Abstract
Background
Natural rubber is a biopolymer with exceptional qualities that cannot be completely replaced using synthetic alternatives. Although several key enzymes in the rubber biosynthetic pathway have been isolated, mainly from plants such as Hevea brasiliensis, Ficus spec. and the desert shrub Parthenium argentatum, there have been no in planta functional studies, e.g. by RNA interference, due to the absence of efficient and reproducible protocols for genetic engineering. In contrast, the Russian dandelion Taraxacum koksaghyz, which has long been considered as a potential alternative source of low-cost natural rubber, has a rapid life cycle and can be genetically transformed using a simple and reliable procedure. However, there is very little molecular data available for either the rubber polymer itself or its biosynthesis in T. koksaghyz.
Results
We established a method for the purification of rubber particles - the active sites of rubber biosynthesis - from T. koksaghyz latex. Photon correlation spectroscopy and transmission electron microscopy revealed an average particle size of 320 nm, and 13C nuclear magnetic resonance (NMR) spectroscopy confirmed that isolated rubber particles contain poly(cis-1,4-isoprene) with a purity >95%. Size exclusion chromatography indicated that the weight average molecular mass ( w) of T. koksaghyz natural rubber is 4,000-5,000 kDa. Rubber particles showed rubber transferase activity of 0.2 pmol min-1 mg-1. Ex vivo rubber biosynthesis experiments resulted in a skewed unimodal distribution of [1-14C]isopentenyl pyrophosphate (IPP) incorporation at a
w) of T. koksaghyz natural rubber is 4,000-5,000 kDa. Rubber particles showed rubber transferase activity of 0.2 pmol min-1 mg-1. Ex vivo rubber biosynthesis experiments resulted in a skewed unimodal distribution of [1-14C]isopentenyl pyrophosphate (IPP) incorporation at a  w of 2,500 kDa. Characterization of recently isolated cis-prenyltransferases (CPTs) from T. koksaghyz revealed that these enzymes are associated with rubber particles and are able to produce long-chain polyprenols in yeast.
w of 2,500 kDa. Characterization of recently isolated cis-prenyltransferases (CPTs) from T. koksaghyz revealed that these enzymes are associated with rubber particles and are able to produce long-chain polyprenols in yeast.
Conclusions
T. koksaghyz rubber particles are similar to those described for H. brasiliensis. They contain very pure, high molecular mass poly(cis-1,4-isoprene) and the chain elongation process can be studied ex vivo. Because of their localization on rubber particles and their activity in yeast, we propose that the recently described T. koksaghyz CPTs are the major rubber chain elongating enzymes in this species. T. koksaghyz is amenable to genetic analysis and modification, and therefore could be used as a model species for the investigation and comparison of rubber biosynthesis.
Background
Natural rubber poly(cis-1,4-isoprene) with a molecular mass of 10-10,000 kDa is one of the most important industrial raw materials in the world, and its sole commercial source is currently the para rubber tree Hevea brasiliensis [1]. Other sources, such as Russian dandelion (Taraxacum koksaghyz Rodin) and Guayule (Parthenium argentatum), could be useful in the event of supply shortages as well as providing a suitable alternative for people with allergies to hevein, a major allergen present in H. brasiliensis latex [2,3].
The biosynthesis of natural rubber takes place in the latex of laticifers or specialized parenchyma cells in the bark [1], where it is stored in rubber particles as an end product. Ultrastructural analysis of rubber particles from different species [4-6] revealed an almost identical globular structure that contains a homogeneous hydrophobic rubber core surrounded by an intact monolayer membrane. The monolayer membrane includes a mixture of lipids, proteins and other molecules with the hydrophilic portions of the phospholipids and glycosylated particle-bound proteins facing the cytoplasm [6-10]. The size of rubber particles ranges from 0.08-2 μm in H. brasiliensis, 0.2-6.5 μm in Ficus species and 1-2 μm in P. argentatum [6,11].
Natural rubber is synthesized by adding activated 2-methyl-1,3-butadiene (isopentenyl diphosphate, IPP) to the growing chain [12,13]. This reaction is catalyzed by specific long-chain cis-prenyltransferases (CPTs, EC 2.5.1.20), which are probably located on the surface of rubber particles. According to their function, CPTs are classified as short-, medium- or long-chain polymerizing enzymes and can be distinguished from trans-prenyltransferases (TPTs) by the presence of five conserved protein motifs [14]. They are found in bacteria [15], yeast [16], animals including humans [17] and plants [18-22].
Recently, two CPTs (RER2 and SRT1) were isolated from Saccharomyces cerevisiae and were shown to be responsible for the biosynthesis of dolichol, a long-chain polyprenol with a saturated alpha-isoprene unit, which serves as a glycosyl carrier for protein glycosylation in the endoplasmic reticulum [23]. The first plant CPT was identified in Arabidopsis thaliana (ACPT), and appears to be required for normal growth and development [24]. The latex of H. brasiliensis contains at least two CPTs, designated HRT1 and HRT2 (for Hevea rubber transferase). The addition of recombinant HRT2 to washed latex particles supplemented with radioactively-labeled IPP resulted in the significant production of a high-molecular-weight labeled rubber product, whereas recombinant HRT1 showed no significant activity [25]. In vitro, initiation of rubber biosynthesis by HRT requires intact particles, isopentenyl diphosphate (IPP), allylic diphosphates such as farnesyl diphosphate (FPP) and divalent metal cations (Mg2+ or Mn2+) as a co-factor [26-28]. However, all attempts to purify a functional rubber transferase from rubber particles have failed, suggesting that the native enzyme needs additional factors for its activity [29].
In this study, we report the comprehensive analysis of rubber particles from T. koksaghyz. The rubber particles contained very pure poly(cis-1,4-isoprene) and retained their capacity to produce natural rubber ex vivo. Immunological analysis revealed that CPTs associated with these particles remain fully functional when expressed as recombinant proteins either in Saccharomyces cerevisiae or tobacco protoplasts.
Results and Discussion
Physical characterization of purified T. koksaghyz rubber particles
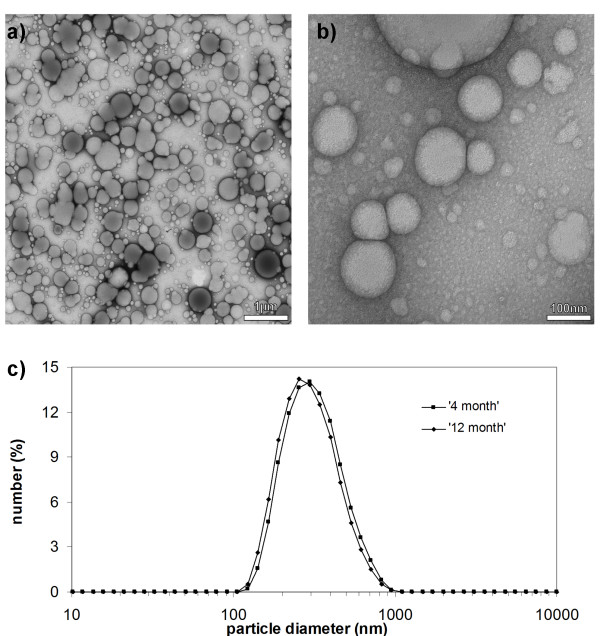
Rubber particles examined by transmission electron microscopy at low magnification appeared approximately spherical, with diameters ranging from 0.2 to almost 1 μm (Figure 1a). At higher magnification, additional small and mostly spherical particles became visible with the smallest barely 10 nm in diameter (Figure 1b). Frequently, those rubber particles touching each other were deformed by the effect of surface tension in the staining solution during air-drying [30]. A similar unimodal particle size distribution ranging from 0.2-0.7 μm was observed by photon correlation spectroscopy, using rubber particles from the latex of 4-month-old and 1-year-old T. koksaghyz plants (Figure 1c). The average size of the rubber particles was 320 nm, and more than 50% of particles were in the size range 250-400 nm. The age of the plants appeared not to affect particle size. Size exclusion chromatography (SEC) analysis of both samples revealed an weight average molecular mass of ~ 4,750 kDa with a unimodal distribution, which again showed no significant variation as a function of plant age (data not shown).
Figure 1.
Morphology of rubber particles from Taraxacum koksaghyz. Transmission electron micrographs at low a) and high b) magnification. c) Rubber particle size distribution measured by photon correlation spectroscopy in latex of 4- and 12-month-old plants. Magnifications in a) and b) are indicated by bar size.
Concentration and chemical properties of T. koksaghyz rubber
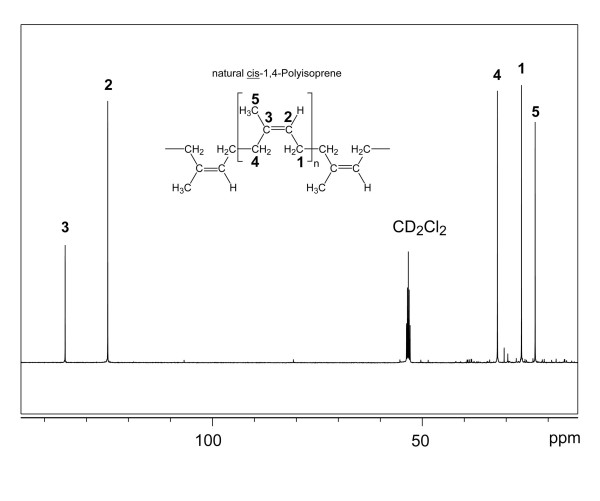
The concentration of rubber in the latex of greenhouse-cultivated T. koksaghyz plants was monitored from 4-18 months after sowing, revealing an increase during the first 8 months after which the concentration leveled off and remained constant at 130-150 mg dry rubber ml-1 latex thereafter. Although the absolute rubber concentration varies in different T. koksaghyz accessions and under different growth conditions, our results agree with earlier investigations of rubber production in field-grown T. koksaghyz plants, where rubber accumulated only during the first growth season and then remained at constant levels [31]. To determine the chemical properties of T. koksaghyz rubber, we performed one- and two-dimensional NMR analysis of isolated rubber particles dissolved in dichloromethane. The 13C NMR spectrum displayed five dominant signals at chemical shifts indicative for poly(cis-1,4-isoprene), but not for poly(trans-1,4-isoprene) (Table 1 and Figure 2) [32]. Notably, the 13C NMR chemical shifts of C-4 and C-5 differ by more than 5 ppm for the cis- and trans-configured compounds (Table 1). Therefore, the observed frequencies leave no doubt that T. koksaghyz rubber is poly(cis-1,4-isoprene). Moreover, all 1H13C and 1H1H correlations detected in two-dimensional HMQC and COSY experiments, respectively, fully agreed with the couplings expected for poly(cis-1,4-isoprene) (Table 1). The NMR spectra contained few low-intensity signals arising from impurities, indicating that the purity of poly(cis-1,4-isoprene) in T. koksaghyz rubber is >95%.
Table 1.
NMR data of poly(cis-1,4-isoprene) from T. koksaghyz.
| Chemical shifts* | Observed correlations in | Published 13C-NMR chemical shifts** | ||||
|---|---|---|---|---|---|---|
| Position | 1H [ppm] | 13C [ppm] | COSY | HMBC | cis-Polyisoprene | trans-Polyisoprene |
| 1 | 2.1 | 27.0 | 2 | 4 | 26.3 | 26.4 |
| 2 | 5.2 | 124.0 | 1, 5(w)*** | 5, 1, 4 | 124.8 | 123.9 |
| 3 | 134.2 | 1, 5 | 134.8 | 134.5 | ||
| 4 | 2.1 | 31.4 | 5, 1 | 31.8 | 39.3 | |
| 5 | 1.7 | 22.3 | 1(w) | 4 | 22.9 | 15.3 |
* referenced to the solvent signals; **[32]; *** (w), weak cross peak
The solvent was CD2Cl2.
Figure 2.
13C NMR spectrum of rubber particles isolated from Taraxacum koksaghyz. The signal arising from the deuterated sample is indicated. The inset displays the structure and the carbon numbering for poly(cis-1,4-isoprene).
In vitro biosynthetic activity of rubber particles
The rubber biosynthetic activity of T. koksaghyz rubber particles was characterized in vitro by assaying the incorporation of the radiolabeled precursor IPP, which should become incorporated into the polymer and thus trapped in the rubber particles [21,33]. The IPP incorporation assay was carried out using intact isolated rubber particles as well as particles pre-treated with proteinase K to destroy particle-associated enzymes and other proteins. The treated and untreated particles were tested by SDS-PAGE and Coomassie Brilliant Blue staining, showing that the protein bands normally found in the particles were eliminated by proteinase K treatment (data not shown). This control allowed us to distinguish between physical and enzymatic incorporation of IPP into the particle, but had no influence either on particle integrity or stability. Earlier investigations into the in vitro activity of rubber particles used boiled particles as a control [33], which was not suitable for T. koksaghyz particles because of their rapid temperature-dependent agglomeration which changes the surface area to which IPP could be attached.
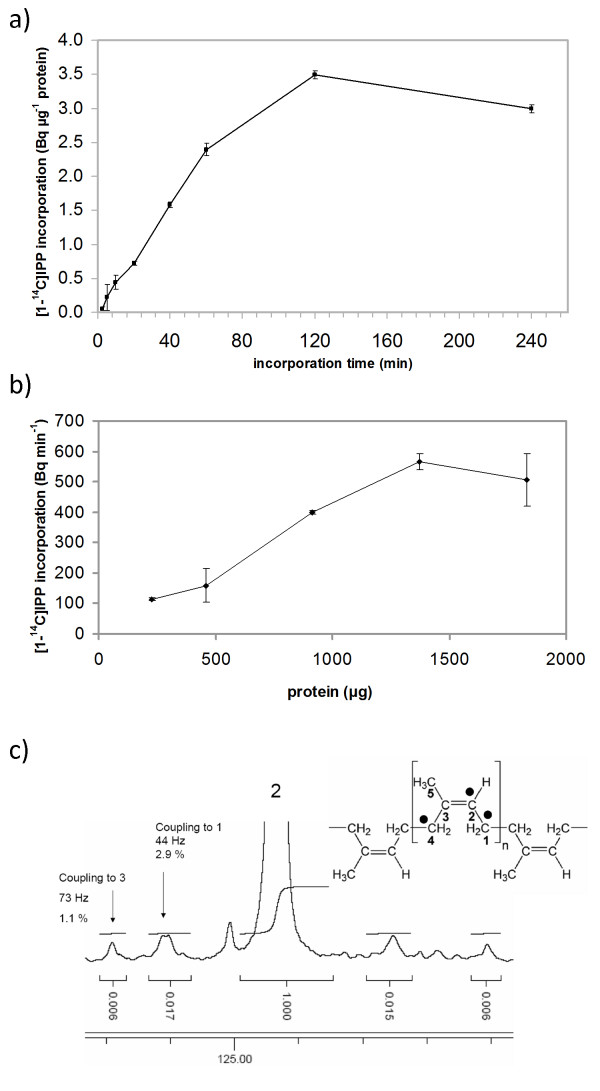
In time course experiments, T. koksaghyz rubber particles enzymatically incorporated increasing amounts of [1-14C]IPP over the incubation time (Figure 3a). Saturation was achieved after 2 h, possibly reflecting the loss of chain elongation activity due to the particles becoming unstable as reported for other rubber-producing plants [29]. The incorporation of [1-14C]IPP also increased as more rubber particles were added to the reaction, giving a sigmoid curve progression that reflected an enzymatic incorporation process. The enzymatic activity of T. koksaghyz rubber particles in vitro was about 0.2 pmol min-1 mg-1.
Figure 3.
Incorporation of IPP into rubber particles. a) Time course-dependent and b) rubber particle protein dependent incorporation of [1-14C]IPP. c) 13C NMR signal of C-2 of poly(cis-1,4-isoprene) from the labeling experiment with [1,2,4-13C3]IPP. The filled circles indicate 13C atoms from [1, 2, 4-13C3]IPP. Values in a) and b) represent mean (± standard deviation) from three biological repetitions.
To calculate the rubber transferase activity of the rubber particles, IPP incorporation was measured as a function of protein concentration (Figure 3b), whereas the mean amount of protein in T. koksaghyz rubber particles remained approximately constant at 70-80 μg mg-1. This allowed us to calculate the enzymatic [1-14C]IPP incorporation activity of 2.6-3 pmol min-1 mg-1 rubber particle protein, which corresponds to the findings for rubber particles from other plants (0.08-13 pmol min-1 mg-1 for H. brasiliensis, 0.03-6 pmol min-1 mg-1 for F. carica and 1.3 pmol min-1 mg-1 for P. argentatum [27,33,34]). The range of rubber particle activity observed within each species could reflect the different assay conditions and particle extraction methods. For example, rubber transferase activity is thought to be Mg2+-dependent [35], and the concentration of Mg2+ may differ between species, whereas optimal concentrations for H. brasiliensis and P. argentatum were 2 mM and 8 mM, respectively [33,36]. We therefore varied the Mg2+ concentrations in the in vitro rubber transferase assay for T. koksaghyz rubber particles, and found the optimal activity at 4-5 mM Mg2+ (Table 2). To verify that rubber chain elongation in T. koksaghyz rubber particles occurred by the transfer of IPP in the cis-configuration, a sophisticated labeling assay was developed in which rubber particles were incubated with [1, 2, 4-13C3]IPP, vacuum dried and the residual material dissolved in CD2Cl2 for 13C NMR analysis. The 13C NMR signal of C-2 poly(cis-1,4-isoprene) (Figure 3c) showed, in addition to the intense central signal, pairs of coupling satellites characterized by coupling constants of 73 Hz (coupling to 13C-3) and 44 Hz (coupling to 13C-1). The C-1 satellite pair was approximately three times more intense than the C-3 pair (Figure 3c), indicating that isotopologues with adjacent 13C-1 and 13C-2 were slightly enriched in accordance with their biosynthetic origin from [1, 2, 4-13C3]IPP (cf. filled circles in Figure 3c). The low enrichment factor can be explained by the large excess of unlabeled poly(cis-1,4-isoprene) at the beginning of the labeling reaction. Nevertheless, the non-stochastic distribution of the satellite pairs provides additional evidence for cis-prenyltransferase activity under these experimental conditions.
Table 2.
Effect of Mg2+ ion on [1-14C]IPP incorporation of T. koksaghyz rubber particles.
| MgCl2 [mM] |
[1-14C]IPP incorporation [Bq μg-1protein] |
|---|---|
| 2 mM | 0.57 (± 0.023) |
| 3 mM | 0.83 (± 0.037) |
| 4 mM | 0.89 (± 0.039) |
| 5 mM | 0.91 (± 0.040) |
| 7.5 mM | 0.52 (± 0.023) |
| 10 mM | 0.04 (± 0.002) |
For the ex vivo rubber biosynthesis experiment 3 mg of freshly prepared rubber particles was incubated for 60 min with [1-14C]IPP in IPP assay buffer containing different concentrations of Mg2+ ion. Values represent mean (± standard deviation) from three biological repetitions.
The distribution of [1-14C]IPP transfer to existing rubber molecules of different molecular masses was investigated by SEC (Figure 4 and Table 3). This revealed a typical skewed unimodal mass, with a weight average molecular mass ( w) of 5,170 kDa (± 134 kDa) and number average molecular mass (
w) of 5,170 kDa (± 134 kDa) and number average molecular mass ( n) of 2,460 kDa (± 142 kDa), indicating a low level polydispersity of 2.1. Again, the incorporation of [1-14C]IPP increased with the incubation time (Figure 4a). After 5 min the incorporation of [1-14C]IPP occurred at a significantly higher rate in untreated particles compared to those treated with proteinase K, whereas the means of labeled material were 2,760 kDa for
n) of 2,460 kDa (± 142 kDa), indicating a low level polydispersity of 2.1. Again, the incorporation of [1-14C]IPP increased with the incubation time (Figure 4a). After 5 min the incorporation of [1-14C]IPP occurred at a significantly higher rate in untreated particles compared to those treated with proteinase K, whereas the means of labeled material were 2,760 kDa for  w and 672 kDa for
w and 672 kDa for  n, respectively. Furthermore, the SEC data indicated that in vitro rubber chain elongation occurred over a broad range (1-5000 kDa), whereas the highest incorporation rate was observed for molecules with
n, respectively. Furthermore, the SEC data indicated that in vitro rubber chain elongation occurred over a broad range (1-5000 kDa), whereas the highest incorporation rate was observed for molecules with  w of 2,000-2,500 kDa. Interestingly, the
w of 2,000-2,500 kDa. Interestingly, the  w and
w and  n of the [1-14C]IPP labeled rubber decreased during the assay (Figure 4b), increasing the polydispersity (Table 3) and indicating both the preferential transfer of [1-14C]IPP to long-chain molecules and the possibility that new rubber molecules are synthesized after the depletion of the long-chain starter molecules. The lower molecular mass distribution of labeled molecules in comparison to the starting material suggested that chain termination by transferase complex might occur preferentially in molecules >2,500 kDa. These data indicate that in vivo rubber biosynthesis can be only partially reconstituted in vitro as also suggested by Tangpakdee et al. [37].
n of the [1-14C]IPP labeled rubber decreased during the assay (Figure 4b), increasing the polydispersity (Table 3) and indicating both the preferential transfer of [1-14C]IPP to long-chain molecules and the possibility that new rubber molecules are synthesized after the depletion of the long-chain starter molecules. The lower molecular mass distribution of labeled molecules in comparison to the starting material suggested that chain termination by transferase complex might occur preferentially in molecules >2,500 kDa. These data indicate that in vivo rubber biosynthesis can be only partially reconstituted in vitro as also suggested by Tangpakdee et al. [37].
Figure 4.
Molecular mass distribution of [1-14C]IPP-labeled rubber synthesized in vitro. a) SEC profile of labeled material. b)  w and
w and  n of the labeled polymer. IPP incorporation assay was performed and stopped after the time points shown. Extracted polymer material was fractionated by SEC and the radioactivity of the resulting fractions was determined by scintillation. Proteinase K (ProtK)-treated particles controlled for IPP trapped on the rubber particles through non-enzymatic mechanisms. Progression lines represent the mean of two measurements.
n of the labeled polymer. IPP incorporation assay was performed and stopped after the time points shown. Extracted polymer material was fractionated by SEC and the radioactivity of the resulting fractions was determined by scintillation. Proteinase K (ProtK)-treated particles controlled for IPP trapped on the rubber particles through non-enzymatic mechanisms. Progression lines represent the mean of two measurements.
Table 3.
Molecular mass and polydispersity of labeled and unlabeled material from ex vivo IPP incorporation.
| rubber sample |
 w (105 Da) w (105 Da)
|
 n (105 Da) n (105 Da)
|
 w w  n-1 n-1
|
|---|---|---|---|
unlabeled  5-120 min 5-120 min
|
51.7 (± 1.34) | 24.6 (± 1.42) | 2.1 |
labeled  5-20 min 5-20 min
|
26.5 (± 1.01) | 7.1 (± 0.45) | 3.7 |
labeled 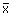 40-120 min 40-120 min
|
22.5 (± 2.27) | 4.1 (± 0.30) | 5.5 |
In an ex vivo rubber biosynthesis experiment 6.5 mg of freshly prepared rubber particles was incubated with [1-14C]IPP for the time periods indicated. Extracted rubber material for each assay was fractionated by size exclusion chromatography and radioactivity levels were determined by scintillation.
Particle associated cis-1,4-polyprenylcistransferases
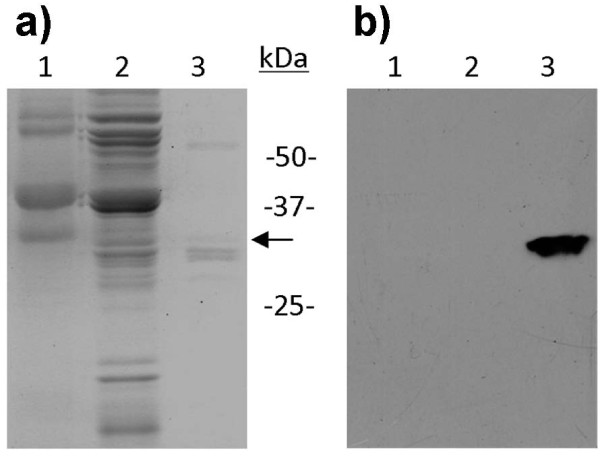
Recently, we identified and isolated three T. koksaghyz CPTs designated TkCPT1-3 [38]. The TkCPT1 and TkCPT2 cDNAs are each 927 bp in length and the TkCPT3 cDNA is 903 bp in length, giving proteins with molecular masses of 32-34 kDa. All three genes are strongly expressed in the latex, but only minimally expressed in other tissues such as leaves, pedicels and roots. Antibodies recognizing the three TkCPTs revealed a strong signal in western blots of the rubber particle fraction of T. koksaghyz latex, with an apparent molecular mass of 34 kDa (Figure 5). Although it has previously been suggested that rubber transferases are associated with rubber particles [39-41], our results for TkCPTs are the first to confirm this theory explicitly.
Figure 5.
Detection of T. koksaghyz CPT in the rubber phase. Latex was harvested and divided into pellet (lane 1), C-serum (lane 2) and rubber phase (lane 3). Proteins were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue a) or transferred to a membrane for western blot analysis using antibodies against TkCPTs b).
Functional analysis of TkCPT1-3
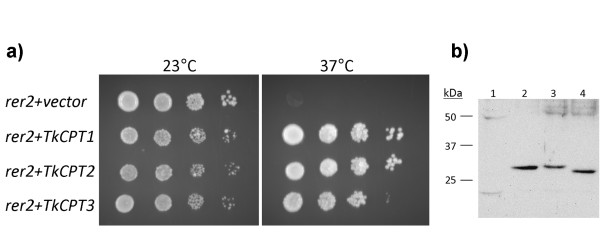
To determine whether the three isolated CPT genes encode functional proteins with long chain cis-prenyltransferase activity in vivo, the cDNAs were expressed in the yeast strain SNH23-7D (Figure 6a), which is a temperature sensitive mutant for the dehydrodolichyl diphosphate (dedol-PP) synthase rer2 [16]. All three T. koksaghyz CPTs were able to suppress the growth deficiency phenotype of the rer2 mutant (Figure 7a), and western blot analysis of the complementing yeast transformants revealed the expression of full length TkCPT1-3 proteins (Figure 7b). The suppression of the temperature sensitive phenotype demonstrated clearly that TkCPT1-3 catalyze the synthesis of the cis-polyprenol dedol-PP, which has a molecular mass of 1.1-1.3 kDa. Similar results were obtained for HRT2, which is believed to be the long-chain rubber producing CPT in H. brasiliensis [25]. The formation of high-molecular-mass polyprenols in yeast is possibly repressed by the rapid de-phosphorylation of the dedol-PP to dolichol, which is not a substrate for the chain elongation process. It has previously been suggested that rubber transferases need co-factors for stability and activity [25,29] but our experiments indicate that, at least for T. koksaghyz CPTs under our experimental conditions, this does not appear to be the case.
Figure 6.
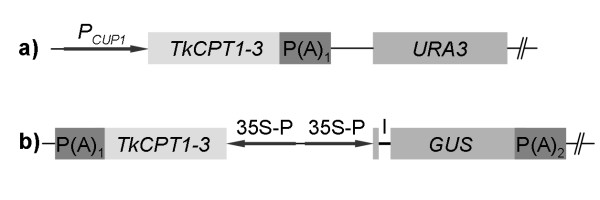
Plasmids for heterologous expression of TkCPT1-3. For heterologous expression of TkCPT1-3 in S. cerevisiae and protoplasts of N. tabacum the corresponding cDNAs were cloned into the plasmids pYEX-BX a) and pCAMBIA-1305.1 b), respectively. PCUP1, S. cerevisiae CUP1 promoter; P(A)1, cauliflower mosaic virus (CaMV) 35S polyadenylation signal sequence; URA3, S. cerevisiae URA3 locus; 35S-P, CaMV 35S promoter; GUS, beta-glucuronidase synthetic construct including catalase intron (I) (GenBank: AAK29426); P(A)2, Agrobacterium tumefaciens D-nopaline synthase polyadenylation signal sequence.
Figure 7.
Functional complementation of the temperature-sensitive dedol-PP synthase rer2 yeast mutant SNH23-D7 by TkCPTs. a) Yeast strain SNH23-D7 transformed with plasmid pYEX-BX, pYEX-TkCPT1, pYEX-TkCPT2 and pYEX-TkCPT3 were dropped on SD-URA plates and incubated for 2 days at the temperatures shown. b) Western blots were performed with antibodies against TkCPTs to detect TkCPT in yeast SNH23-D7 (rer2) transformants pYEX-BX (vector control) (1), pYEX-TkCPT1 (2), pYEX-TkCPT2 (3) and pYEX-TkCPT3 (4).
More detailed analysis of TkCPT activity was carried out using N. tabacum mesophyll protoplasts transfected with each of the cDNAs (Figure 6b, Table 4). In order to avoid qualitative or quantitative differences in protoplast and/or plasmid DNA preparations interfering with the direct comparison of CPT activities between transfection experiments, beta-glucuronidase was co-expressed from the same plasmid as an internal standard as described by Lepetit et al. [42]. Protein extracts from protoplasts expressing each of the TkCPTs showed strong IPP transferase activity, with no significant difference between the three cDNAs. It is therefore most likely that all three enzymes act cooperatively in T. koksaghyz rubber biosynthesis.
Table 4.
Heterologous expression of TkCPTs in Nicotiana tabacum protoplasts.
| Enzymes | GUS activity (μmol min-1 μg-1) | CPT activity (pmol min-1 mg-1) | CPT/GUS ratio |
|---|---|---|---|
| GUS/- | 1.78 (± 0.10) | - | - |
| GUS/TkCPT1 | 1.34 (± 0.16) | 3.48 (± 0.38) | 2.73 (± 0.50) |
| GUS/TkCPT2 | 0.97 (± 0.15) | 2.05 (± 0.37) | 2.28 (± 0.70) |
| GUS/TkCPT3 | 1.22 (± 0.21) | 2.90 (± 0.64) | 2.50 (± 0.16) |
Protoplasts were transfected with plasmids containing both GUS and TkCPTs or GUS alone in all cases controlled by the CaMV 35S promoter. Protoplast protein extracts were used for measurement of enzyme activities, whereas GUS activity served as the internal standard for normalization of transfection experiments. Values represent mean (± standard deviation) from six biological repetitions. No significant differences existed in enzyme activity between TkCPT1, TkCPT2 and TkCPT3, as calculated using Student's t-test (P, 0.01).
Conclusion
We have investigated the physical properties and developmental profile of rubber particles from T. koksaghyz latex and have shown that they possess intrinsic cis-1,4-polyprenylcistransferase (rubber transferase) activity of 0.2 pmol min-1 mg-1 which can be partially reconstituted in vitro without further co-factors. We are the first to demonstrate conclusively that CPTs are an intrinsic part of the rubber particle and we have demonstrated a correspondence between the CPT activity in isolated rubber particles and the recently identified CPT genes TkCPT1-3 in T. koksaghyz, which are able to complement a yeast strain deficient in CPT activity and maintain their activity in tobacco protoplasts. In vitro rubber biosynthesis experiments resulted in a skewed unimodal distribution of [1-14C]isopentenyl pyrophosphate (IPP) incorporation at a weight average molecular mass of 2,500 kDa. Our data indicate that TkCPT1-3 are responsible for the rubber chain elongation that occurs in T. koksaghyz rubber particles and that their roles in this regard may be redundant. Our experiments provide crucial background information that will allow the development of T. koksaghyz as a potential alternative commercial source of rubber.
Methods
Plant material and cultivation conditions
Taraxacum koksaghyz plants were obtained from the Botanical Gardens Karlsruhe (Karlsruhe, Germany) and cultivated at 18°C with a 16-h photoperiod (20 klx) in controlled growth chambers or in the greenhouse. Plants were cultivated in a prefertilized 1:1 mixture of standard soil (ED73 Einheitserde, Fröndenberg, Germany) and garden mold (Botanical Garden Münster, Germany) and fertilized every 4 weeks with a commercial fertilizer according to the manufacturer's recommendations (Hakaphos Plus, Compo GmbH, Münster, Germany).
Rubber preparation and determination
For the isolation of native and functional rubber particles we followed the general procedure described for H. brasiliensis [27,43] with the following modifications. Latex was harvested from petioles or roots of 20 week old T. koksaghyz plants, if not stated otherwise, by dissecting the tissue with a razor blade and transferring the expelling latex into an equal volume of ice-cold rubber extraction buffer (100 mM Tris.Cl (pH 7.8), 350 mM sorbitol, 10 mM NaCl, 5 mM MgCl2, 5 mM DTT) and centrifuging (12,000 × g, 20 min, 4°C). The latex separated into three fractions (pellet, C-serum and rubber phase) the latter two of which were transferred to a new tube and centrifuged as above. The rubber phase containing the rubber particles was transferred to a fresh tube and washed with 800 μl rubber extraction buffer and then dissolved in rubber extraction buffer and stored briefly at 4°C. To determine the dry rubber content, 20 μl of latex was transferred to a fresh tube and gently overlaid with 20 μl glacial acetic acid to coagulate the rubber particles. Afterwards, the coagulum was air-dried for 24 h.
Photon correlation spectroscopy
Freshly prepared rubber particles were dispersed by brief ultrasonication and then filtered with a 5 μm syringe filter prior to dynamic light scattering with a Zetasizer Nano ZS (Malvern Instruments GmbH, Herrenberg, Germany) containing a He-Ne laser (4.0 mW at 633 nm) and an Avalanche photodiode detector with a Q.E. >50% at 633 nm. Measurements were performed according to the manufacturer's instructions.
Transmission electron microscopy
Freshly prepared T. koksaghyz latex rubber phase was diluted 1:3 with rubber extraction buffer containing 1 mM DTT, and a small droplet was placed for 1 min on a freshly glow-discharged carbon layer (thickness ~ 10 nm) onto a Pioloform film supported by a commercial Cu-mesh grid. After removing excess liquid, the grid was washed with double-distilled water and stained with 2% aqueous uranyl acetate for 30 s before air drying. Transmission electron microscopy (TEM) was carried out using a Philips EM 410 (acceleration voltage 80 kV) in the bright-field mode, and micrographs were recorded on Imaging Plates (Ditabis, Pforzheim, Germany).
IPP-incorporation assay
To measure the incorporation of IPP into rubber, the latex rubber phase was mixed with assay buffer to a final concentration of 100 mM Tris-HCl (pH 7.5), 2.5 mM CaCl2, 10 mM DTT, 1 mM sodium azide, 0.05% Triton X-100, 5 mM MgCl2, 2.8 μM E,E- farnesyl diphosphate (Sigma-Aldrich) and 7.2 μM [1-14C]- or [1, 2, 4-13C3]isopentenyl pyrophosphate (IPP) (GE Healthcare). The reaction mixture was incubated at 30°C for 2 h and then stopped by heating to 95°C for 5 min. The reaction products were hydrolyzed to corresponding alcohols using 2 μl of potato acid phosphatase (1 U μl-1 dissolved in double-distilled water) and 398 μl phosphatase buffer (50 mM sodium acetate (pH 4.7), 0.1% Triton X-100, 60% (v/v) methanol) for 2 h at 37°C as described [44]. The products were extracted by shaking with 600 μl n-hexane for 1 h, air dried and resuspended in 300 μl n-hexane. Radioactivity was measured using a scintillation counter (Beckman Scintillation Counter LS6500) after mixing 50 μl of the extracts with 4 ml Rotiszint® eco plus (Roth, Karlsruhe, Germany). Extracts were air dried and resolved in tetrahydrofuran. Size-exclusion chromatography to determine the molecular size distribution of natural rubber and rubber produced in radioactive assays was carried out as previously described [45].
NMR spectroscopy
One-dimensional 1H and 13C NMR spectra were measured at 500 and 125 MHz, respectively, using a DRX500 or AVANCE 500 spectrometer (Bruker, Rheinstetten, Germany). Two-dimensional HMQC, HMBC and COSY spectra were measured with the AVANCE 500 spectrometer using an inverse probe-head and standard parameter sets implemented in TOPSPIN 1.1. The solvent was deuterated dichloromethane and the temperature was 27°C. The experimental time for 13C NMR spectra was typically 15 h (corresponding to more than 10,000 scans). Data were processed using TOPSPIN 1.1 or MestReNova.
Construction of expression vectors for heterologous expression
RNA from T. koksaghyz latex was isolated as described previously [45] and cDNA synthesis was carried out using the SuperScript II™ Reverse Transcriptase Kit (Invitrogen, Karlsruhe, Germany) with an oligo(dT) primer. For all three TkCPTs (TkCPT1, TkCPT2 and TkCPT3) cDNA was generated using the primer combination cpt-TK_EcoRI (5'-AAA GAA TTC ATG CAA GTG AAT CCA ATC ATT ACT AC-3') and cpt-TK-rev_SalI (5'-AAA GTC GAC TTA TGC CTG CTT CTT CTT CTT CTC C-3'). The products were inserted into the pCRII-TOPO® vector (Invitrogen, Karlsruhe, Germany), sequenced and then transferred using EcoRI and SalI restriction sites into the expression vector pGEX4-T1 (GE Healthcare Europe GmbH, Freiburg, Germany). This allowed the expression of TkCPTs as N-terminal fusions to glutathione-S-transferase (GST) for heterologous expression in Escherichia coli BL21-cells and downstream antibody generation.
For heterologous expression of TkCPT1-3 in Nicotiana tabacum var. SR1 protoplasts, the corresponding cDNAs were amplified with the primers TkCPT_PciI (5'-AAA ACA TGT TAC AAG TGA ATC CAA TCA TTA CTA C-3') and TkCPT-rev_XbaI (5'-AAA ACA TGT TAC AAG TGA ATC CAA TCA TTA CTA C-3') and transferred using the NcoI and XbaI sites into the pUC18-based pAM vector containing the CaMV 35S promoter and polyadenylation sequences from pRT104 [46]. The cassette was then transferred using the KpnI and EcoRI sites into pCAMBIA-1305.1 (GenBank: AF354045), which already contains the gusA reporter gene including a catalase intron under the control of the CaMV 35S promoter.
For expression in yeast, the TkCPT cDNAs were released from pAM using XhoI and EcoRI and introduced into vector pYEXBX (Clontech Laboratories Inc., Saint-Germain-en-Laye, France), which had been digested with SalI and EcoRI.
Generation of antibodies against TkCPTs
The three TkCPT clones in pGEX4-T1 were overexpressed in 300-ml cultures of E. coli strain BL21, induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). Purified TkCPT1 was sequenced by MALDI-MS and administered to rabbits by EUROGENTEC (Seraing, Belgium). The pre-immune and antibody sera were tested for specificity by western blot against the recombinant proteins TkCPT1, TkCPT2 and TKCPT3.
SDS-PAGE and western blots
SDS-PAGE was carried out using 15 μg protein per lane from the pellet, C-serum and rubber phases of fresh T. koksaghyz latex. Protein concentrations were determined using the Bradford method [47]. Proteins were separated on SDS-PAGE gels and either stained with Coomassie Brilliant Blue or transferred to nitrocellulose membranes as described [48]. The membranes were incubated with the primary antibody (1:500 dilution) for 1 h at room temperature, washed and then incubated with a mouse anti-rabbit IgG conjugated to horseradish peroxidase (Sigma, Munich, Germany) according to the manufacturer's instructions. Membranes with HRP-coupled secondary antibodies were imaged on X-ray films by chemiluminescence detection.
Expression of TkCPTs in yeast
Yeast strain SNH23-D7 (MATa rer2-2 mfa1::ADE2 mfa2::TRP1 bar1::HIS3 ade2 trp1 his3 leu2 ura3 lys2) [16] was cultivated in standard YPD medium for 48 h at 20°C, then transformed [49] with pYEXBX-TkCPT1-3, and the pYEXBX base vector as a control. After regeneration, cells were plated on SD-URA agar, colonies for each construct were transferred to 0.9% NaCl and adjusted to different optical densities (1, 0.1, 0.01 and 0.001) and 5-μl droplets were spotted onto SD-URA plates. After 48 h incubation in SD-URA liquid medium, denatured protein extracts were prepared from 20 OD600 units [50] and used for SDS-PAGE and western blot analysis.
Expression in protoplasts and GUS assay
Protoplasts were isolated from Nicotiana tabacum var. SR1 and Ca(NO3)2 polyethylene glycol-mediated DNA transfer was performed as described [51] using 3.3 × 105 protoplasts and 10 μg of pCambia1305.1-TkCPT1-3 DNA per transfection. Frozen protoplasts were sheared in IPP assay buffer, briefly centrifuged and the supernatants pooled from six replicate transfections. Protein concentration was determined using the Bradford method [47] and bovine serum albumin as a standard. Approximately 100 μg of protein extract was used in the IPP incorporation assay and 1 μg in the glucuronidase activity assay with 4-methylumbelliferylglucuronide (4-MUG) as the substrate [52].
Abbreviations
4-MUG: 4-methylumbelliferylglucuronide; ACPT: Arabidopsis thaliana cis-prenyltransferase; COSY: correlation spectroscopy; CPT: cis-1,4-polyprenylcistransferases; FPP: farnesyl pyrophosphate; GUS: beta-glucuronidase; HMBC: heteronuclear multiple bond correlation; HMQC: heteronuclear multiple quantum coherence; HRT: Hevea brasiliensis rubber transferase; IPP: isopentenyl pyrophosphate; NMR: nuclear magnetic resonance; RER2: Saccharomyces cerevisiae dehydrodolichyl diphosphate synthase; SEC: size exclusion chromatography; SRT1: Saccharomyces cerevisiae cis-1,4-polyprenylcistransferases; TEM: transmission electron microscopy; TkCPT: Taraxacum koksaghyz cis-1,4-polyprenylcistransferases; TPT: trans-prenyltransferase
Authors' contributions
TS contributed to results presented in figures 1, 3 and 4. ML generated antibodies and performed experiments for figures 5 and 7. AH, NvD and OM performed heterologous expression of TkCPT in tobacco protoplasts (Table 3) and yeast (Figure 7). RR did the transmission electron microscopy (Figure 1) and WE the nuclear magnetic resonance spectroscopy (Figures 2 and 3c). RF helped with the interpretation of data. DP and CSG set up the experimental outline and prepared the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Ministry of Science and Education of Germany (grant no. FKZ 0313712), by EVONIK Industries AG, by the Hans-Fischer Gesellschaft Munich (to WE), by the Deutsche Bundesstiftung Umwelt (to ML) and the federal state North-Rhine Westphalia co-financed by the European Union.
Contributor Information
Thomas Schmidt, Email: thoschmi@uni-muenster.de.
Malte Lenders, Email: m_lend01@uni-muenster.de.
Andrea Hillebrand, Email: a_hill05@uni-muenster.de.
Nicole van Deenen, Email: nicki.v.d@uni-muenster.de.
Oliver Munt, Email: o.munt@uni-muenster.de.
Rudolf Reichelt, Email: reichru@uni-muenster.de.
Wolfgang Eisenreich, Email: wolfgang.eisenreich@ch.tum.de.
Rainer Fischer, Email: fischer@molbiotech.rwth-aachen.de.
Dirk Prüfer, Email: pruefer@molbiotech.rwth-aachen.de.
Christian Schulze Gronover, Email: c.sg@uni-muenster.de.
Acknowledgements
The authors would like to thank Ursula Malkus (Institut für Medizinische Physik und Biophysik, Münster) for carrying out the TEM analysis of rubber particles. The technical assistance of Sandra Ponanta (Institut für Biochemie und Biotechnologie der Pflanzen, Münster), Raphael Soeur (Fraunhofer Institut für Molekularbiologie und Angewandte Ökologie, Aachen) and Karin Wacker (Fraunhofer Institut für Chemische Technologie, Pfinztal) is gratefully acknowledged. We would like to thank Prof. Dr. Thomas Hirth (Fraunhofer Institut für Grenzflächen und Bioverfahrenstechnik, Stuttgart) and Dr. Ulrich Fehrenbacher (Fraunhofer Institut für Chemische Technologie, Pfinztal) for providing SEC and PCS facilities. We are grateful to Dr Akihiko Nakano and Dr Ken Sato (RIKEN, Japan) for kindly providing the yeast strain SNH23-D7.
References
- van Beilen JB, Poirier Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol. 2007;25:522–529. doi: 10.1016/j.tibtech.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Ray DT. In: New Crops. Janick J, Simon JE, editor. Wiley New York, New York; 1993. Guayule: A source of natural rubber; pp. 338–343. [Google Scholar]
- Yagami A, Suzuki K, Saito H, Matsunaga K. Hev b 6.02 is the most important allergen in health care workers sensitized occupationally by natural rubber latex gloves. Allergol Int. 2009;58:347–355. doi: 10.2332/allergolint.08-OA-0046. [DOI] [PubMed] [Google Scholar]
- Gomez JB, Hamzah S. Particle size distribution in Hevea latex - some observations on the electron microscopic method. J Nat Rubber Res. 1989;4:204–211. [Google Scholar]
- Yeang HY, Yip E, Hamzah S. Characterization of zone 1 and zone 2 rubber particles in Hevea brasiliensis latex. J Nat Rubber Res. 1995;10:108–123. [Google Scholar]
- Wood DF, Cornish K. Microstructure of purified rubber particles. Int J Plant Sci. 2000;161:435–445. doi: 10.1086/314269. [DOI] [PubMed] [Google Scholar]
- Hasma H, Subramaniam A. Composition of lipids in latex of Hevea brasiliensis clone RRIM 501. J Nat Rubber Res. 1986;1:30–40. [Google Scholar]
- Hasma H. Lipids associated with rubber particles and their possible role in mechanical stability of latex concentrates. J Nat Rubber Res. 1991;6:105–114. [Google Scholar]
- Siler DJ, Goodrich-Tanrikulu M, Cornish K, Stafford AE, McKeon TA. Composition of rubber particles of Hevea brasiliensis, Parthenium argentatum, Ficus elastica, and Euphorbia lactiflua indicates unconventional surface structure. Plant Physiol Biochem. 1997;35:881–889. [Google Scholar]
- Cornish K, Wood DF, Windle JJ. Rubber particles from four different species, examined by transmission electron microscopy and electron-paramagnetic- resonance spin labeling, are found to consist of a homogeneous rubber core enclosed by a contiguous, monolayer biomembrane. Planta. 1999;210:85–96. doi: 10.1007/s004250050657. [DOI] [PubMed] [Google Scholar]
- Cornish K, Siler DJ, Grosjean OK, Godman N. Fundamental similarities in rubber particle architecture and function in three evolutionarily divergent plant species. J Nat Rubber Res. 1993;8:275–285. [Google Scholar]
- Poulter CD, Rilling HC. Prenyltransferase - mechanism of reaction. Biochemistry. 1976;15:1079–1083. doi: 10.1021/bi00650a019. [DOI] [PubMed] [Google Scholar]
- Poulter CD, Rilling HC. The prenyl transfer-reaction. Enzymatic and mechanistic studies of 1'-4 coupling reaction in the terpene biosynthetic-pathway. Acc Chem Res. 1978;11:307–313. doi: 10.1021/ar50128a004. [DOI] [Google Scholar]
- Kharel Y, Koyama T. Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep. 2003;20:111–118. doi: 10.1039/b108934j. [DOI] [PubMed] [Google Scholar]
- Kharel Y, Zhang Y, Fujihashi M, Miki K, Koyama T. Identification of significant residues for homoallylic substrate binding of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase. J Biol Chem. 2001;276:28459–28464. doi: 10.1074/jbc.M102057200. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K, Nishikawa S, Hirata A, Kato J, Nakano A. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol Cell Biol. 1999;19:471–483. doi: 10.1128/mcb.19.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Viswanathan K, Krag SS, Betenbaugh MJ. Polyprenyl lipid synthesis in mammalian cells expressing human cis-prenyltransferase. Biochem Biophys Res Commun. 2005;331:379–383. doi: 10.1016/j.bbrc.2005.03.181. [DOI] [PubMed] [Google Scholar]
- Light DR, Dennis MS. Purification of a prenyltransferase that elongates cis-polyisoprene rubber from the latex of Hevea brasiliensis. J Biol Chem. 1989;264:18589–18597. [PubMed] [Google Scholar]
- Siler DJ, Cornish K. A protein from Ficus elastica rubber particles is related to proteins from Hevea brasiliensis and Parthenium argentatum. Phytochemistry. 1993;32:1097–1102. doi: 10.1016/S0031-9422(00)95072-6. [DOI] [Google Scholar]
- Cornish K, Siler DJ, Grosjean O. Immunoinhibition of rubber particle-bound cis-prenyltransferases in Ficus elastica and Parthenium argentatum. Phytochemistry. 1994;35:1425. doi: 10.1016/S0031-9422(00)86868-5. [DOI] [Google Scholar]
- Cornish K, Siler DJ. Characterization of cis-prenyltransferase activity localized in a buoyant fraction of rubber particles from Ficus elastica latex. Plant Physiol Biochem. 1996;34:334–377. [Google Scholar]
- Kang H, Soo Kim Y, Chung GC. Characterization of natural rubber biosynthesis in Ficus benghalensis. Plant Physiol Biochem. 2000;38:979–987. doi: 10.1016/S0981-9428(00)01204-3. [DOI] [Google Scholar]
- Sato M, Fujisaki S, Sato K, Nishimura Y, Nakano A. Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations. Implication for their distinct physiological roles in dolichol synthesis. Genes Cells. 2001;6:495–506. doi: 10.1046/j.1365-2443.2001.00438.x. [DOI] [PubMed] [Google Scholar]
- Oh SK, Han K, Ryu SB, Kang H. Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana. J Biol Chem. 2000;275:18482–18488. doi: 10.1074/jbc.M002000200. [DOI] [PubMed] [Google Scholar]
- Asawatreratanakul K, Zhang Y, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Rattanapittayaporn A, Koyama T. Molecular cloning, expression and characterization of cD3A encoding cis-prenyltransferases from Hevea brasiliensis . Eur J Biochem. 2003;270:4671–4680. doi: 10.1046/j.1432-1033.2003.03863.x. [DOI] [PubMed] [Google Scholar]
- Berndt J. The biosynthesis of rubber. US Government Res Rep AD-601. 1963. p. 729.
- Cornish K, Backhaus RA. Rubber transferase activity in rubber particles of guayule. Phytochemistry. 1990;29:3809–3813. doi: 10.1016/0031-9422(90)85337-F. [DOI] [Google Scholar]
- Cornish K, Siler DJ. Effect of different allylic diphosphates on the initiation of new rubber molecules and on cis-1,4-polyisoprene biosynthesis in guayule (Parthenium argentatum Gray) J Plant Physiol. 1995;147:301–305. [Google Scholar]
- Singh AP, Wi SG, Chung GC, Kim YS, Kang H. The micromorphology and protein characterization of rubber particles in Ficus carica, Ficus benghalensis and Hevea brasiliensis. J Exp Bot. 2003;54:985–992. doi: 10.1093/jxb/erg107. [DOI] [PubMed] [Google Scholar]
- Matzelle T, Reichelt R. Review: Hydro, micro- and nanogels studied by complementary measurements based on SEM and SFM. Acta Microscopica. 2008;17:45–61. [Google Scholar]
- Suomela H. On the possibilities of growing Taraxacum kok-saghyz in Finland. National Agricultural Experimental Publications (valtion maatalous koetoiminnan julkaisuja), Helsinki; 1950. [Google Scholar]
- Duch MW, Grant DM. Carbon-13 chemical shift studies of the 1,4-polybutadienes and the 1,4-polyisoprenes. Macromolecules. 1970;3:165–174. doi: 10.1021/ma60014a010. [DOI] [Google Scholar]
- Kang H, Kang MY, Han K. Identification of natural rubber and characterization of rubber biosynthetic activity in fig tree. Plant Physiol. 2000;123:1133–1142. doi: 10.1104/pp.123.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish K. Similarities and differences in rubber biochemistry among plant species. Phytochemistry. 2001;57:1123–1134. doi: 10.1016/S0031-9422(01)00097-8. [DOI] [PubMed] [Google Scholar]
- da Costa BMT. Regulation of rubber biosynthetic rate and molecular weight in Hevea brasiliensis by metal cofactor. Biomacromolecules. 2005;6:279. doi: 10.1021/bm049606w. [DOI] [PubMed] [Google Scholar]
- Scott DJ, da Costa BMT, Espy SC, Keasling JD, Cornish K. Activation and inhibition of rubber transferases by metal cofactors and pyrophosphate substrates. Phytochemistry. 2003;64:123–134. doi: 10.1016/S0031-9422(03)00266-8. [DOI] [PubMed] [Google Scholar]
- Tangpakdee J, Tanaka Y, Ogura K, Koyama T, Wititsuwannakul R, Wititsuwannakul D. Rubber formation by fresh bottom fraction of Hevea latex. Phytochemistry. 1997;45:269–274. doi: 10.1016/S0031-9422(96)00838-2. [DOI] [Google Scholar]
- Schmidt T Hillebrand A Wurbs D Wahler D Lenders M Schulze Gronover C Prüfer D Molecular cloning and characterization of rubber biosynthetic genes from Taraxacum koksaghyz Plant Mol Biol Rep doi.org/10.1007/s11105-009-0145-911542597
- Archer BL, Audley BG, Cockbain EG, McSweeny GP. The biosynthesis of rubber - incorporation of mevalonate and isopentenyl pyrophosphate into rubber by Hevea brasiliensis -latex fractions. Biochem J. 1963;89:565–574. doi: 10.1042/bj0890565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen AI, McSweeny GP. The biosynthesis of rubber: Incorporation of isopentenyl pyrophosphate into purified rubber particles by a soluble latex serum enzyme. Biochem J. 1966;101:42. doi: 10.1042/bj1010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynen F. Biosynthetic pathways from acetate to natural products. Pure Appl Chem. 1967;14:137–167. doi: 10.1351/pac196714010137. [DOI] [PubMed] [Google Scholar]
- Lepetit M, Ehling M, Gigot C, Hahne G. An internal standard improves the reliability of transient expression studies in plant protoplasts. Plant Cell Rep. 1991;10:401–405. doi: 10.1007/BF00232611. [DOI] [PubMed] [Google Scholar]
- Wititsuwannakul D, Rattanapittayaporn A, Koyama T, Wititsuwannakul R. Involvement of Hevea latex organelle membrane proteins in the rubber biosynthesis activity and regulatory function. Macromol Biosci. 2004;4:314–323. doi: 10.1002/mabi.200300080. [DOI] [PubMed] [Google Scholar]
- Fujii H, Koyama T, Ogura K. Efficient enzymatic hydrolysis of polyprenyl pyrophosphates. Biochim Biophys Acta - Lipids and Lipid Metabolism. 1982;712:716–718. doi: 10.1016/0005-2760(82)90304-6. [DOI] [PubMed] [Google Scholar]
- Wahler D, Schulze Gronover C, Richter C, Foucu F, Twyman RM, Moerschbacher BM, Fischer R, Muth J, Prüfer D. Polyphenoloxidase silencing affects latex coagulation in Taraxacum spp. Plant Physiol. 2009;151:334–346. doi: 10.1104/pp.109.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss H. A set of plant expression vectors for transcriptional and translational fusions. Nucl Acids Res. 1987;15:5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucl Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJS, Faber KN, Kram AM, Kiel JAKW, Klei IJ van der, Veenhuis M. A stretch of positively charged amino acids at the 3 terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem. 2000;275:9986–9995. doi: 10.1074/jbc.275.14.9986. [DOI] [PubMed] [Google Scholar]
- Negrutiu I, Shillito R, Potrykus I, Biasini G, Sala F. Hybrid genes in the analysis of transformation conditions. Plant Mol Biol. 1987;8:363–373. doi: 10.1007/BF00015814. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions - beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]