SUMMARY
Combinatorial interactions among transcription factors are critical to directing tissue-specific gene expression. To build a global atlas of these combinations, we have screened for physical interactions among the majority of human and mouse DNA-binding transcription factors (TFs). The complete networks contain 762 human and 877 mouse interactions. Analysis of the networks reveals that highly connected TFs are broadly expressed across tissues, and that roughly half of the measured interactions are conserved between mouse and human. The data highlight the importance of TF combinations for determining cell fate, and they lead to the identification of a SMAD3/FLI1 complex expressed during development of immunity. The availability of large TF combinatorial networks in both human and mouse will provide many opportunities to study gene regulation, tissue differentiation, and mammalian evolution.
INTRODUCTION
Tissue specificity is enabled by spatial and temporal patterns of gene expression which, in turn, are driven by transcriptional regulatory networks (Naef and Huelsken, 2005; Zhang et al., 2004). Such networks involve assemblies of control proteins, such as DNA-binding transcription factors (TFs), connected to the sets of promoters of genes they induce or repress (Tan et al., 2008b). Typically, TFs do not act independently, but form complexes with other TFs, chromatin modifiers, and co-factor proteins, which bind together and assemble upon the regulatory regions of DNA to affect transcription (Fedorova and Zink, 2008). Mapping the combinatorial interactions among TFs would represent a significant leap forward in our understanding of how tissue specificity is determined.
In recent years, a variety of genome-scale technologies have been introduced which allow mammalian transcriptional regulatory networks to be investigated at high resolution and depth. Many such studies have inferred transcriptional networks through mRNA expression profiling combined with genome-wide active promoter mapping and promoter motif analysis (Suzuki et al., 2009). These data have been supplemented with Fluorescence Activated Cell Sorting (FACS) (Shachaf et al., 2008) or Reverse Transcriptase Quantitative Polymerase Chain Reaction (qRT-PCR) (Roach et al., 2007; Wen et al., 1998).
Another technology that has revolutionized the study of transcriptional networks is Chromatin Immuno-Precipitation (ChIP) which, when coupled with microarrays or high-throughput sequencing (Johnson et al., 2007), enables genome-wide measurements of TF binding locations in vivo. A complementary approach is the Protein Binding Microarray (PBM) (Berger et al., 2008), which rapidly characterizes the complete DNA sequence repertoire bound by a TF in vitro. ChIP and PBMs have been applied to map transcriptional networks in a variety of human cell types, including stem cells (Cole et al., 2008; Lee et al., 2006) and lymphocytes (Marson et al., 2007; Schreiber et al., 2006), and to characterize the binding motifs of many mammalian TF families (Berger et al., 2008).
Although these studies have led to the construction of very large models of transcriptional networks, they are based on experiments that largely treat each TF in isolation: for instance, ChIP-chip measures binding locations for one TF at a time, although separate profiles for several TFs can be later combined into networks (Mathur et al., 2008). However, it is well known that the transcriptional output of a gene is due to the joint activity of many TFs whose binding and activation are highly interdependent. This cooperativity is often mediated by direct physical contact between two or more TFs, forming homodimers, heterodimers, or larger transcriptional complexes. In fact, it has been estimated that approximately 75% of all metazoan TFs heterodimerize with other factors (Walhout, 2006). Newman and Keating used protein arrays to reveal a network of several hundred domain interactions among the bZIP TF family alone (Grigoryan et al., 2009). Other studies have successfully assembled large networks of protein interactions using technologies such as co-immunoprecipitation and two-hybrid screening (Park et al., 2005; Yu et al., 2008), but to date these have not been systematically applied to map networks of transcription factors. Thus, a clear and immediate task is to map which combinations of TFs act together, and how these combinations lead to modes of regulation that are not evident when each factor is considered separately.
Towards this goal, we have pursued an integrative approach to systematically map combinatorial interactions among mammalian TFs. Our approach draws from two systems-wide data sets generated in both human and mouse: Physical protein-protein interaction among TFs measured using the Mammalian Two Hybrid (M2H) system, and quantitative TF expression levels measured using qRT-PCR across tissues. Analysis of these data identifies a database of TF complexes and networks, which can be used to elucidate the regulatory programs behind developmental processes and disease. Chief among these results is a network of homeobox TFs which we show can predict tissue type in mammals.
RESULTS
Mammalian transcription factor protein-protein interaction networks
We compiled a list of 1988 human and 1727 mouse DNA-binding transcription factors using information from public gene databases (Supplementary Table 1). Of these, 1222 and 1112 cDNA clones were captured in human and mouse, respectively, that could be verified to express full-length protein (Supplementary Table 1). All pair-wise combinations of TF cDNAs were systematically screened for protein-protein interaction using the M2H system (Suzuki et al., 2001). Bait and prey constructs were co-transfected in CHO-K1 cells, and the interaction of the expressed proteins was monitored by luciferase reporter activity. This process identified 762 and 877 high-stringency TF-TF interactions in human and mouse, respectively (Supplementary Tables 2,3). The use of M2H meant that the human and mouse TF interactions were measured in near-physiological conditions including mammalian post-translational and other modifications. The web-accessible atlas of all pairwise TF interactions mapped by M2H is available at http://fantom.gsc.riken.jp/4/tf-ppi. This resource is searchable by gene ID or function and provides network visualizations as well as raw lists of interactions.
To estimate the sensitivity of the screening approach (the percentage of all true TF-TF interactions that are identifiable by M2H), we assembled a gold-standard set of high confidence TF-TF dimers reported in previous literature. To obtain this gold standard, a set of 289 mouse TF-TF interactions were downloaded from public databases and further curated to select 91 interactions supported by two or more independent lines of evidence or primary experimental reports (Supplementary Information and Supplementary Table 3). We found that M2H recovered protein-protein interactions for 23 of these heterodimers, yielding a sensitivity of 25%. Apart from sensitivity we were also interested in precision (the percentage of reported interactions that are true, equal to 1 – false discovery rate). Precision is more difficult to estimate than sensitivity, because it requires a gold standard that contains not only known interactions but also a large number of protein pairs that are known to be non-interacting. Since such data are not available, we sought to confirm the M2H positives using in-vitro pull down assays as a second technology. Of 34 randomly chosen mouse M2H positives, 18 (53%) were detected by in-vitro pull down (Supplementary Table 4). This second assay is not a gold standard, such that failure to confirm an M2H positive by in-vitro pull down does not negate the corresponding protein-protein interaction, which might be transient or unstable under conditions of the pull-down. However, this analysis does show that the M2H network recovers approximately one quarter of known TF heterodimers and that the majority of M2H interactions can be replicated by a second technology. These figures are consistent with high quality interaction networks published recently elsewhere (Yu et al., 2008).
We now describe four case studies that use the atlas to address questions of how transcriptional control contributes to tissue specificity in mammals. These case studies cover: (1) Integration of the atlas with quantitative TF abundance levels across human and mouse tissues, revealing a prominent relationship between TF connectivity and expression— (2) Identification of a subnetwork of homeobox factors that is highly discriminative and predictive of tissue type— (3) A proteome-wide map of conserved transcriptional complexes in mammals, many of which have tissue-specific expression patterns that are also highly conserved— and (4) Examples of how the atlas can be used to recognize and further explore TF heterodimers in control of tissue differentiation.
Integration of TF interaction and expression reveals insights into network structure
In order to physically interact, TFs must be co-expressed in the same tissue or cell type. To investigate the tissue specificity of TF interactions, we obtained quantitative mRNA profiles of all TFs using qRT-PCR across a panel of 34 human and 20 mouse tissues (Supplementary Table 5). For each TF we computed a Tissue Specificity Score (TSPS), which uses relative entropy to quantify the extent to which the observed TF expression pattern departs from the null distribution of uniform expression across all tissues (Experimental Procedures, Supplementary Tables 1,5). Examination of tissue specificity over all TFs suggested a mixture of two distinct TF populations, with one population of TFs having widespread tissue expression (TSPS < 1) and a second smaller population at higher tissue specificity (TSPS ≥ 1, Figures 1A–B). We called TFs with widespread expression “facilitators”, based on the hypothesis that they facilitate transcriptional programs across many different tissues, and those with high specificity tissue “specifiers”. For example, the TFs JUN and FOS, which form the AP-1 heterodimer, were classified as strong facilitators owing to low TSPS (average around 0.6, Supplementary Table 5). This score is consistent with the classical view of AP-1 as a broad activator of expression in major cellular processes including differentiation, proliferation, and apoptosis (Ameyar et al., 2003). In contrast, many TFs with known roles in tissue differentiation were classified as “specifiers”, such as MYOD1, which regulates muscle development and members of the Paired box (Pax) TF family involved in tissue morphogenesis. The observed bimodal distribution of TF expression is in agreement with recent findings from a meta-analysis of publicly-available expression profiles in humans (Vaquerizas et al., 2009).
Figure 1. TF expression versus connectivity.
(A) Distribution of tissue specificity for all TFs. The green curves fit the bi-modal distribution as a mixture of two Gaussian. (B) Scatterplot of tissue specificity (y-axis) versus number of neighbors (x-axis). Red points are defined as specifier hubs and blue points as facilitator hubs (Supplementary Table 1). (C) TFs are binned into four groups of approximately equal size based on their number of interactions (x-axis). The tissue specificity distribution of each bin is represented by stacks of colored segments. Segment height represents the fraction of TFs in an expression group (left y-axis), and segment color represents the number of tissues in which TFs in that group are expressed. The black line displays the median TSPS of each group (right y-axis). Among TFs with six or more interactions, 70% are expressed in more than half of tissues. Among TFs with fewer than six interactions, this number falls to 45%. The results shown are for human M2H interactions supplemented with TF-TF interactions downloaded from literature (Supplementary Table 2); similar results are obtained for mouse interactions or for M2H interactions only (Supplementary Table 3. See also Supplementary Table 4 for confirmation of the M2H positives using in-vitro pull down assays as a second technology).
Examining the relationship between expression and interaction, we observed a strongly negative Pearson correlation of −0.79 between a TF’s number of protein interactions and its TSPS. That is, we found that TFs with few interactions tend to be expressed in a tissue-specific pattern while TFs with many interactions—so called network “hubs” (Jin et al., 2007; Yu et al., 2006)— tend to be expressed across many tissues (Figure 1C). The observed correlation was highly significant, as assessed by 10,000 random trials in which the assignment of expression values to TFs was permuted (r = 0.00 ± 0.03). Such widespread expression of TF hubs bears some similarity to previous studies of TF-DNA (transcriptional) interactions, in which the number of promoters bound by a TF was found to correlate with the number of growth conditions in which it is expressed (Luscombe et al., 2004; Zhou et al., 2008).
A homeobox network associated with specification of tissue type
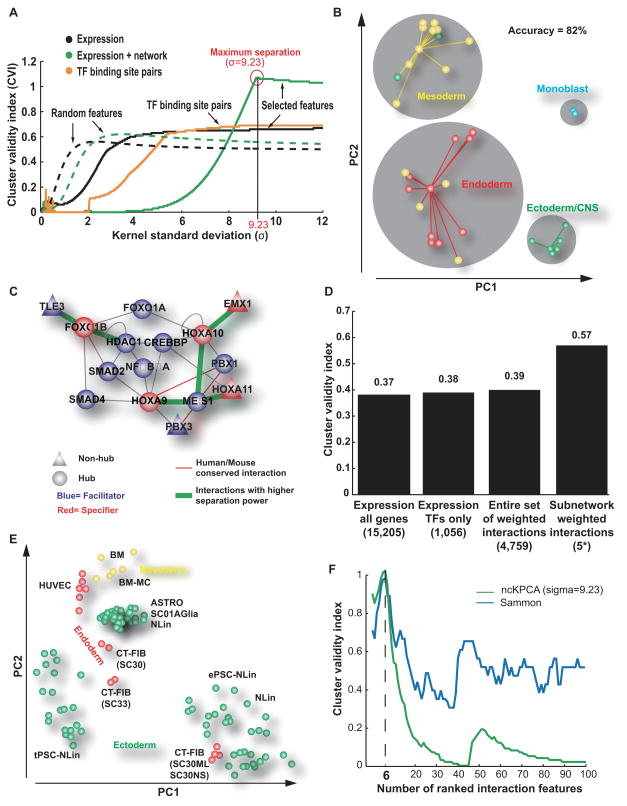
Combinatorial interaction among transcription factors is critical for differentiation of tissues (Davidson et al., 2002). To identify TF interaction networks involved in tissue development, we clustered the TF expression profiles across the 34 human tissues (see above) using two approaches: a basic tissue separation approach using expression levels only, and a “network-transformed” approach in which we exploited as features the differences in expression level across TF-TF interactions, as suggested by a recent study (Taylor et al., 2009). We found that network transformation resulted in an increased separation of tissues into four well-formed clusters (a 38% increase, Figures 2A,B and Supplementary Figure 1). These corresponded to well-defined tissue classes according to embryonic origin: ectoderm (including Central Nervous System or CNS), mesoderm, endoderm, and cell lines. Strikingly, only six TF interactions were sufficient to classify tissue type with a high accuracy of 82% (Figures 2B,C). Moreover, we found that these interactions fell into the same small network neighborhood defined by a subnetwork of 15 proteins (Figure 2C). This subnetwork was highly enriched for homeobox factors (7/15 proteins) many of which have, at least individually, known roles in tissue type specification during development (Duverger and Morasso, 2008). Although we expected that many of these TFs would be tissue specifiers, we found that 10 of the 15 were in fact facilitators expressed broadly across most tissue types. These results support the notion that it is the interactions among transcription factors, more than their expression levels alone that help to determine tissue identity.
Figure 2. A homeobox network associated with tissue differentiation.
(A) Performance of tissue separation with (green solid curve) or without (black solid curve) information about TF protein-protein interactions (Supplementary Table 2). The Bezdek cluster validity index (CVI, y-axis) is a measure of separation between the four tissue classes. CVI is plotted for increasing kernel standard deviation (x-axis), the only tuning parameter of the ncKPCA algorithm used for tissue separation. Performance was also evaluated for TF pairs predicted to cooperate based on co-occurrence of TF binding sites (yellow curve) (Yu et al., 2006) as well as for random features (dashed curves). (B) Tissue dimensionality reduction by ncKPCA into the first two Principle Components (PCs), considering features derived from the six most informative TF-TF interactions. Points represent tissues derived from ectoderm (green), mesoderm (yellow), or endoderm (red), or a monocyte cell line (blue). Gray circles denote four clusters obtained by affinity propagation in the (PC1, PC2) space, with each point connected to its cluster exemplar. This figure is related to Figure S1. (C) Informative subnetwork containing six interactions (green) used to generate features for tissue separation. Also shown are the immediate network neighbors of the interacting TFs. (D) CVI for the separation of stem cells (Supplementary Table 6) using Sammon Mapping. Four feature sets are shown: the original expression values from Muller et al, the expression of the TFs only, the entire set of TF protein-protein interactions, or the features corresponding to the six interactions in panel C (5* indicates that the interaction HOXA9-MEIS1 was not considered because HOXA9 expression was not measured in the stem cell investigation of Muller et al). (E) Stem cell dimensionality reduction obtained by Sammon Mapping using the panel C interaction set. Points represent stem cell lines derived from ectoderm (green), mesoderm (yellow), or endoderm (red). (F) Good performance of tissue separation observed with two different algorithms. ncKPCA (green curve) and Sammon Mapping (blue curve). CVI (y-axis) is plotted against the number of PC2-ranked interactions used to separate tissues (x-axis). In both cases, the maximum performance is observed using the first six PC2-ranked interactions to separate tissues.
Given the ability of the homeobox-related subnetwork to separate tissues based on their embryological origin, we sought to test whether this subnetwork was also able to discriminate the embryological origin of different types of stem cells. Understanding the transcriptional events that commit stem cells to different tissue lineages is one of the major goals of stem cell research (Jaenisch, 2009). For this purpose, we downloaded the publicly-available gene expression profiles of 219 stem cell lines derived from a variety of different tissue types (Muller et al., 2008) (Supplementary Table 6 lists the tissue origin of each cell line). As shown in Figures 2D–E, the homeobox-related subnetwork was indeed able to separate these stem cell expression profiles by ectoderm, mesoderm, and endoderm origin. This separation was 33% better than that achieved using other methods (Figure 2D). This analysis suggests that the good performance of the homeobox-related subnetwork (Figure 2C) is not the result of overfitting to a specific set of tissue expression profiles. Moreover, it provides further evidence that the combinatorial interactions revealed in this subnetwork play an important role in cell commitment to different tissue lineages.
Conservation of TF complexes across mammalian evolution
A strong line of evidence that a particular TF interaction is functional is to observe conservation of that interaction across species. For each human TF, we used the InParanoid algorithm (O’Brien et al., 2005) to identify its set of amino-acid sequence orthologs in mouse. We then identified pairs of TFs for which the orthologs were observed to interact in both species. In total, 80 conserved interactions were identified between the M2H data of human and mouse—this number rose to 305 conserved interactions when supplementing M2H data with literature (Supplementary Tables 2,3). Considering this number together with the M2H sensitivity and precision estimates above, we computed the fraction of conserved TF-TF interactions between human and mouse to be in the range of 34 – 64% (depending on the value one uses for the precision of M2H screening, see Supplementary Information).
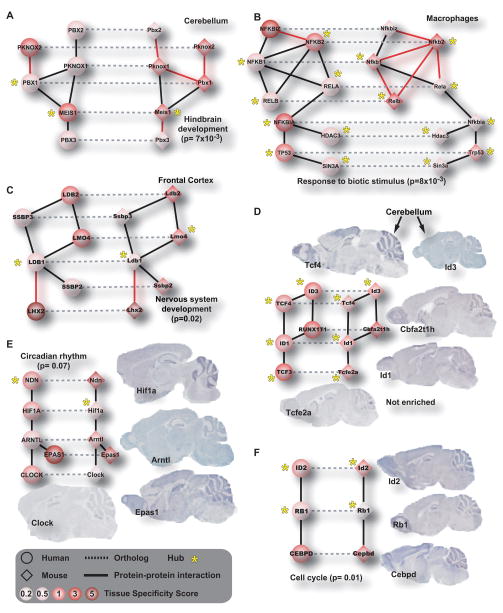
We next used NetworkBLAST (Kalaev et al., 2008) to examine how these conserved interactions clustered within the network, i.e. whether they fell within common subnetworks suggestive of conserved transcriptional complexes. In total, 68 conserved complexes were identified which contained approximately six TFs on average. Examples of conserved complexes are shown in Figures 3A–F; the complete set is included as part of the atlas at http://fantom.gsc.riken.jp/4/tf-ppi. Eighty percent of the conserved complexes were enriched for Gene Ontology Biological Process annotations. These conserved TF complexes provide a first-draft map of the combinatorial regulatory circuits common to mammals.
Figure 3. TF subnetworks conserved across human and mouse.
(A–F) Examples of TF subnetworks conserved in specific tissues. Human proteins are circles and mouse proteins are diamonds, colored in increasing shades of red representing increasing tissue specificity (TSPS), (Supplementary Table 1). Stars indicate hubs. Horizontal dashed links indicate protein orthology relationships across species, whereas solid links indicate protein-protein interactions within species (red links are newly-discovered, black links are literature-curated). (E–F) Conserved TF subnetworks that are specific to cerebellum, as first indicated by qRT-PCR (red nodes and Supplementary Table 5) and subsequently confirmed by in-situ hybridization to mouse brain tissue samples. All conserved subnetworks are available at http://fantom.gsc.riken.jp/4/tf-ppi.
The conserved complexes also suggest combinations of heterodimers in specific biological contexts for future investigation. Figure 3C shows a conserved complex of six TFs, in which five are broadly expressed across all tissues in both species, and one TF (LHX2) is restricted to frontal cortex also in both species (Supplementary Table 5). Figures 3D–F show three conserved TF complexes consisting of proteins co-expressed in cerebellum. Messenger RNA in-situ hybridization analysis of mouse cerebellum, obtained from the Allen Brain Atlas (Lein et al., 2007), confirms that the interacting TFs are indeed expressed in cerebellum and that this localization is cerebellum-specific at single-cell resolution.
FLI1 and SMAD3 form a heterodimeric complex associated with monocyte development
The vast majority of TF-TF interactions recorded in the atlas represent new combinations not yet documented in the literature. Thus, an important question is how particular interactions of interest should be carried forward in the laboratory to identify new transcriptional heterodimers and to study their regulatory functions. As an example use of the atlas to identify tissue-restricted heterodimers, four interactions were selected for which at least one TF had moderate to high tissue specificity (Figure 4A). For example, Peroxisome Proliferator-Activated Receptor Gamma (PPARG) is expressed in adipose, skin, lung, and breast, with little or no expression in other tissues. Although its interaction partner, Retinoid X Receptor Beta (RXRB), is expressed ubiquitously the interaction requires the presence of both TFs and thus remains tissue restricted (Supplementary Table 5).
Figure 4. Physical and functional exploration of tissue-restricted heterodimers.
(A) Four heterodimers that display combinatorial logic across tissues. The heatmap shows the mRNA copy number of each heterodimeric TF across tissues measured by qRT-PCR (Supplementary Table 5). (B) In-vitro pull down experiment shows clear bi-directional physical interaction for each of the four heterodimers as detected originally by M2H assay (Supplementary Table 2). (C) mRNA levels of FLI1 and SMAD3 during THP-1 differentiation induced by PMA, as measured by qRT-PCR. (D) Graphical representation of FLI1/SMAD3 control during myeloid differentiation.
Given these tissue-restricted TF combinations, a first step was to characterize and further establish their physical interaction. We used bidirectional in-vitro pull-down assays to examine whether each TF pair could exhibit strong, stable, and direct physical binding under the conditions of the pull-down, independent of other proteins or factors. As shown in Figure 4B, all four TF interactions were recapitulated as in-vitro pull-downs, making them strong candidates for functional transcriptional complexes.
Next, we sought detailed information on the dynamic expression of a TF combination in the tissue(s) in which both TFs were active. One of the identified TF interactions was between Friend Leukemia virus Integration 1 (FLI1) and SMAD family member 3 (SMAD3), in which FLI1 was restricted primarily to macrophage-related tissues (THP-1, spleen, lymph) while SMAD3 was found to be expressed more generally (Figure 4A and Supplementary Table 5). Thus, we investigated the role of the FLI1/SMAD3 interaction in macrophage differentiation, using qRT-PCR to record a time-course of expression of both TFs during differentiation of THP-1 monoblasts to monocytes following stimulation by PMA. Strikingly, both TFs were coordinately down-regulated at early time points during differentiation (Figure 4C). These data are supported by previous findings, in which SMAD3 has been shown to regulate cell proliferation through TGF-β1 signaling (Meran et al., 2008), and FLI1 has been shown to re-activate NOTCH pathways resulting in p53-dependent cell cycle arrest (Ban et al., 2008). A hypothesis for future work is that FLI1/SMAD3 may function together as a repressor complex that controls cell proliferation during differentiation (Figure 4D).
DISCUSSION
In this study, we have mapped an atlas of combinatorial interactions among the majority of human and mouse TFs. This work makes available a number of significant resources for the biomedical community, including a database of over 1,600 human or mouse TF-TF interactions (Supplementary Tables 2,3) and quantitative TF expression measurements across human and mouse tissues (Supplementary Table 5). The data highlight conserved TF subnetworks whose patterns of interaction and tissue specificity suggest transcriptional complexes in control of tissue identity.
Our analysis, derived by the integration of these datasets, supports a model whereby the transcriptional network structure is dominated by facilitator TFs expressed broadly across tissues (Figure 1 and Supplementary Table 1). The implication is that tissue identity is not determined by tissue-restricted TFs, but relies on tissue-restricted interaction among TFs. Each TF may be expressed in a variety of tissues, but it is only where two TFs are co-expressed and co-localized that an interaction, and its functional consequences, may occur. In this model, tissues restricted TFs (specifiers) tend to interact with TFs that are broadly-expressed (Figure 1), increasing the number of possible combinatorial events only in certain tissues or during tightly-regulated developmental processes. In support of this interaction-centric model, we identified a subnetwork of just 15 TFs that was sufficient to confer maximal separation of tissues and stem cell lines into the three germ layers associated with embryogenesis (Figure 2). This network significantly outperformed tissue separation based on the expression of individual factors alone. Two thirds of these “germ layer” factors were facilitator TFs expressed in the majority of tissues. .
The theme of “specificity through interaction” is also evident among the conserved TF subnetworks (Figure 3). The majority of TFs in these networks are broadly expressed, and it is the minority of TFs that confer tissue specificity. Further evidence comes from the four identified TF complexes we validated and placed into biological contexts (Figure 4 and Supplementary Table 5). Although they were not selected on this basis, at least three of these complexes involve combination of a tissue restricted TF (i.e., NR3C1, PPARG, FLI1) with a partner whose expression pattern is more widespread (RXRB, RXRB, SMAD3).
The availability of large TF-TF combinatorial interaction networks in both human and mouse will provide many opportunities to study network conservation and divergence over the course of mammalian evolution. Debate is still ongoing regarding the rate at which various types of molecular networks evolve. Here, we found that conservation between human and mouse TF-TF interactions was moderate (Figure 3), in the range of 34 to 64 percent. In contrast, a recent comparison of transcriptional (protein-DNA) interactions reported that this type of network is highly divergent over even very short evolutionary timescales (Tuch et al., 2008). A comparison of genetic networks (synthetic lethal and epistatic interactions) also found extreme rates of divergence (Roguev et al., 2008). On the other hand, protein-protein interactions, especially those that form major structural and functional components of the eukaryotic cell, were found to be highly conserved (Tan et al., 2008a). Protein-protein interactions forming transcriptional complexes, as we have studied here, appear to be conserved at an intermediate level somewhere between the extremes. That is, TF-TF complexes are likely more mutable than the major complexes of cell structure and central metabolism, but much less so than the rapid rewiring that appears to take place in networks of transcription factor/promoter binding.
It has long been appreciated that gene regulation involves combinatorial interactions among transcription factors. The contribution of the present work is to map, on a global scale, precisely what many of these connections are. With few exceptions, almost all of the uncovered connections are undocumented in the existing literature. Future work will dissect more precisely how each of these combinations contributes to developmental programs and to an individual’s relative state of health or disease.
EXPERIMENTAL PROCEDURES
Mammalian two-hybrid assays
Following PCR amplification of full-length TFs, M2H was carried out as previously described (Usui et al., 2005). To assess potential for self-activation each BIND TF fragment (bait) was transfected into CHO-K1 cells containing the luciferase reporter plasmid pG5luc. Reporter activity was measured after 20h and BIND samples with high self-activation (more than 5-fold larger than average) were removed. For non-self-activating baits, eight BIND TF fragments (baits) and two ACT TF fragments (preys) were co-transfected into CHO-K1 cells with pG5luc2, and luciferase reporter activity was measured after 20h. The screen was also performed using two BIND TFs combined with two ACT TFs. For transfections with positive reporter activity, the assay was repeated using all 2×2 or 8×2 BIND/ACT combinations to identify the interacting TF pairs. Positive interactions were scored as those that showed at least three times higher luciferase activity than background (measured using transfection of either an ACT-TF or BIND-TF alone). For more details see Supplementary Information and Supplementary Tables 2,3.
In vitro pull-down assay
PCR products encoding the TF coding sequence and the SV40LPAS fragment were used to construct a template for in vitro transcription/translation. The products were combined by overlapping PCR using the primer pair T7-RBS-KOZAK (5′-GAGCGCGCGTAATACGACTCACTATAGGGGAAGGAGCCGCCACCATG-3′) and LGT10L (5′-AGCAAGTTCAGCCTGGTTAAG-3′), yielding a final template encoding a 5′ T7 RNA polymerase promoter. In vitro pull-down assays were carried out as previously described (Suzuki et al., 2004). Briefly, biotinylated or [35S]-labeled TF was synthesized in vitro from the template using Transcend™ Biotinylated lysine-tRNA (Promega) or Redivue L-[35S]-methionine (Amersham Biosciences) in combination with the TNT T7 Quick Coupled Transcription/Translation System (Promega). After confirmation of [35S]-labeled protein synthesis by SDS–PAGE and autoradiography, biotinylated protein and [35S]-labeled protein were mixed 1:1 and incubated on ice for one hour. Control reactions containing [35S]-labeled protein alone were conducted in parallel. The reaction was then incubated with streptavidin Dynabeads (Dynal Biotech, Milwaukee, WI) for 30 min at 4°C on a rotary shaker. Dynabeads were isolated with a magnet and washed 5 times with ice-cold TBST buffer (50 mM Tris-HCl pH 8.0, 137 mM NaCl, 2.68 mM KCl, 0.1% Tween 20). The amount of radio-labeled protein co-precipitated with the biotinylated protein was measured by scintillation counting or was detected by SDS-PAGE. The ratio of scintillations with and without biotinylated protein was calculated to measure the interaction between the two proteins (Supplementary Table 4).
Tissue specificity score (TSPS)
The value , the fractional expression level of TF i in tissue j, was computed as the ratio of the TF expression level in tissue j (qRT-PCR) to its sum total expression level across all tissues. Tissue specificity TSPSi was then computed using relative entropy:
where qi is the fractional expression of TFi under a null model assuming uniform expression across tissues. According to this definition, a minimal TSPS = 0 would be reported for TFs expressed uniformly across all tissues, while a maximal TSPS ≌ 5 would be reported for TFs expressed only in a single tissue. The threshold chosen for classifying TFs as tissue “specifiers” (TSPS ≥ 1) was based on the observed bimodal distribution of expression over all TFs and tissues (Figure 1A). This threshold is conservative, as it selects TFs with roughly a 20-fold expression difference or greater across tissues (Supplementary Tables 1 and 5).
Unsupervised tissue separation
Two different feature sets were considered for tissue separation: (1) TF expression values and (2) TF-TF interaction values. For both feature sets the raw qRT-PCR expression values were normalized so that each tissue had the same average value over all TFs, then log transformed (Supplementary Tables 1,5). Following (Taylor et al., 2009) interaction values were computed for each interaction between a hub and any other TF, with hubs taken as TFs with > 12 interactions (Figure 1C Supplementary and Tables 2,3). Separations were performed using a hybrid two-phase procedure. The first phase was non-centered Principal Components Analysis (ncPCA), in which the second principal component resulting from this analysis (PC2) was found to be the main direction informative for tissue separation (either feature set). The features were then ranked according to their absolute PC2 loadings and a second phase of dimensionality reduction was performed using the ranked features. For this second phase, non-centered Kernel PCA (ncKPCA) was used with two parameters: (1) the standard deviation of the Gaussian kernel and (2) the number of top-ranked features selected for separation. Performance of separation into the tissue classes was measured by the Bezdek cluster validity index (CVI) considering the first two dimensions (PC1, PC2). Further details are provided in Supplementary Information.
We also examined the dependence of tissue specification on the particular network used. Although the M2H network reported here (Supplementary Tables 2,3) is the first large-scale experimental screen for TF-TF interactions, previous studies have sought to predict relevant TF combinations based on co-occurrence of TF binding sites within gene promoters (Yu et al., 2006). However, we found that a network of TF pairs predicted using binding site co-occurrence did not perform as well as the network of physical TF interactions elucidated by M2H and previous literature (Figure 2A). We also found that the performance of network-based tissue specification was not dependent on the particular algorithm used for separation. Both ncKPCA and Sammon Mapping approaches yielded very similar performance with Cluster Validity Index (CVI) ≌ 1, and in both cases CVI was maximized for exactly six interactions (Figure 2F).
Supplementary Material
Acknowledgments
The work for the RIKEN Omics Science Center was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) through the Genome Network Project and for the RIKEN Omics Science Center (YH, Principal Investigator). Members of the FANTOM Consortium were supported by grant MH062261 from the US National Institute of Mental Health (TR, KT, TI), the King Abdullah University of Science and Technology (TR, VBB), the Max Planck Society for the Advancement of Science (AK), the SA National Bioinformatics Network (SS, AR, VBB, WAH), the Claude Leon Foundation (MK), a CJ Martin Fellowship from the Australian NHMRC (ARRF), and the Scuola Interpolitecnica di Dottorato (CVC). The authors gratefully acknowledge S. Choi for critical feedback on the manuscript.
The FANTOM Consortium:
Timothy Ravasi*1,2, Carlo Vittorio Cannistraci*1,2,3,4,5, Shintaro Katayama*6, Vladimir B. Bajic*1,7, Kai Tan2#, Altuna Akalin8, Sebastian Schmeier7, Mutsumi Kanamori-Katayama6, Nicolas Bertin6, Piero Carninci6, Carsten O. Daub6, Alistair R. R. Forrest6,9, Julian Gough10, Sean Grimmond11, Jung-Hoon Han12, Takehiro Hashimoto6, Winston Hide7,13, Oliver Hofmann7, Hideya Kawaji6, Atsutaka Kubosaki6, Timo Lassmann6, Erik van Nimwegen14, Chihiro Ogawa6, Rohan D. Teasdale11, Jesper Tegnér15, 16, Boris Lenhard8, Sarah A. Teichmann12, David A. Hume17, Trey Ideker†2,18
Riken Omics Science Center:
Takahiro Arakawa6, Noriko Ninomiya6, Kayoko Murakami6, Michihira Tagami6, Shiro Fukuda6, Kengo Imamura6, Chikatoshi Kai6, Ryoko Ishihara6, Yayoi Kitazume6, Jun Kawai6
General Organizers:
Harukazu Suzuki*6, Yoshihide Hayashizaki†6
Affiliations:
1. Red Sea Integrative Systems Biology Laboratory, Division of Chemical & Life Sciences and Engineering, Computational Bioscience Research Center (CBRC), King Abdullah University for Science and Technology (KAUST), Jeddah, Kingdom of Saudi Arabia.
2. Departments of Medicine and Bioengineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093 USA.
3. Department of Mechanics, Politecnico di Torino, Turin, Italy
4. Proteome Biochemistry, San Raffaele Scientific Institute, Milan, Italy
5. CMP Group Microsoft Research, Politecnico di Torino, Turin, Italy.
6. RIKEN Omics Science Center, RIKEN Yokohama Institute, 1-7-22 Suehiro-cho Tsurumi-ku Yokohama, Kanagawa, 230-0045 Japan.
7. South African National Bioinformatics Institute, University of the Western Cape, Private Bag X17, Bellville, 7535 South Africa
8. Bergen Center for Computational Science, Høyteknologisenteret Thormøhlensgate 55, N-5008 Bergen, Norway.
9. The Eskitis Institute for Cell and Molecular Therapies, Griffith University, QLD 4111, Australia.
10. Department of Computer Science, University of Bristol, Merchant Venturers Building, Woodland Road, Bristol, BS8 1UB, UK
11. Australian Research Council (ARC) Special Research Centre for Functional and Applied Genomics, Institute for Molecular Bioscience, The University of Queensland, St. Lucia, QLD 4072, Australia
12. MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK
13. Biostatistics Department, Harvard School of Public Health, 655 Huntington Avenue, Boston, Massachsetts 02115, USA
14. Biozentrum, University of Basel, and Swiss Institute of Bioinformatics, Klingelbergstrasse 50/70, CH-4056 Basel, 4056, Switzerland
15. Computational Medicine Group, Atherosclerosis Research Unit, Center for Molecular Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Solna SE- 171 76 Stockholm, Sweden
16. Department of Physics, Chemistry and Biology, Linköping University, SE-581 83 Linköping, Sweden
17. The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Roslin, EH259PS, UK
18. The Institute for Genomic Medicine, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093 USA
Footnotes
The data and analysis results of the paper are available from: http://fantom.gsc.riken.jp/4/tf-ppi.
Competing interests’ statement: The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameyar M, Wisniewska M, Weitzman JB. A role for AP-1 in apoptosis: the case for and against. Biochimie. 2003;85:747–752. doi: 10.1016/j.biochi.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Ban J, Bennani-Baiti IM, Kauer M, Schaefer KL, Poremba C, Jug G, Schwentner R, Smrzka O, Muehlbacher K, Aryee DN, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 2008;68:7100–7109. doi: 10.1158/0008-5472.CAN-07-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Duverger O, Morasso MI. Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J Cell Physiol. 2008;216:337–346. doi: 10.1002/jcp.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783:2174–2184. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Stem cells, pluripotency and nuclear reprogramming. J Thromb Haemost . 2009;7(Suppl 1):21–23. doi: 10.1111/j.1538-7836.2009.03418.x. [DOI] [PubMed] [Google Scholar]
- Jin G, Zhang S, Zhang XS, Chen L. Hubs with network motifs organize modularity dynamically in the protein-protein interaction network of yeast. PLoS ONE. 2007;2:e1207. doi: 10.1371/journal.pone.0001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kalaev M, Smoot M, Ideker T, Sharan R. NetworkBLAST: comparative analysis of protein networks. Bioinformatics. 2008;24:594–596. doi: 10.1093/bioinformatics/btm630. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meran S, Thomas DW, Stephens P, Enoch S, Martin J, Steadman R, Phillips AO. Hyaluronan facilitates transforming growth factor-beta1-mediated fibroblast proliferation. J Biol Chem. 2008;283:6530–6545. doi: 10.1074/jbc.M704819200. [DOI] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef F, Huelsken J. Cell-type-specific transcriptomics in chimeric models using transcriptome-based masks. Nucleic Acids Res. 2005;33:e111. doi: 10.1093/nar/gni104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Lee S, Bolser D, Schroeder M, Lappe M, Oh D, Bhak J. Comparative interactomics analysis of protein family interaction networks using PSIMAP (protein structural interactome map) Bioinformatics. 2005;21:3234–3240. doi: 10.1093/bioinformatics/bti512. [DOI] [PubMed] [Google Scholar]
- Roach JC, Smith KD, Strobe KL, Nissen SM, Haudenschild CD, Zhou D, Vasicek TJ, Held GA, Stolovitzky GA, Hood LE, et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc Natl Acad Sci U S A. 2007;104:16245–16250. doi: 10.1073/pnas.0707757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci U S A. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Gentles AJ, Elchuri S, Sahoo D, Soen Y, Sharpe O, Perez OD, Chang M, Mitchel D, Robinson WH, et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 2008;68:5132–5142. doi: 10.1158/0008-5472.CAN-07-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Fukunishi Y, Kagawa I, Saito R, Oda H, Endo T, Kondo S, Bono H, Okazaki Y, Hayashizaki Y. Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 2001;11:1758–1765. doi: 10.1101/gr.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ogawa C, Usui K, Hayashizaki Y. In vitro pull-down assay without expression constructs. Biotechniques 37. 2004;918:920. doi: 10.2144/04376BM06. [DOI] [PubMed] [Google Scholar]
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci U S A. 2008a;105:2934–2939. doi: 10.1073/pnas.0708670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Tegner J, Ravasi T. Integrated approaches to uncovering transcription regulatory networks in mammalian cells. Genomics. 2008b;91:219–231. doi: 10.1016/j.ygeno.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- Usui K, Katayama S, Kanamori-Katayama M, Ogawa C, Kai C, Okada M, Kawai J, Arakawa T, Carninci P, Itoh M, et al. Protein-protein interactions of the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Genome Biol. 2005;6:R98. doi: 10.1186/gb-2005-6-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Walhout AJ. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Wen X, Fuhrman S, Michaels GS, Carr DB, Smith S, Barker JL, Somogyi R. Large-scale temporal gene expression mapping of central nervous system development. Proc Natl Acad Sci U S A. 1998;95:334–339. doi: 10.1073/pnas.95.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34:4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, et al. The functional landscape of mouse gene expression. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Ma X, Sun F. The effects of protein interactions, gene essentiality and regulatory regions on expression variation. BMC Syst Biol. 2008;2:54. doi: 10.1186/1752-0509-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.