Abstract
Aromatase is an enzyme required for the conversion of androgens to estrogens. Estrogens are female sex hormones involved in the development and growth of breast tumors. It has been of significant interest to investigate the structure-function relationship of aromatase since its inhibitors have shown great promise in fighting breast cancer. Aromatase belongs to the cytochrome P450 family, and forms an electron-transfer complex with its partner, NADPH-cytochrome P450 reductase (CPR), during the aromatization reaction. Aromatase is found to be widely expressed in vertebrates with unique substrates androstenedione and testosterone, but with various catalytic capacities reflecting species differences in Km, Vmax, and etc. This report will summarize current progress in sequence-function correlation analysis of the aromatase protein family and molecular characterization of the interaction between aromatase and CPR. These studies may lead to a novel field for the development of new inhibitors which interfere with the interaction between aromatase and CPR in order to inhibit the aromatization reaction.
1. Introduction
Aromatase is the rate-limiting enzyme in estrogen biosynthesis. Its important roles in breast cancer and reproductive dysfunction have led to a tremendous interest by investigators worldwide to study this enzyme. Aromatase, cytochrome P450 19A1, is found to be expressed within the gonads and brain of vertebrates, with additional expression in the placenta of primates and artiodactyls, and broad expression in humans [1–3]. To date, the aromatase protein family has been found to have a single form in amphibians, reptiles, birds, and most mammals, except for pigs having ovarian, embryonic, and placental forms and fish having brain and ovarian forms. All these forms are active with the same substrates: testosterone and androstenedione [1, 2, 4–7]. The amino acid sequence of aromatase is well conserved among all the vertebrates from a phylogenetic analysis [7]. However, the aromatase family shows a marginal divergence of functional characteristics, reflecting differences in Km, Vmax, binding affinity for the androgen substrate, and response to aromatase inhibitors [5, 8–13]. The correlation between sequence and function in the aromatase family is unclear.
Cytochrome P450 enzyme forms an electron-transfer complex with NADPH-cytochrome P450 reductase (CPR). CPR is composed of four domains: the FMN-binding domain, connecting domain, FAD-binding domain, and the NADP-binding domain, as revealed by the crystal structure of CPR solved in 1997 [14]. During the aromatization reaction, electrons are transferred from NADPH, through FAD and FMN, to the heme of aromatase, then to the androgen substrate. Upon receiving electrons from reductase, aromatase converts androgens, including androstenedione and testosterone, to estrogens estrone and estradiol, respectively. The membrane binding site of CPR is situated around the residue V64 and near some hydrophobic patches of the surface [14]. These membrane binding sites enable a CPR molecule to sit on the membrane surface. Aromatase is also a transmembrane protein with an N-terminal transmembrane helix. The crystal structure of aromatase presented at the IXth International Aromatase Conference by Dr. Debashis Ghosh reveals residues associated with the membrane, including the N terminus up to the A helix, and other loops near the C terminus [15]. Thus, the interactions between CPR and aromatase should include the interaction of their hydrophobic membrane binding portions. In addition, electrostatic attraction through cytoplasmic domains could also contribute to their interaction. However, the detailed mechanism of how aromatase and CPR interact is not yet fully understood.
2. Correlation of amino acid sequence with function in the aromatase protein family
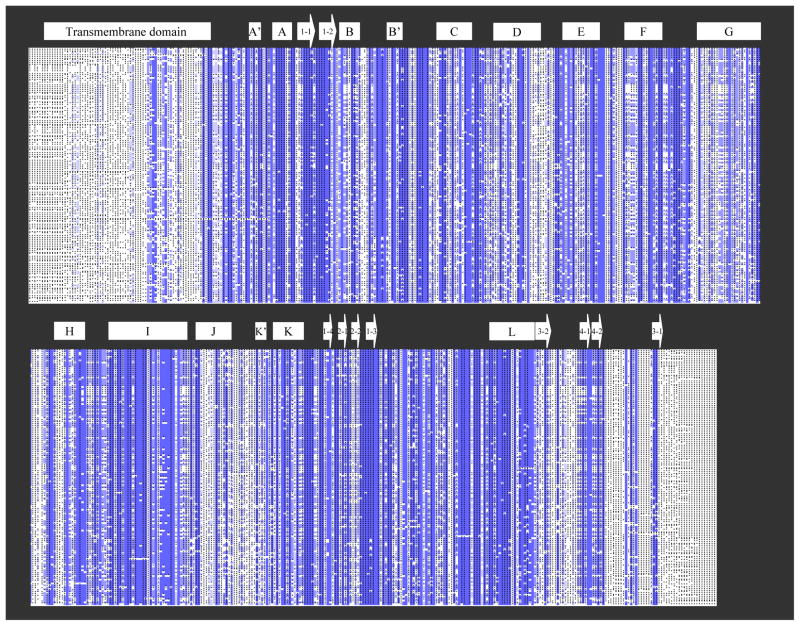
To better understand the mechanism responsible for function divergence, members of the aromatase protein family, about one hundred and fifty members, were applied for multiple sequence alignments at ensembl (www.ensembl.org). These members are from fish, amphibians, reptiles, birds, and mammals. The sequence alignments (Fig. 1) show the most diverse sequences are at the N-terminal transmembrane domain, and the C-terminus. The major internal sequences are well conserved, including several important regions, the B′-C loop, I helix, and the b-4 sheet. The F–G loop, which is diverse among P450s, is also well conserved among aromatase family members. Human aromatase active site residues I133, F134, E302, D309, T310, and S478, predicted from previous studies [16, 17] and confirmed by the newly solved crystal structure of aromatase [15], are highly conserved. Their percentage identities are 99.4%, 100%, 100%, 98.2%, 98.8%, and 95.9%, respectively.
Fig. 1. Multiple amino acid sequence alignments of 150 aromatase family members.
Blue indicates well-conserved residues, and grey indicates residues with low conservation. The secondary structure of human aromatase is indicated by box (α-helix) and arrow (β-sheet) above the alignments. The sequences were aligned using Ensembl BLAST Server (www.ensembl.org).
Although the physiological significance of sequence divergence at the N- and C-termini of the aromatase family members is not well understood, it suggests that the enzyme can tolerate sequence modification at the N- and C-termini for its expression and purification. The N-terminal transmembrane domain serves a structural role (anchored to the ER membrane) rather than a functional role, thus deletion of the N-terminal transmembrane sequence of human aromatase for its expression in E. coli doesn’t affect its catalytic activity [17–19]. Attachment of a His-tag to the C-terminus of aromatase to facilitate purification also maintains its activity [17, 19]. Unexpectedly, the β4 sheet, which has been proposed to be located in the active site of aromatase and confirmed by the recently solved crystal structure [15], exhibits sequence divergence. This may actually contribute to differences in functional characteristics of the aromatase family members. Interestingly, active site residue serine in the β4 sheet (human aromatase S478) remains highly conserved among all the species. Another active site residue, histidine (human aromatase H480), exhibits species divergence: histidine in amphibians, reptiles, birds, and mammals, while glutamine at this position in fish. In a previous study, we have found that the human aromatase mutant H480Q has lower Km and Vmax values than the wild type enzyme [20]. To fully understand the relationship between amino acid sequence divergence and function divergence among the aromatase family, extensive enzyme functional studies and bioinformatics studies on sequence-function correlation will be needed.
3. Protein-protein interaction between aromatase and reductase
Aromatase forms an electron-transfer complex with CPR. Studies of the interaction between these two proteins will help us to understand the mechanism of the aromatization reaction. With two available purified proteins in our laboratory, recombinant aromatase and full-length CPR, we performed kinetic analysis. The kinetic analysis demonstrates the binding of CPR and androstenedione to aromatase is non-competitive, indicating that CPR and the androgen substrate bind to different sites of aromatase [21]. CPR binds to aromatase with a much smaller Km value than androstenedione, which demonstrates that CPR binds more strongly to aromatase than to the androgen substrate [21].
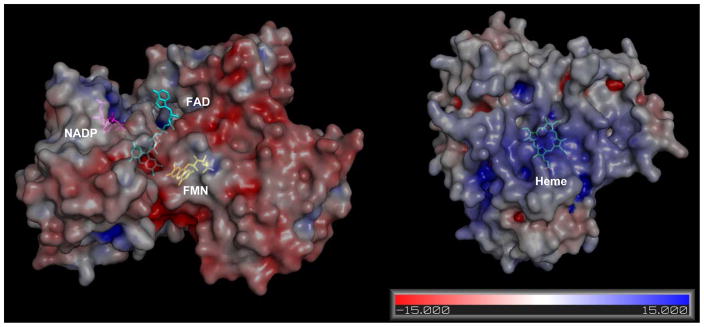
What is the mechanism for the strong interaction between aromatase and CPR? The interactions between CPR and aromatase may include the interaction of two hydrophobic membrane binding portions and electrostatic attraction. Literatures have shown that the FMN domain of CPR contains multiple carboxylate groups, including aspartate and glutamate, which would form ion pairs with basic residues on P450s [22–24]. To check for potential electrostatic interaction between aromatase and CPR, we calculated the surface electrostatic potential of the crystal structure of aromatase (PDB: 3EQM) and CPR (PDB: 1AMO) by PYMOL. As we expected, the FMN binding domain onto which aromatase binds is mainly negatively charged, and the proximal surface of aromatase is highly positively charged, which agrees with the fact that the androgen substrate binds to the distal side of aromatase (Fig. 2).
Fig. 2. Electrostatic potential surface of CPR and aromatase.
The electrostatic potential surface mapped onto the surface of the crystal structure of CPR (left) and aromatase (right). The positive potential is shown in blue, and the negative potential is shown in red. NADP, FAD, FMN, and HEME are shown in stick representation, and colored in purple, cyan, yellow, and green, respectively. The electrostatic potential was calculated with the APBS Plugin in PyMOL [28].
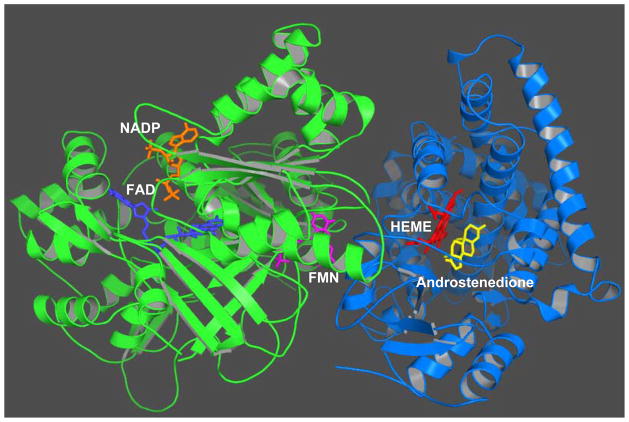
Previous direct rigid docking of the 3-D structure model of aromatase into the crystal structure of CPR generated an initial contact between aromatase and CPR, and suggested that CPR may adopt a structural rearrangement when it forms a complex with aromatase [25]. Recently, we decided to use a flexible step-wise docking approach. Firstly, the crystal structure of aromatase was docked with the FMN domain of CPR. The docking model with the highest score was picked from 20,000 decoys produced from ZDOCK [26] software version 3.0.1. This docking model allows electrostatic interaction between two acidic residues, E115 and D147, of CPR, and a basic residue, K108, of aromatase. The distance between the N5 atom of FMN and the heme iron is 22 Å, which is similar to the distance in the crystal structure of P450BM3, a self-sufficient enzyme with the heme domain and the reductase domain linked together on a single polypeptide [27]. The FAD domain and the NADPH domain were then docked into the complex. The model with the highest docking score is shown in Fig. 3. This final docking model allows the N-terminal transmembrane domains and the membrane binding portions of two proteins to face the same orientation. Our analysis has also revealed that the FMN-binding domain of CPR undergoes a structural rearrangement, allowing the proximal surface of aromatase to fit in the cleft between the FMN- and FAD-binding domains of CPR, and forms an electrostatic interaction. A flexible hinge between the FMN and FAD domains may facilitate the conformational changes of the FMN domain [14, 27].
Fig. 3. A binding model of aromatase with CPR by step-wise docking.
The aromatase-CPR complex is displayed as secondary structure cartoon and colored in green (CPR), and blue (aromatase). NADP, FAD, FMN, HEME, and androstenedione are shown in stick representation, and colored in orange, blue, purple, red, and yellow, respectively. The docking model was produced from ZDOCK [26] software version 3.0.1.
This docking model predicts that residue K108 of aromatase is involved in the electrostatic interaction with CPR. This residue is well conserved among the aromatase family. Mutagenesis data confirm a basic residue at this position is important for aromatization [21]. Future studies will be needed to determine whether residue K108 is indeed involved in the electrostatic interaction with reductase.
4. Conclusion
The crystal structure of aromatase in complex with androstenedione, presented at the IXth International Aromatase Conference by Dr. Ghosh [15], provides an insight into the active site of aromatase. However, a crystal structure only captures a single state of a protein and provides a static snapshot of one conformation. Thus, additional biochemical studies are required to address unsolved mysteries about this enzyme. In this report, we analyzed the relationship between amino acid sequence and function of the aromatase protein family, and presented a binding model of aromatase with its electron-transfer partner CPR. Future directions include bioinformatics studies on sequence-function correlation and validation of the aromatase-CPR binding model.
Acknowledgments
This research was supported by the California Breast Cancer Research Program [11GB-0125 (YH)] and National Institutes of Health [CA44735 (SC), ES08528 (SC), and CA33572 (the City of Hope Cancer Center Grant)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conley A, Hinshelwood M. Mammalian aromatases. Reproduction. 2001;121(5):685–695. doi: 10.1530/rep.0.1210685. [DOI] [PubMed] [Google Scholar]
- 2.Simpson ER, Clyne C, Rubin G, Boon WC, Robertson K, Britt K, Speed C, Jones M. Aromatase--a brief overview. Annu Rev Physiol. 2002;64:93–127. doi: 10.1146/annurev.physiol.64.081601.142703. [DOI] [PubMed] [Google Scholar]
- 3.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15(3):342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 4.Choi I, Simmen RC, Simmen FA. Molecular cloning of cytochrome P450 aromatase complementary deoxyribonucleic acid from periimplantation porcine and equine blastocysts identifies multiple novel 5′-untranslated exons expressed in embryos, endometrium, and placenta. Endocrinology. 1996;137(4):1457–1467. doi: 10.1210/endo.137.4.8625924. [DOI] [PubMed] [Google Scholar]
- 5.Corbin CJ, Trant JM, Walters KW, Conley AJ. Changes in testosterone metabolism associated with the evolution of placental and gonadal isozymes of porcine aromatase cytochrome P450. Endocrinology. 1999;140(11):5202–5210. doi: 10.1210/endo.140.11.7140. [DOI] [PubMed] [Google Scholar]
- 6.Graddy LG, Kowalski AA, Simmen FA, Davis SL, Baumgartner WW, Simmen RC. Multiple isoforms of porcine aromatase are encoded by three distinct genes. J Steroid Biochem Mol Biol. 2000;73(1–2):49–57. doi: 10.1016/s0960-0760(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 7.Wilson JY, McArthur AG, Stegeman JJ. Characterization of a cetacean aromatase (CYP19) and the phylogeny and functional conservation of vertebrate aromatase. Gen Comp Endocrinol. 2005;140(1):74–83. doi: 10.1016/j.ygcen.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Corbin CJ, Trant JM, Conley AJ. Porcine gonadal and placental isozymes of aromatase cytochrome P450: sub-cellular distribution and support by NADPH-cytochrome P450 reductase. Mol Cell Endocrinol. 2001;172(1–2):115–124. doi: 10.1016/s0303-7207(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 9.Pompon D, Liu RY, Besman MJ, Wang PL, Shively JE, Chen S. Expression of human placental aromatase in Saccharomyces cerevisiae. Mol Endocrinol. 1989;3(9):1477–1487. doi: 10.1210/mend-3-9-1477. [DOI] [PubMed] [Google Scholar]
- 10.Ackerman GE, Smith ME, Mendelson CR, MacDonald PC, Simpson ER. Aromatization of androstenedione by human adipose tissue stromal cells in monolayer culture. J Clin Endocrinol Metab. 1981;53(2):412–417. doi: 10.1210/jcem-53-2-412. [DOI] [PubMed] [Google Scholar]
- 11.Corbin CJ, Graham-Lorence S, McPhaul M, Mason JI, Mendelson CR, Simpson ER. Isolation of a full-length cDNA insert encoding human aromatase system cytochrome P-450 and its expression in nonsteroidogenic cells. Proc Natl Acad Sci U S A. 1988;85(23):8948–8952. doi: 10.1073/pnas.85.23.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou DJ, Pompon D, Chen SA. Stable expression of human aromatase complementary DNA in mammalian cells: a useful system for aromatase inhibitor screening. Cancer Res. 1990;50(21):6949–6954. [PubMed] [Google Scholar]
- 13.Stresser DM, Turner SD, McNamara J, Stocker P, Miller VP, Crespi CL, Patten CJ. A high-throughput screen to identify inhibitors of aromatase (CYP19) Anal Biochem. 2000;284(2):427–430. doi: 10.1006/abio.2000.4729. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94(16):8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh D. X-ray Structure of Human Placental Aromatase: Molecular Basis for Androgen Specificity and Estrogen Synthesis. Presented at the IXth International Aromatase Conference; Shanghai, China. October 13–16, 2008. [Google Scholar]
- 16.Hong Y, Cho M, Yuan YC, Chen S. Molecular basis for the interaction of four different classes of substrates and inhibitors with human aromatase. Biochem Pharmacol. 2008;75(5):1161–1169. doi: 10.1016/j.bcp.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol. 2007;21(2):401–414. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- 18.Kagawa N, Hori H, Waterman MR, Yoshioka S. Characterization of stable human aromatase expressed in E. coli. Steroids. 2004;69(4):235–243. doi: 10.1016/j.steroids.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, Zhou D, Kao YC, Ye J, Chen S. Expression and purification of a recombinant form of human aromatase from Escherichia coli. Biochem Pharmacol. 2002;64(9):1317–1324. doi: 10.1016/s0006-2952(02)01361-8. [DOI] [PubMed] [Google Scholar]
- 20.Kao YC, Korzekwa KR, Laughton CA, Chen S. Evaluation of the mechanism of aromatase cytochrome P450. A site-directed mutagenesis study. Eur J Biochem. 2001;268:243–251. doi: 10.1046/j.1432-1033.2001.01886.x. [DOI] [PubMed] [Google Scholar]
- 21.Hong Y, Li H, Ye J, Miki Y, Yuan YC, Sasano H, Evans DB, Chen S. Epitope characterization of an aromatase monoclonal antibody suitable for the assessment of intratumoral aromatase activity. PloS ONE. 2009 doi: 10.1371/journal.pone.0008050. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nisimoto Y. Localization of cytochrome c-binding domain on NADPH-cytochrome P-450 reductase. J Biol Chem. 1986;261(30):14232–14239. [PubMed] [Google Scholar]
- 23.Bernhardt R, Pommerening K, Ruckpaul K. Modification of carboxyl groups on NADPH-cytochrome P-450 reductase involved in binding of cytochromes c and P-450 LM2. Biochem Int. 1987;14(5):823–832. [PubMed] [Google Scholar]
- 24.Nadler SG, Strobel HW. Role of electrostatic interactions in the reaction of NADPH-cytochrome P-450 reductase with cytochromes P-450. Arch Biochem Biophys. 1988;261(2):418–429. doi: 10.1016/0003-9861(88)90358-x. [DOI] [PubMed] [Google Scholar]
- 25.Hong Y, Li H, Yuan YC, Chen S. Molecular characterization of aromatase. J Steroid Biochem Mol Biol. 2008 In press. [Google Scholar]
- 26.Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Integrating statistical pair potentials into protein complex prediction. Proteins. 2007;69(3):511–520. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 27.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci U S A. 1999;96(5):1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]