBcAg: how to make the chimeric particle assemble. Amino Acids. 2000;18(4):329–337. doi: 10.1007/s007260070072. This group analyzed the features of HBcAg foreign inserts that affect the ability of the chimeric particles to assemble. [DOI] [PubMed] [Google Scholar]
- 39.Kratz PA, Bottcher B, Nassal M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc Natl Acad Sci USA. 1999;96(5):1915–1920. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skamel C, Ploss M, Bottcher B, et al. Hepatitis B virus capsid-like particles can display the complete, dimeric outer surface protein C and stimulate production of protective antibody responses against Borrelia burgdorferi infection. J Biol Chem. 2006;281(25):17474–17481. doi: 10.1074/jbc.M513571200. [DOI] [PubMed] [Google Scholar]
- 41.Nassal M, Skamel C, Kratz PA, Wallich R, Stehle T, Simon MM. A fusion product of the complete Borrelia burgdorferi outer surface protein A (OspA) and the hepatitis B virus capsid protein is highly immunogenic and induces protective immunity similar to that seen with an effective lipidated OspA vaccine formula. Eur J Immunol. 2005;35(2):655–665. doi: 10.1002/eji.200425449. [DOI] [PubMed] [Google Scholar]
- 42.Hoofnagle JH, Dusheiko GM, Seeff LB, Jones EA, Waggoner JG, Bales ZB. Seroconversion from hepatitis B e antigen to antibody in chronic type B hepatitis. Ann Intern Med. 1981;94(6):744–748. doi: 10.7326/0003-4819-94-6-744. [DOI] [PubMed] [Google Scholar]
- 43.Maruyama T, McLachlan A, Iino S, Koike K, Kurokawa K, Milich DR. The serology of chronic hepatitis B infection revisited. J Clin Invest. 1993;91(6):2586–2595. doi: 10.1172/JCI116497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 45.Milich DR, McLachlan A, Moriarty A, Thornton GB. Immune response to hepatitis B virus core antigen (HBcAg): localization of T cell recognition sites within HBcAg/HBeAg. J Immunol. 1987;139(4):1223–1231. [PubMed] [Google Scholar]
- 46••.Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234(4782):1398–1401. doi: 10.1126/science.3491425. First description of how HBcAg could act as a T-cell-independent antigen. [DOI] [PubMed] [Google Scholar]
- 47.Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science. 1993;262(5138):1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 48.Milich DR, Chen M, Schodel F, Peterson DL, Jones JE, Hughes JL. Role of B cells in antigen presentation of the hepatitis B core. Proc Natl Acad Sci USA. 1997;94(26):14648–14653. doi: 10.1073/pnas.94.26.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fehr T, Skrastina D, Pumpens P, Zinkernagel RM. T cell-independent type I antibody response against B cell epitopes expressed repetitively on recombinant virus particles. Proc Natl Acad Sci USA. 1998;95(16):9477–9481. doi: 10.1073/pnas.95.16.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper A, Tal G, Lider O, Shaul Y. Cytokine induction by the hepatitis B virus capsid in macrophages is facilitated by membrane heparan sulfate and involves TLR2. J Immunol. 2005;175(5):3165–3176. doi: 10.4049/jimmunol.175.5.3165. [DOI] [PubMed] [Google Scholar]
- 51.Vanlandschoot P, Van Houtte F, Serruys B, Leroux-Roels G. The arginine-rich carboxy-terminal domain of the hepatitis B virus core protein mediates attachment of nucleocapsids to cell-surface-expressed heparan sulfate. J Gen Virol. 2005;86(Pt 1):75–84. doi: 10.1099/vir.0.80580-0. [DOI] [PubMed] [Google Scholar]
- 52.Vanlandschoot P, Van Houtte F, Serruys B, Leroux-Roels G. Contamination of a recombinant hepatitis B virus nucleocapsid preparation with a human B-cell activator. J Virol. 2007;81(5):2535–2536. doi: 10.1128/JVI.02507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanlandschoot P, Van Houtte F, Ulrichts P, Tavernier J, Leroux-Roels G. Immunostimulatory potential of hepatitis B nucleocapsid preparations: lipopolysaccharide contamination should not be overlooked. J Gen Virol. 2005;86(Pt 2):323–331. doi: 10.1099/vir.0.80605-0. [DOI] [PubMed] [Google Scholar]
- 54.Lee BO, Tucker A, Frelin L, et al. Interaction of the hepatitis B core antigen and the innate immune system. J Immunol. 2009;182(11):6670–6681. doi: 10.4049/jimmunol.0803683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28(6):799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Schodel F, Wirtz R, Peterson D, et al. Immunity to malaria elicited by hybrid hepatitis B virus core particles carrying circumsporozoite protein epitopes. J Exp Med. 1994;180(3):1037–1046. doi: 10.1084/jem.180.3.1037. An HBcAg-based vaccine candidate is shown to elicit 100% protection against a murine malarial infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pumpens P, Grens E. HBV core particles as a carrier for B cell/T cell epitopes. Intervirology. 2001;44(2–3):98–114. doi: 10.1159/000050037. [DOI] [PubMed] [Google Scholar]
- 58.Herrington DA, Clyde DF, Losonsky G, et al. Safety and immunogenicity in man of a synthetic peptide malaria vaccine against Plasmodium falciparum sporozoites. Nature. 1987;328(6127):257–259. doi: 10.1038/328257a0. [DOI] [PubMed] [Google Scholar]
- 59.Zavala F, Tam JP, Hollingdale MR, et al. Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science. 1985;228(4706):1436–1440. doi: 10.1126/science.2409595. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980;207(4426):71–73. doi: 10.1126/science.6985745. [DOI] [PubMed] [Google Scholar]
- 61.Zavala F, Tam JP, Barr PJ, et al. Synthetic peptide vaccine confers protection against murine malaria. J Exp Med. 1987;166(5):1591–1596. doi: 10.1084/jem.166.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schodel F, Peterson D, Milich DR, Charoenvit Y, Sadoff J, Wirtz R. Immunization with hybrid hepatitis B virus core particles carrying circumsporozoite antigen epitopes protects mice against Plasmodium yoelii challenge. Behring Inst Mitt. 1997;(98):114–119. [PubMed] [Google Scholar]
- 63.Milich DR, Hughes J, Jones J, Sallberg M, Phillips TR. Conversion of poorly immunogenic malaria repeat sequences into a highly immunogenic vaccine candidate. Vaccine. 2001;20(5–6):771–788. doi: 10.1016/s0264-410x(01)00400-5. [DOI] [PubMed] [Google Scholar]
- 64.Birkett A, Lyons K, Schmidt A, et al. A modified hepatitis B virus core particle containing multiple epitopes of the Plasmodium falciparum circumsporozoite protein provides a highly immunogenic malaria vaccine in preclinical analyses in rodent and primate hosts. Infect Immun. 2002;70(12):6860–6870. doi: 10.1128/IAI.70.12.6860-6870.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langermans JA, Schmidt A, Vervenne RA, et al. Effect of adjuvant on reactogenicity and long-term immunogenicity of the malaria vaccine ICC-1132 in macaques. Vaccine. 2005;23(41):4935–4943. doi: 10.1016/j.vaccine.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 66.Gregson AL, Oliveira G, Othoro C, et al. Phase I trial of an alhydrogel adjuvanted hepatitis B core virus-like particle containing epitopes of Plasmodium falciparum circumsporozoite protein. PLoS One. 2008;3(2):e1556. doi: 10.1371/journal.pone.0001556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nardin EH, Oliveira GA, Calvo-Calle JM, et al. Phase I testing of a malaria vaccine composed of hepatitis B virus core particles expressing Plasmodium falciparum circumsporozoite epitopes. Infect Immun. 2004;72(11):6519–6527. doi: 10.1128/IAI.72.11.6519-6527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliveira GA, Wetzel K, Calvo-Calle JM, et al. Safety and enhanced immunogenicity of a hepatitis B core particle Plasmodium falciparum malaria vaccine formulated in adjuvant Montanide ISA 720 in a Phase I trial. Infect Immun. 2005;73(6):3587–3597. doi: 10.1128/IAI.73.6.3587-3597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walther M, Dunachie S, Keating S, et al. Safety, immunogenicity and efficacy of a pre-erythrocytic malaria candidate vaccine, ICC-1132 formulated in Seppic ISA 720. Vaccine. 2005;23(7):857–864. doi: 10.1016/j.vaccine.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 70.Ito T, Gorman OT, Kawaoka Y, Bean WJ, Webster RG. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65(10):5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zebedee SL, Lamb RA. Nucleotide sequences of influenza A virus RNA segment 7: a comparison of five isolates. Nucleic Acids Res. 1989;17(7):2870. doi: 10.1093/nar/17.7.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5(10):1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 73.De Filette M, Min Jou W, Birkett A, et al. Universal influenza A vaccine: optimization of M2-based constructs. Virology. 2005;337(1):149–161. doi: 10.1016/j.virol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 74.Jennings GT, Bachmann MF. Immunodrugs: therapeutic VLP-based vaccines for chronic diseases. Annu Rev Pharmacol Toxicol. 2009;49:303–326. doi: 10.1146/annurev-pharmtox-061008-103129. [DOI] [PubMed] [Google Scholar]
- 75.Ionescu RM, Przysiecki CT, Liang X, et al. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J Pharm Sci. 2006;95(1):70–79. doi: 10.1002/jps.20493. [DOI] [PubMed] [Google Scholar]
- 76.Murata Y, Lightfoote PM, Rose RC, Walsh EE. Antigenic presentation of heterologous epitopes engineered into the outer surface-exposed helix 4 loop region of human papillomavirus L1 capsomeres. Virol J. 2009;6:81. doi: 10.1186/1743-422X-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar S, Ochoa W, Singh P, et al. Tomato bushy stunt virus (TBSV), a versatile platform for polyvalent display of antigenic epitopes and vaccine design. Virology. 2009;388(1):185–190. doi: 10.1016/j.virol.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 78.Denis J, Acosta-Ramirez E, Zhao Y, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26(27–28):3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]