Abstract
Drug repositioning refers to alternative drug use discoveries which differ from the original intent of the drug. One challenge in these efforts lies in choosing which indication to prospectively test a drug of interest. We systematically evaluated a drug treatment-based view of diseases in order to address this challenge. Suggested novel drug uses were generated using a guilt-by-association approach. Compared with control drug uses, the suggested novel drug uses were significantly enriched in clinical trials.
Introduction
The discovery of a new use of a drug for a new condition can be a haphazard process, as illustrated by many past examples: the use of zinc acetate for the treatment of Wilson’s disease[1]; arsenic for acute promyelocytic leukemia[2]; amphotericin B for leishmaniasis[3], and thalidomide for multiple myeloma[4]. These “alternative” drug use discoveries, which differ from the original intent of the drug development process, are referred to as drug repurposing or repositioning[5]. Drug repositioning efforts have many advantages. Since the drug pharmacokinetics and pharmacodynamics are known, drug repositioning “discoveries” are less costly and quicker than traditional discovery efforts[5], which typically takes 10–15 years[6] and upwards of $1 billion[7]. While physicians and pharmaceutical/biotechnology companies have manual methods and prior knowledge that enable repositioning clinical trials, these occurrences are often serendipitous and rare. One challenge in drug repositioning, therefore, lies in choosing which (therapeutic) indication to prospectively test a drug of interest. To address this challenge and to enable more connections between diseases and drugs, we systematically explored a treatment-based view that includes off-label (not approved by the Food and Drug Administration [FDA]) drug uses from a network perspective in order to suggest novel drug uses.
RESULTS
Novel drug use suggestions via guilt-by-association
We systematically captured the FDA-approved (FDA approved drug and indications) and practiced (FDA-approved drugs, but off-label drug uses) views from the DRUGDEX system[8] into the Drug-Disease Knowledge Base (DrDKB) consisting of 726 diseases and 2,022 drugs. Given that the treatment profiles between a small subset of disease pairs can be quite different between the FDA-approved and practiced views (Figure 1), we reasoned that we can use these discrepancies to suggest novel drug uses. This is based on the idea that if two diseases share some similar therapies, then other drugs used for only one of the two may also be therapeutic for the other. While this is an over-simplification of drug-based therapies of diseases, it is informed from many past examples such as rivastigmine for the treatment of Alzheimer’s disease and Parkinson’s disease dementia and bevacizumab for the treatment of colorectal cancer and non-squamous non-small-cell lung cancer. Furthermore, as the full mechanism of action of many drugs is not always known and as application of drugs across therapeutic classes is likely under-explored, a systemic evaluation of all pairwise comparisons may yield interesting drug use suggestions.
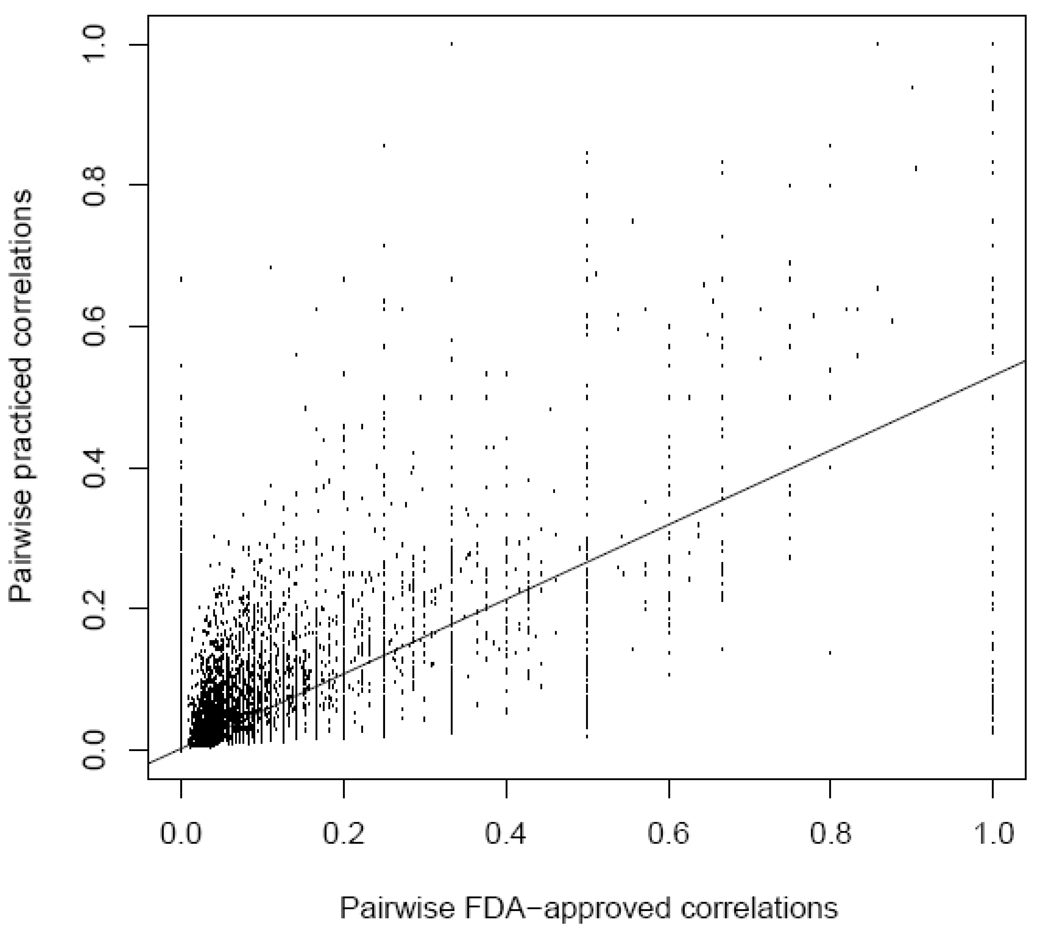
Figure 1.
Pairwise disease correlations under the FDA-approved and practiced views. Each point represents a pairing of two of the 726 diseases in DrDKB that share at least one FDA-approved drug. Each pair of diseases was scored for similarity based on binary correlation calculated using shared FDA approved drug indications (x-axis) and combined FDA- and non-FDA drug uses (y-axis). Zero on either axis indicates no drugs in common between the two diseases, while one indicates an exactly matching set in the pair. The regression line indicates an overall similarity in FDA and combined indication data (r = 0.68, p = 2.2×10−16). However, many pairs of diseases share many FDA approved drugs, and few non-FDA drugs; we exploit these to suggest novel drug uses across the pair of diseases.
To do this, we implemented a network-based, guilt-by-association method for those 5,549 disease pairs that shared at least one FDA approved drug in common under the FDA-approved view. Novel drug use suggestions were made based on shared treatment profile from any disease pairs. Using this methodology, we made 156,279 unique drug use suggestions. We also filtered out weakly suggested drug uses (those with only one suggestion), yielding a final total of 57,542 unique novel drug use suggestions. In order to assess the robustness of the 57,452 suggested drug uses, we randomly removed 10% of the newly suggested drug uses separately 10,000 times and determined what percentage of the removed uses were still suggested by other diseases. On average, 98.3% of the removed drug uses were still suggested. Even after 10,000 random simulations with removal of 20% of the suggested drug uses, 85.82% of the suggested drug uses were recovered, indicating robustness of our method.
Suggested novel drug uses are highly enriched in clinical trials
To externally validate our suggestions, we tested whether a subset of our suggested novel drug uses is already under evaluation in clinical trials, as clinical trials reflect an externally-determined treatment predication between the drug and disease. Importantly, we found that our drug use suggestions are 12 times more likely to be in a clinical trial than those drug uses not suggested by our guilt-by-association approach (Chi-square p-value < 2.2×10−16).
We further investigated novel drug uses for two drugs: rituximab and atorvastatin. Rituximab is approved to treat non-Hodgkin’s lymphoma and rheumatoid arthritis. We suggest 107 novel diseases that could be treated with rituximab, 96 of which map to diseases in clinical trials. Of the 59 diseases undergoing clinical trials with rituximab, 6 of them are already known and in practice (found in DrDKB). Of the remaining 53 diseases, clinical trials for 13 were among our 96 suggestions, resulting in a statistically significant over-enrichment (hypergeometric p = 0.0346). As expected, nearly a third of the clinical trials on rituximab involve autoimmune disorders such as multiple sclerosis and lupus erythematosus, which corresponds very well with our suggested drug uses. However, we found no ongoing clinical trials for cataracts, gastric ulcers, and stomach cancers, which we suggest as novel drug uses (Table 1).
TABLE 1.
Sample drug use suggestions for five drugs, some of which already have past/ongoing clinical trials.
| Suggested novel drug uses | ||
|---|---|---|
| Drug | In clinical trials.gov | Not in clinicaltrials.gov |
| Atorvastatin | Asthma, Crohn’s disease, myocardial infarction | Breast cancer, Hodgkin’s lymphoma, osteosarcoma |
| Bevacizumab | Mantle cell lymphoma, multiple myeloma, stomach cancer | AIDS, mycosis, nephroblastoma |
| Doxycycline | Chronic obstructive lung disease, endometritis, multiple sclerosis | Asthma, lupus erythematosus, osteosarcoma |
| Magnesium | Alcohol withdrawal syndrome, anemia, chronic obstructive lung disease | Crohn’s disease, glaucoma, rheumatoid arthritis |
| Rituximab | Mantle cell lymphoma, multiple sclerosis, sarcoidosis | Cataract, gastric ulcer, stomach cancer |
Seventy-six novel drug uses were suggested for atorvastatin which 65 mapped to diseases in clinical trials. Of the 37 diseases found in atorvastatin clinical trials, 6 are already known and in practice (found in DrDKB). Among the remaining 31 drug uses, 8 matched our suggestions, yielding a significant hypergeometric p-value of 0.022. As expected, myocardial infarction and acute coronary syndrome were the top two indications for novel use of atorvastatin. We also found a similar trend to be true of other statins in the mapped clinical trials set. Interestingly, we suggest atorvastatin to be therapeutic in breast cancer, osteosarcoma and Hodgkin’s lymphoma, of which there are currently no ongoing clinical trials (Table 1). Table 1 also lists suggested novel uses for three additional drugs: bevacizumab, doxycycline, and magnesium.
DISCUSSION
The pharmaceutical industry is under pressure to develop blockbuster drugs at a time when the cost of validating indications is skyrocketing. Together, combined with the soaring costs due to the high failure rate of de novo drug discovery, a renewed interest in drug repositioning is gathering momentum. One of the first challenges of repositioning efforts lies in choosing the right therapeutic areas to prospectively test drugs of interest. We specifically address this challenge by first systematically compiling a drug-disease knowledge base (DrDKB) to capture the 3,517 FDA approved drug indications and 8,130 off-label uses of 2,022 distinct drugs used to treat each of 726 diseases.
The disparity in prescription patterns between what is approved by the FDA and how physicians actually practice is well-known, and what we exploit to find our novel suggested drug uses. Our guilt-by-association approach generated approximately 57,000 robust novel drug uses. A considerable fraction was significantly enriched in clinical trials, indicating external validation of our suggestions. We examined suggested novel drug uses for two drugs: rituximab and atorvastatin. We suggest the anticancer rituximab to treat cataract, gastric ulcer, and stomach cancer, of which there are no ongoing clinical trials. While no published work shows a relationship between rituximab and these diseases, it is known that prolonged steroid therapy can result in side effects such as cataract and gastric ulcer, and, rituximab has been shown to alleviate these side effects[9]. We suggest that atorvastatin may also be therapeutic in treating some cancers, such as Hodgkin’s lymphoma, breast cancer, and osteosarcoma. Currently, there are no ongoing clinical trials studying the role of statins in these cancers, however, there are some promising preclinical studies. Statins have been used to inhibit Myc-induced lymphomagenesis[10]. In addition, atorvastatin, together with other anticancer drugs, can enhance inhibitory effects of anticancer drugs[11].
Though there have been other networks of drugs created [12, 13], none have been used for suggesting novel drug uses. Our network-based, guilt-by-association method demonstrates that even a high-level systematic analysis based on drug-diseases relations, and not structural, clinical or other pharmacological data, can yield many reasonble suggestions.
We acknowledge that some of our suggested novel drug uses will always seem obvious, particularly to experts; however, they are not so obvious as to be currently used in clinical practice, as defined by DRUGDEX. For example, the use of infliximab for actinic keratosis or eczema might seem like an obvious extension of its use in psoriasis. However, we cannot find evidence that clinicians have made this connection. Furthermore, our method systematically suggests uses across the pharmacopeia, not just for a select few drugs.
Given the design of our proof-of-principle study, we were unable to evaluate those diseases that did not have even a single FDA-approved drug. Moreover, we acknowledge that the method is biased toward older drugs that have diverse drug uses and diseases with diverse treatments. Also, we were unable to examine potential confounding factors, such as drug-drug interactions. Nonetheless, based on our success in finding enrichment of these suggested drug uses within clinical trials, we believe that our automated method for suggesting novel drug uses, in conjunction with existing clinical discovery approaches, such as in vitro and in vivo models of diseases and clinical data, can potentially enable pharmaceutical companies and medical associations to drive more testing of existing drugs and help expedite future drug repositioning efforts.
METHODS
Compilation of Drug-Disease Knowledge Base (DrDKB)
In order to capture both the FDA approved and off-label uses, we chose the DRUGDEX System[8], as our comprehensive pharmacopeia resource. We extracted 83,338 SNOMED-CT concept terms from Unified Medical Language System[14] (UMLS) whose semantic type fall under the UMLS semantic subtree of Pathological Functions, Injury/Poisoning, or Abnormalities. 4,068 concept terms had associated drug information when queried against the DRUGDEX System (Feb. 2007). We then performed a series of filtering steps (see supplementary methods) to generate a final set of 726 diseases and 2,022 drugs in the Drug-Disease Knowledge Base (DrDKB). We represented the DrDKB in two binary, two-dimensional matrices: FDA-approved Matrix and Practiced Matrix (see supplementary methods).
Network-based, guilt-by-association method
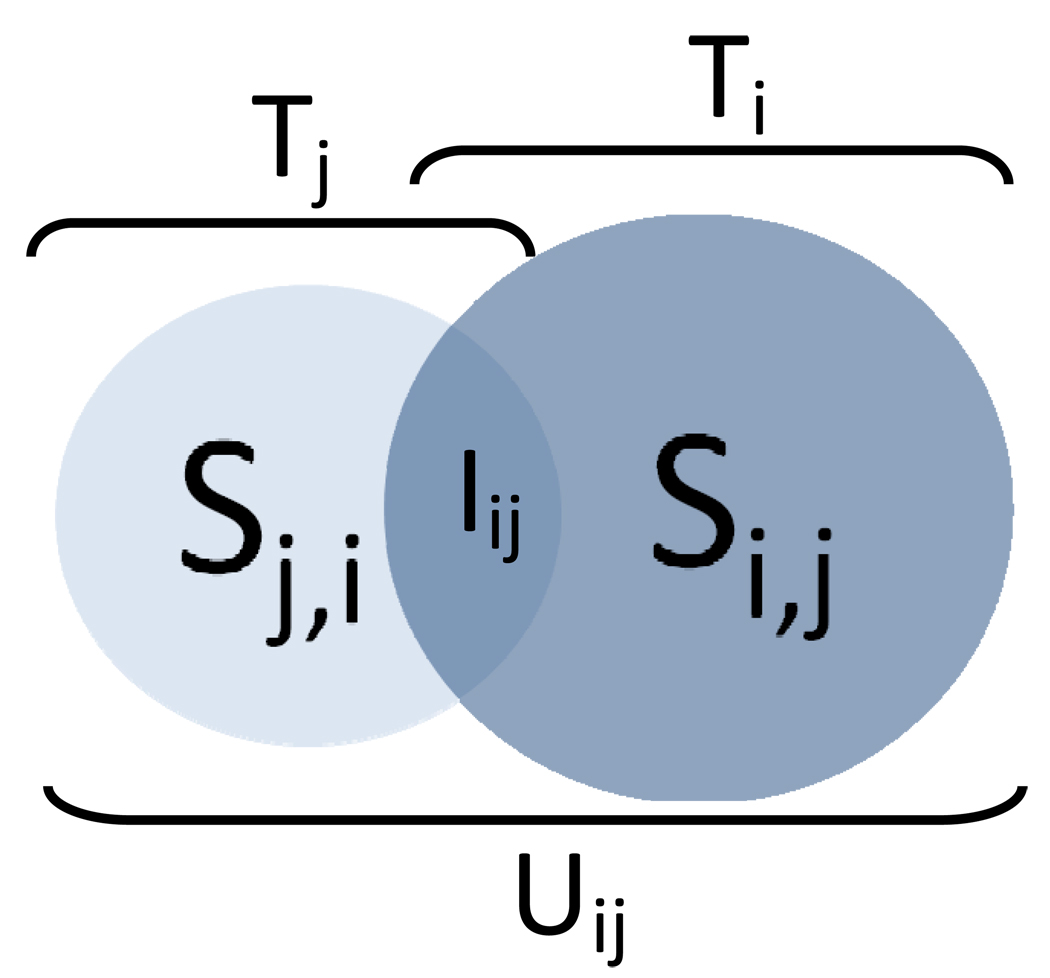
The network-based, guilt-by-association method is based on the idea that if two diseases share some similar therapies, then other drugs used for only one of the two may also be therapeutic for the other. We represent any two diseases in DrDKB as i, j and their treatment profiles as Ti and Tj, respectively (Figure 2). The set of drugs common between disease i and j is expressed by Iij where Iij = Ti ∩ Tj while Uji (Uij= Ti ∪ Tj) contains the union set of drugs used to treat i, j or both. First, we restricted to those 5,549 disease pairs that shared at least one FDA approved drug in common under the FDA-approved view. Then, using the practiced view containing both the FDA-approved and off-label drugs, we generate two sets of suggested novel drug use sets Si,j and Sj,i. The suggested drug use set Si,j contains the set of drugs suggested to be used for disease i by disease j and is generated from subtracting Ti from Uij (Si,j = Uij-Ti). Conversely, the drug use set Sj,i contains the set of drugs suggested to be used for disease j by disease i and is generated from subtracting Tj from Uij (Sj,i=Uij-Tj). See supplementary methods for an example of this approach.
Figure 2.
Schematic of the guilt-by-association method of suggesting leads for novel drug uses. Given two diseases i, j and their corresponding treatment profiles Ti and Tj, respectively, the suggested novel drug uses for i based on shared treatment profile with j is Si,j or the difference of the union treatment profile (Uij) and Ti. Conversely, suggested novel drug uses for j based on i (Sj,i) are obtained from subtracting Tj from Uij.
Clinical trials
We downloaded the complete list (~48,000 records) of clinical trials from http://clinicaltrials.gov in Dec. 2007. We excluded those trials that involved behavioral, biological, medical device, or procedural interventions. Roughly 30,000 records remained. We then looked for exact term matching between both the disease and drug names. Those with exact disease and drug term matches comprise our mapped clinical trial set.
Statistical analyses
Correlation values between disease pairs in DrDKB were calculated (separately) from binary distances of the two matrices: FDA-approved Matrix and Practiced Matrix. Further, for each disease pair, correlations between the FDA-approved and Practiced matrices were calculated. Overall Pearson’s correlations for the 5,549 disease pairs that shared at least one FDA drug was 0.68 (p-value < 2.2×10−16). Hypergeometric distributions were calculated using the HYPGEODIST function in Microsoft Excel. Remaining calculations were performed using R[15].
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the Lucile Packard Foundation for Children’s Health, National Library of Medicine (K22 LM008261 and T15 LM007033), National Institute of General Medical Sciences (R01 GM079719), Howard Hughes Medical Institute, The Hewlett-Packard Company Foundation, and the Pharmaceutical Research and Manufacturers of America Foundation. The authors would like to thank K. Sachs and Y. Liu for useful discussions. We thank A. Skrenchuk and B. Oskotsky from Stanford University for computer support
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Scheindlin S. Rare diseases, orphan drugs, and orphaned patients. Molecular interventions. 2006 Aug;6(4):186–191. doi: 10.1124/mi.6.4.2. [DOI] [PubMed] [Google Scholar]
- 2.Soignet SL, Maslak P, Wang ZG, Jhanwar S, Calleja E, Dardashti LJ, et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. The New England journal of medicine. 1998 Nov 5;339(19):1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 3.Yardley V, Croft SL. Activity of liposomal amphotericin B against experimental cutaneous leishmaniasis. Antimicrobial agents and chemotherapy. 1997 Apr;41(4):752–756. doi: 10.1128/aac.41.4.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singhal S, Mehta J, Desikan R, Ayers D, Roberson P, Eddlemon P, et al. Antitumor activity of thalidomide in refractory multiple myeloma. The New England journal of medicine. 1999 Nov 18;341(21):1565–1571. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 5.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews. 2004 Aug;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 6.Dimasi JA. New drug development in the United States from 1963 to 1999. Clinical pharmacology and therapeutics. 2001 May;69(5):286–296. doi: 10.1067/mcp.2001.115132. [DOI] [PubMed] [Google Scholar]
- 7.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. Journal of health economics. 2003 Mar;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 8.DRUGDEX System. Greenwood Village, Colo: Thomson Healthcare; 2007. [Google Scholar]
- 9.Saito K, Nawata M, Nakayamada S, Tokunaga M, Tsukada J, Tanaka Y. Successful treatment with anti-CD20 monoclonal antibody (rituximab) of life-threatening refractory systemic lupus erythematosus with renal and central nervous system involvement. Lupus. 2003;12(10):798–800. doi: 10.1191/0961203303lu450xx. [DOI] [PubMed] [Google Scholar]
- 10.Shachaf CM, Perez OD, Youssef S, Fan AC, Elchuri S, Goldstein MJ, et al. Inhibition of HMGcoA reductase by atorvastatin prevents and reverses MYC-induced lymphomagenesis. Blood. 2007 Oct 1;110(7):2674–2684. doi: 10.1182/blood-2006-09-048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromigue O, Hamidouche Z, Marie PJ. Statin-induced inhibition of HMG-CoA reductase sensitizes human osteosarcoma cells to anticancer drugs. The Journal of pharmacology and experimental therapeutics. 2008 Feb 5; doi: 10.1124/jpet.108.136127. [DOI] [PubMed] [Google Scholar]
- 12.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature biotechnology. 2007 Oct;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 13.Salanti G, Kavvoura FK, Ioannidis JP. Exploring the geometry of treatment networks. Annals of internal medicine. 2008 Apr 1;148(7):544–553. doi: 10.7326/0003-4819-148-7-200804010-00011. [DOI] [PubMed] [Google Scholar]
- 14.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research. 2004 Jan 1;32(Database issue):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Development Core Team R. Vienna, Austria: 2008. A Language and Environment for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.