Abstract
The detection of genetically modified (GM) materials in food and feed products is a complex multi-step analytical process invoking screening, identification, and often quantification of the genetically modified organisms (GMO) present in a sample. “Combinatory qPCR SYBR®Green screening” (CoSYPS) is a matrix-based approach for determining the presence of GM plant materials in products. The CoSYPS decision-support system (DSS) interprets the analytical results of SYBR®GREEN qPCR analysis based on four values: the C t- and T m values and the LOD and LOQ for each method. A theoretical explanation of the different concepts applied in CoSYPS analysis is given (GMO Universe, “Prime number tracing”, matrix/combinatory approach) and documented using the RoundUp Ready soy GTS40-3-2 as an example. By applying a limited set of SYBR®GREEN qPCR methods and through application of a newly developed “prime number”-based algorithm, the nature of subsets of corresponding GMO in a sample can be determined. Together, these analyses provide guidance for semi-quantitative estimation of GMO presence in a food and feed product.
Keywords: GMO, Screening, Prime number, qPCR, SYBRGreen
Introduction
At a global level, more than 100 genetically modified organisms (GMO) have received an authorization for commercial usage as food or feed (http://www.gmo-compass.org, http://www.agbios.com/dbase.php). As such, GMO represent an important component of the global food and feed market, and enforcement legislation has been put into place to verify compliance with the local regulations. As an example, testing of materials derived from food and feed materials is regulated in Europe through the EC Regulations EC/2003/1829 and 1830 [1, 2]. Due to existence of differences in local GMO legislations and the global nature of the food and feed market, asynchronously authorized GMO are more and more appearing on the market, invoking specific decisions at the local level in case of infringement (e.g., LL601 and Bt63 rice in the EC) [3, 4]. With respect to the market introduction of GMO, many countries, including the EC, have installed a government policy supporting a strong commitment to consumer protection and freedom of choice [1, 2]. For this, the traceability and/or labeling of GM products along the food chain are critical. At the EC level, two particular concepts have been elaborated to support such policy: (1) the amount of GMO is to be calculated per ingredient (defined as a “taxon”, e.g. soy, maize, etc.), and (2) product labeling is mandatory when exceeding a threshold level of GMO (above 0.9% GMO per ingredient) [2]. In view of the diversity of the GM-market (from grain to pizzas), the EC recommended to apply the “Haploid Genome Equivalent” (HGE) as the standard DNA detection unit for all GM food/feed products [5].
Due to the broad range of authorized GMO on the market, the first step in GMO analysis consists in general of screening for presence of GMO. The establishment of a GMO screening approach requires the availability of (1) appropriate detection methods (with adequate performance, scope, and legal basis), (2) suitable reference materials, and (3) a DSS. Moreover, such an approach should preferentially apply a familiar technology in GMO analysis (such as “Polymerase Chain reaction” (PCR) [6]) and be suited for flexible adaptation to the market reality and the client needs in a cost-efficient way. The enforcement activities are, to date, primarily focusing on DNA-based approaches of which the vast majority applies PCR. Several GMO screening approaches, mostly based on real-time PCR (qPCR) or micro-arrays, have already been developed aiming at covering such broad range of GMO with a minimal number of analyses. For a recent critical review of those technologies, we refer to the article of Querci et al. [7]. To date, only the so-called Dualchip array is readily commercially available [8–10]. Most of the enforcement laboratories have today already a (costly) qPCR platform in place, but many of these novel technologies may require additional investments in new equipment, in technology validation, and in logistic support. It is thus considered the most appropriate to exploit further qPCR-based screening approaches (see also Querci et al. [7] for a more elaborate discussion).
In setting up a GMO screening that meets the above criteria, it was considered that SYBR®Green qPCR could offer a good alternative with a number of advantages over the other PCR-based approaches: (1) SYBR®Green qPCR monitors the increase in total fluorescence throughout the amplification, allowing to estimate the presence of non-specific amplification, (2) the melting temperature analysis allows post-PCR verification of the amplification of the expected target and any closely related target(s), (3) the SYBR®Green technology is more cost-effective as no dye-labeled oligonucleotide probes are required [11].
For this, the “Combinatory SYBR®GREEN qPCR Screening” (abbreviated as “CoSYPS”) was developed for the commonly applied 96-well plate qPCR format, supported by a matrix-based interpretation of the analytical results. Here, we present the theoretical background to the key concepts as applied in CoSYPS: the “GMO Universe”, the matrix-based screening, and the use of “prime-number GMO tagging” in facilitating selection of the possible GMO present in the sample. Next, the combinatory interpretation of the analytical results based on four analytical values (the C t, the T m, the “LOD6” and the “LOQ6”) is explained. Each of these concepts is documented by using the RoundUp Ready® soy GTS40-3-2 as a model. Finally, future perspectives on the use, the development, and the integration of CoSYPS into a complete GMO detection platform are discussed.
General description of the field of application
GMO analysis for enforcement is a complex process addressing the compliance of food and feed products with the legal requirements for GMO use. To formally describe the CoSYPS approach, the following elements needed either to be newly developed, either to be described within the context of the CoSYPS approach:
The formal description of a GMO in terms of their detection targets,
The concept of a “Universe” for GMO,
An illustration of the matrix-based screening approach,
A mathematical GMO tagging, based on prime numbers,
The decision criteria applied in CoSYPS (C t, T m, LOD, and LOQ)
The principles in combinatory decision taking
The first three items are considered to represent key concepts that are driving the general understanding of the principles in a matrix-based screening approach. These concepts are presented in the glossary part below. The fourth topic represents a novel developed mathematical way of identifying the possible GMO present in a sample based on the outcome of the screening results. In the last two points, the experimental parameters used specifically as decision criteria in CoSYPS are outlined and the principles in the combinatory interpretation within CoSYPS are explained.
Glossary to general concepts in GM plants (GMP) screening analysis
Description of a GMP in function of the constituents of the genetic modification
All EU-authorized GMO for food and feed use, represent plant or microbial transformation events that resulted from the introduction of a specific DNA sequence into the genome of a host species (1, 2, and 3). Here, we will only discuss the genetically modified plants (GMP), but a similar reasoning can hold for any other set of GMO (such as bacteria or viruses). In the GMP engineering process, a certain transformation vector with known molecular DNA structure is being constructed and used for the introduction of new traits in a host plant. The precise organization and content of the newly introduced DNA sequences in the GMP are determined through detailed molecular analysis of each GMP (e.g., by Southern blot analysis, PCR analysis, and ultimate DNA sequence analysis). All this information is comprehensively documented in the official registration files required to obtain an authorization of use of the GMP in the EC (1, 2, and 3).
The DNA introduced after transformation (designated as “TransDNA”) can be described in a “1 × Zi” linear matrix format in which Zi represents the respective (relevant) DNA elements of the transformation vector and a
i the number of copies of that element (see left panel):

As an example, the TransDNA for the RoundUp Ready® soy GTS40-3-2 is described as a function of three vector elements (here the p35S, tNOS, and CP4-EPSPS; see right panel). Typical relevant TransDNA elements can thus in general be transcription regulatory sequences (e.g. the CaMV 35S and Agrobacterium NOS promoter and terminator …), “Open Reading Frames” (ORF) (e.g. CryIAB, PAT/bar…) or so-called vector-specific elements comprising sequences from two different kinds of elements (such as the 35S-bar [3] or CryIAb-Nos [4]).
When combined with host plant taxon-specific elements (e.g. lectin (soy), alcohol dehydrogenase (maize),…) and so-called event-specific elements (e.g. junction fragments between transformation vector DNA and ‘flanking’ plant genome DNA (http://gmo-crl.jrc.ec.europa.eu; CRL-GMFF)), a unique linear matrix can be developed for each GMP.

-
Concept 2:
Definition of the “EU-GMP Universe” in time and space
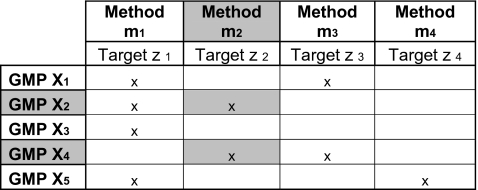
Next, the set of GMP to be analyzed can be defined by the mathematical concept “Universe”, being “a class that contains (as elements) all the entities under consideration in a given situation”. In Table 1, a general representation in a table format is shown. The different GMP are represented by the rows, the constituents are listed in the columns. In this example, the GMP Universe comprises five GMP(X1–5), while the targets (z 1–4) represent DNA sequences within the corresponding constituents Z 1–4 detected by the methods m1–4.
Table 1.
GMP analysis interpretation according to a matrix-based approach
The formal logical decision path when screening a sample potentially containing GMP X1–5 with "Method m2 is indicated in gray (“x”: target is present in GMP Xi; “ ”: target is not present in GMP Xi)
With respect to the EU enforcement world, the most relevant “Universe” would be the authorized GM plants to be placed on the market (EC directive EC/2001/18) [12], and authorized as GM food/feed (EC regulation EC/2003/1829) [1]. This set of GMP will be further denoted as the “EU-GMP Universe” (EU-GMP). An example of a representation of this Universe is shown in Table 2. Herein, the following constituents are listed: the species/taxon in column A, the event-specific element in column B and generic recombinant elements (such as transcription elements, vector-specific elements…) and GM-trait DNA elements in columns C to S. Each GMP is then represented as a single row in the table by a linear ‘1 × 19’ matrix and the EU-authorized GM plant Universe-Anno 2009 (including the legal decisions on some unauthorized GMP) comprising 29 GMP (omitting any of the stacked events), can be described by a “29 × 19” matrix.
-
Concept 3:
“Matrix-based” GMP screening approach by establishing a formal relation between analytical results and GMP presence through validated detection methods
Table 2.
The EU-GMP Universe of authorized and “legally regulated” GMP in the EU, anno 2009 (except stacked events)
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | GMP | p35S | tNOS | pFGW | t35S | pNOS | rice actin | tOCS | nptII | CP4 EPSPS | mEPSPS | PAT/pat | PAT/bar | barnase | Cry1Ab | CryIAc | Cry1F | Cry3Bb1 |
| Soybean | GTS 40/3/2 | X | X | X | ||||||||||||||
| Soybean | A 2704-12 | X | X | X | ||||||||||||||
| Soybean | A 5547-127 | X | X | X | ||||||||||||||
| Soybean | MON 89788 | |||||||||||||||||
| Maize | Bt 11 | X | X | X | X | |||||||||||||
| Maize | Bt 176 | X | X | X | X | |||||||||||||
| Maize | MON 810 | X | X | X | ||||||||||||||
| Maize | GA 21 | X | X | X | ||||||||||||||
| Maize | T25 | X | X | X | ||||||||||||||
| Maize | NK 603 | X | X | X | X | X | ||||||||||||
| Maize | MON 863 | X | X | X | X | |||||||||||||
| Maize | TC1507 | X | X | X | X | |||||||||||||
| Maize | DAS59122 | X | X | |||||||||||||||
| Maize | Bt10 | X | X | X | X | |||||||||||||
| Canola | GT73 | X | X | |||||||||||||||
| Canola | MS1/RF2/MS1xRF2 | X | X | X | X | X | ||||||||||||
| Canola | MS1/RF1/MS1xRF1 | X | X | X | X | X | ||||||||||||
| Canola | MS8/RF3/MS8xRF3 | X | X | X | ||||||||||||||
| Canola | TOPAS 19/2 | X | X | X | X | X | X | |||||||||||
| Canola | T45 | X | X | |||||||||||||||
| Canola liberator | L62 | X | X | X | ||||||||||||||
| CANOLA falcon | pHoe6/AC | X | X | X | ||||||||||||||
| Canola | Oxy235 | |||||||||||||||||
| Cotton | MON 1445 | X | X | X | ||||||||||||||
| Cotton | MON 531 | X | ||||||||||||||||
| Cotton | LLCOTTON25 | X | ||||||||||||||||
| Rice | LLRICE601 | X | X | |||||||||||||||
| Rice | Bt63 | X | X | X | ||||||||||||||
| Sugar beet | RUR H7-1 | X | X | |||||||||||||||
| Frequency | 20 | 11 | 2 | 8 | 3 | 3 | 3 | 3 | 5 | 1 | 11 | 5 | 3 | 5 | 1 | 1 | 1 | |
| Ranking | 1 | 2 | 6 | 3 | 5 | 5 | 5 | 5 | 4 | 7 | 2 | 4 | 5 | 4 | 7 | 7 | 7 |
GMP genetically modified plant, x target is present in the GMP
Markers/targets: “p35S” and t35S: promoter (p) and terminator (t) of the 35Sgene from Cauliflower Mosaic Virus; “pFGW”: promoter of the 35S gene of Figwort Mosaic Virus; “pNOS” & “tNOS”: promoter (p) and terminator (t) of Nopaline Synthetase gene of Agrobacterium; “rice actin”: promoter of the rice actin gene; “nptII”: coding region of the neomycine-phosphotransferase gene; CP4-EPSPS: coding region of the 5-Enol-pyruvylshikimate-3-phosphate synthase RoundUp Ready® herbicide resistance gene; mEPSPS: mutated coding region of the 5-Enol-pyruvylshikimate-3-phosphate synthase gene of maize; “PAT/pat” & “BAR/bar”: coding regions of the phosphinotricine acetyl-transferase Liberty-link herbicide resistance; “CryIAb”, “CryAc”, “CryIF” and Cry3Bb1: coding regions of Bt insect resistance genes
In general, the first step in GMP analysis consists of a generic screening for the presence of GMP in a sample (for a review see, [13]). In the ideal situation, such screening should allow to detect all GMP of the GMP Universe. In setting up a minimal format (being the least number of methods required to detect the most of the GMP in the best-discriminating way), the matrix description of the GMP Universe can serve in determining (1) the frequency of presence of the different targets in the GMP (= “coverage” power) and (2) the relative distribution of the respective targets in the GMP (= “discriminative” power).
In GMP, the most commonly present GM targets are the CaMV 35S promoter (p35S) and the Agrobacterium NOS terminator (tNOS; see Table 2), which have accordingly been very often used in GMP screening. Limiting the initial screening to those common targets only, has the disadvantage that the presence/absence of large numbers of GMP needs to be confirmed in a second GM identification analysis. Adding GMP discriminating targets (e.g. endogenous targets, GM-trait targets) to the initial screening can however greatly reduce the number of possible GMP.
Having defined a set of targets suited to detect the GMP in a particular Universe, the basic principle in matrix-based screening analysis requires establishing a formal relation D which stipulates the detection of a particular target in a GMP by an analytical method (see Box below).
The formal relation D may contain, in principle, any detection method used in GMP analysis that meets the validation criteria set by the ISO norm 5275 [14] and/or, at least in the EU, the ENGL guidelines [15]. Here, as an example, the EU-GMPsoy Universe is described, which to date contains three authorized GMP in casu GTS40-3-2, A2704-12, and MON89788 (see Table 2).

When applying such relationship in a GMP analysis (see Table 1), any positive signal (target per definition present “≥LOD”) obtained with a validated method for a particular target indicates that GMP comprising this target could be present in the sample, as indicated by the x correlation in the matrix. If the target cannot be detected (<LOD), an “empty” is annotated. When multiple GMP comprise the same target (see, e.g. target z 2 in Table 1), a positive result generated by screening with method m2, indicates that the GMP X2 and X4 could be present in that sample (see gray fields in Table 1). This principle represents the key algorithm to the matrix-based approaches developed in GMP screening. A similar reasoning holds when multiple methods are being used in a screening approach (see example below).
Mathematical GMP “tagging” and selection through “Gödel Prime Product” modulation
The aim of COSYPS analysis is to obtain sufficient information allowing predicting the set of GMP present in a sample. Given the constant increasing number of GMP, a numerical tag of each GMP would be an asset in developing a simple mathematical way of identifying each GMP. In most models, the presence of a target is mathematically converted into “1”, while absence is indicated by “0”. Although very simple and directly linked to the binary computer system and logical truth functions, the use of such description is limited to binary interpretation requiring handling large data arrays by informatics programs for interpretation of the outcomes. In the mathematical model supporting CoSYPS, a novel target identifying concept based on prime numbers has been introduced. Prime numbers have by definition only two dividers, the prime number itself and “1”. As such, not only can these numbers be used as a true/false function similar to the binary “1/0” combination, the choice of a different prime number for each element/target/method provides a specific representation of each of them.
In CoSYPS analysis, a unique prime number P mK is thus assigned to each particular screening qPCR method/target combination (see Table 3). When the target is present in the GMP and a positive signal is obtained when applying qPCR method m k, instead of a x, the corresponding prime number P mK is assigned to that relationship. When the qPCR/target combination is absent in the GMP (“empty” in Table 1), a “1” is assigned, “1” being the neutral element in the multiplication.
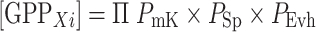 |
1 |
Wherein
- PmK
Prime number of the method mk recognizing a vector element z k in Xi
- PSp
Prime number of the method recognizing the species DNA element in Xi
- PEvh
Prime number of the method recognizing an event-specific element in Xi
When a certain set of screening methods is then applied (in the example in Table 3, the methods m1–4), each GMP can be represented by product of the different P mK of the respective methods that recognize a target comprised in this GMP (see last column in Table 3). This product is designated the “Gödel Prime Number” of the GMP Xi (=GPPXi) and represents a “mathematical tag” for this GMP in a particular GMP Universe [16]. From this GPPXi the different elements present in a GMP Xi can be obtained through factorization into it’s the dividers (= Central theorem of natural numbers). Vice versa, dividing the GPPXi by the primes of the respective qPCR/target combinations used to compile the GMP Universe will, as a consequence of the nature of prime numbers, always yield an integer number whenever the target is present in that GMP. If the target is not present, a non-integer number will be obtained as the quotient. Thus, the presence of a target in a GMP can be mathematically traced as generating an integer quotient by the following division:
 |
2 |
Table 3.
Conversion of “x”/“ ” Matrix into “Prime Number” Matrix (as a basis the matrix in Table 1 was taken)
| Method m1 | Method m2 | Method m3 | Method m4 | ||
|---|---|---|---|---|---|
| Target z 1 | Target z 2 | Target z3 | Target z4 | ||
| 3 | 5 | 7 | 11 | GPP Xi | |
| GMO X1 | 3 | 1 | 7 | 1 | 21 |
| GMO X2 | 3 | 5 | 1 | 1 | 15 |
| GMO X3 | 3 | 1 | 1 | 1 | 3 |
| GMO X4 | 1 | 5 | 7 | 1 | 35 |
| GMO X5 | 3 | 1 | 1 | 11 | 33 |
GPP Xi Gödel prime product of the GMP Xi
In a similar way, when using multiple methods in a screening approach, the product of the prime numbers of the positively scored screening outcomes for a sample can be represented by “GPPSample” product (see Table 4). This number comprises the targets that are detectable (≥LOD) in the sample, and the GPPSample can be used as a divider to search for the GMP present in the sample in a similar way as in Eq. 2:
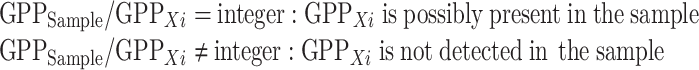 |
3 |
Thus, based on the GPPXi of the respective GMP present in the EU-GMP and the outcome of the screening analysis, expressed as the GPPSample, the possible presence of a GMP can be easily determined by a simple division function.
Table 4.
Mathematical “Prime number"-based GMP tracing in samples S1–4
| Sample | GPPsample | GMP X1 | GMP X2 | GMP X3 | GMP X4 | GMP X5 |
|---|---|---|---|---|---|---|
| 21 | 15 | 3 | 35 | 33 | ||
| S1 | 15 | 0.71428571 | 1 | 5 | 0.42857143 | 0.45454545 |
| S2 | 21 | 1 | 1.4 | 7 | 0.6 | 0.63636364 |
| S3 | 77 | 3.66666667 | 5.13333333 | 25.6666667 | 2.2 | 2.33333333 |
| S4 | 165 | 7.85714286 | 11 | 55 | 4.71428571 | 5 |
S1 possibly contains GMP X2 and 3
S2 possibly contains GMP X1 and 3
S3 does not contain any of the GMPs
S4 possibly contains GMP X2, 3 and 5
In Table 4, the outcome of screening four different samples S1–4 the methods m1–4 is shown. In S1, 2, and 4, the GPPSample/GPPXi quotient yields an integer number for some GMP, indicating that these GMP may be present in that sample. In sample S3, none of the GPPSample/GPPXi quotients yield an integer, indicating that none of these GMP is detectable in this sample. Such a “prime number”-based tagging of the respective GMP allows thus for an easy interpretation of the analytical results in terms of GMP present in a sample. An example is shown in Tables 5 and 6 for the RR soy GTS40-3-2 using the {qPCR methods} = {LEC, p35S, tNOS, CP4-EPSPS, PAT}. The EU-GMPsoy is used as universe (see above). In the upper panel, a prime description of this Universe is presented; in the lower panel, the outcome of a “prime number” tracing analysis on three (theoretical) samples is shown. In two samples, the outcome shows that RRsoy can be present (integer quotient; S2 and S3), in one sample no GMsoy can be detected (S1).
Table 5.
Formal detection of RR soy GTS40-3-2 presence in a sample by “Prime Number” tagging
| Target/Method | RBCL | LEC | P35S | CP4-EPSPS | PAT | |
| Prime | 3 | 5 | 7 | 11 | 13 | GPP XI |
| GTS40-3-2 | 3 | 5 | 7 | 11 | 1 | 1155 |
| A2704-12 | 3 | 5 | 7 | 1 | 13 | 1365 |
| MON89788 | 3 | 5 | 1 | 11 | 1 | 165 |
Table 6.
Formal detection of RR soy GTS40-3-2 presence in a sample by “Prime Number” tracing
| RBCL | LEC | P35S | EPSPS | PAT | GTS40-3-2 | A2704-12 | MON89788 | ||
| Primes | 3 | 5 | 7 | 11 | 13 | GGP | 1155 | 1365 | 165 |
| Sample | GPPsample | ||||||||
| S1 | 3 | 5 | 7 | 1 | 1 | 165 | 0.14285714 | 0.12087912 | 0.0008658 |
| S2 | 3 | 5 | 7 | 7 | 1 | 1155 | 1 | 0.84615385 | 0.00606061 |
| S3 | 3 | 5 | 7 | 1 | 13 | 15015 | 13 | 11 | 0.07878788 |
The four CoSYPS decision parameters: the Tm value, the Ct value, and the LOQ6 and LOD6 values
Next to providing a mathematical tool for formal interpretation of the outcome of a screening analysis, the criteria allowing assigning a particular decision value to a measurement are to be defined. CoSYPS analysis determines the presence of GMP, based on the presence of (preferentially) multiple elements comprised in a GMP by qPCR analysis. As these elements are linked in the transformation event, each of the elements comprised in a GMP should be positive in the qPCR profile in order to conclude the presence of that particular GMP. In CoSYPS, four values for each target are taken into consideration: the T m value, the C t value, and the LOQ6 and LOD6 values.
Taking a correct T m value as the primary criterion, a qPCR measurement in a sample analysis is considered to be matching the relationship D for a certain target in a sample only when: (1) a peak upon melting analysis is obtained with a T m value corresponding to the nominal T m value obtained with the reference material as template DNA (with an acceptable SD ± 1 °C), and (2) an (exponential) amplification above the threshold level can be measured (=a C t value). The nominal T m value for each target has been determined by melting curve analysis upon cloning the amplicons into a uniform plasmid background, the “Sybricon” plasmids [17]. In this way, the influence of background surroundings is considered to be minimized in comparison to genomic DNA backgrounds.
Next, the amount of target present in the sample, represented by the C t value in qPCR analysis, is taken as the second decision criterion in CoSYPS. In this respect, two quantity-based decision levels are defined: the “LOD6” and the “LOQ6”. The lower level of detection (here designated as the “LOD6”), is estimated essentially according to the former AFNOR Norm XP V03-020-2 [18] and the IUPAC guidelines [19]. The “LOD6” of a qPCR method represents the minimal number of targets required in a sample to obtain six of six positive signals at a sixfold repetition of the qPCR analysis on that sample. This LOD6 represents, as such, the (estimated) HGE (measured in qPCR by the C t value), at which level, within a linear serial dilution analysis, each of the 6 repeats provides a positive signal (n = 6; six of six positive signals). All qPCR methods applied in a CoSYPS approach should (preferentially) yield an LOD6 of about two to ten target copies in order to meet the requirements of the ENGL [15].
In view of the necessity to obtain quantitative data on the GMO presence for labeling purposes, an additional level of quantity was determined the so-called “LOQ6”. In this case, the minimal amount of copies of target (designated “LOQ6”) present in 5 μl DNA extract in order to obtain a consistent analytical result (accepted SD ± 0.5 C t) for six repeated measurements was determined for each qPCR method. This LOQ6-based decision criterion applied in CoSYPS is, as such, a more quantity-based threshold. The LOQ6 was determined through SYBR®Green qPCR analysis on (low-bias) single-copy target genomic DNA (e.g. gDNA from leaf tissue of homozygous/hemizygous seeds with certified HGE for the target). All qPCR methods should (preferentially) have an LOQ6 of about 20 target copies in the CoSYPS set-up [15].
The C t values corresponding to the LOD and LOQ6 as determined on the single-copy target genomic DNA, are retained per se as decision values in the CoSYPS decision-support system.
Combinatory interpretations in CoSYPS analysis
In practice, sampling for enforcement purposes on GMO presence is performed by government officials and a laboratory sample is prepared according to standardized protocols (such as described in [5]). This laboratory sample, typically about 500 g to 1 kg, is at first homogenized by the analyzing laboratory and at least two representative analytical samples (between 250 mg to 1 g, depending on the nature of the sample) are extracted and analyzed using a set of SYBR®Green qPCR methods. Each subsample is herein analyzed once for the presence of the respective “CoSYPS” targets in the sample.
The combinatory data interpretation within CoSYPS lays at three levels: (1) a combined evaluation of the T m and C t values for the presence of a certain target in each analysis of a subsample, (2) the combined evaluation of the outcome for the presence of a certain target in two subsamples, and (3) the combined evaluation of the presence of multiple targets for the determination of the presence of a particular GMP.
Evaluating the T m and C t value for each target in each subsample
The primary criterion in CoSYPS is the T m value obtained upon melting analysis of the PCR amplification products. Any T m signals falling out of the range of the acceptance criteria for a particular qPCR method (set at the a priori determined nominal value ± 1 °C), will not be retained for further analysis. Applying the C t values corresponding to the LOQ6 and LOD6 (semi-quantitative) scores for each method, allows establishing a semi-quantitative decision for each target with an acceptable T m value.
-
Level 2:
Combining the analytical results of for each target in the two subsamples
The possible outcomes of the analysis of two subsamples for each qPCR method using the CoSYPS decision criteria LOD6 and LOQ6 are listed in Table 7. In principle, each target that is present “≥LOD6” will be retained in the CoSYPS evaluation and the corresponding prime number of that target will be comprised in the GPPSample (see Table 8 (upper panel)). For each qPCR method, three types of outcome can thus be obtained based by the combined interpretation of the analysis of the two subsamples: the target is present in both subsamples “above or equal to LOQ6” (≥LOQ), “above or equal to the LOD6” (≥LOD), or “Below the LOD6” (<LOD). The interpretation of the combined screening results may require additional verification/confirmation if different outcomes are obtained for different targets comprised in the same GMP (e.g. “≥LOQ” in subsample A and “≥LOD” for the same marker). Such verification is especially important when the result may have an impact on the final legal decision (e.g. labeling requirement (at LOQ), GMP presence (at LOD)). To date, all samples with diverging decisions on combined targets are re-analyzed and a decision is taken according ISO-standard 24276 [20].
-
Level 3:
Combinatory interpretation of the analytical results for multiple targets present in a GMP
Table 7.
qPCR result “Level 2” evaluation in CoSYPS for a single target in the two subsamples
| Sample | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 |
|---|---|---|---|---|---|---|
| Subsample A | ≥LOQ | ≥LOQ | ≥LOQ | ≥LOD | ≥LOD | <LOD |
| Subsample B | ≥LOQ | ≥LOD | <LOD | ≥LOD | <LOD | <LOD |
| Outcome | ≥LOQ | ≥LOD | ≥LOD | ≥LOD | <LOD | <LOD |
Table 8.
Combinatory “Level 3” evaluation according to CoSYPS analyses for Sample 2 in Table 4
| Sample 1 | Method m1 | Method m2 | Method m3 | Method m4 | GPP | GGPSAMPLE1/GPP | Conclusion |
| Prime | 3 | 5 | 7 | 11 | |||
| Screening outcome | Target z1 | Target z2 | Target z3 | Target z4 | |||
| SB1 | >LOQ | <LOD | >LOQ | <LOD | |||
| SB2 | >LOQ | <LOD | >LOQ | <LOD | |||
| Conclusion | >LOQ | <LOD | >LOQ | <LOD | |||
| PRIME CONVERSION | 3 | 1 | 7 | 1 | |||
| GGPSAMPLE1 | 21 | ||||||
| GPP EVALUATION: | |||||||
| GMP X1 | 21 | 1 | Possibly present | ||||
| GMP X2 | 15 | 1.4 | No match | ||||
| GMP X3 | 3 | 7 | Possibly present | ||||
| GMP X4 | 35 | 0.6 | No match | ||||
| GMP X5 | 33 | 0.63636364 | No match |
Finally, the combined outcome of the CoSYPS screening is compared to the respective GMP of the EU-GMP Universe, applying the “Prime number”-based GMP-tagging algorithm. As all targets in a particular GMP are linked, the outcome in each subsample sample should be coherent for each target comprised in that GMP. “Coherence” here means that for one GMP to be present, all targets comprised in that GMP should be at least detectable at the level of the least represented target in the sample. Indeed, as some targets are present in multiple GMP or in multiple copies in the same GMP, overrepresentation of certain targets in a combined evaluation may occur.
For this, the CoSYPS algorithm retains all prime numbers of the targets wherein all constituents are present “≥LOD6”, and multiplies these to obtain the so-called “GPPSample”. Dividing this GPPSample by the GPP of each of the GMP of the EU-GMP Universe, allows assessing the presence of the GMP based on the integer nature of the quotient (see Table 8 (lower panel)). The mathematical and logical combination of the outcome of the respective qPCR methods based on the presence (T m value) and the amount (C t value) can be organized in a two-worksheet Excel-based model, that can be (relatively) easily adapted to cover different scopes (at species level), new GM-events and additional GM-screening elements (data not shown).
The presence of each of the matching GMP is then in a next step of the process to be confirmed by event-specific identification (ID). All GMP detected in the ID analysis are then to be tested for conformity with the labeling requirements whenever all targets of a GMP are scored “≥LOQ” These conditions are considered reflecting to the best the requirements imposed by the EC legislations to date. If none of the GMP present in the universe can explain the presence of one or more targets, it can be concluded that a sample contains unassigned quantities/presence of that particular target. Further analysis will then have to be conducted as to identify the origin or the nature of the materials generating these unassigned signals (e.g. originating from donor organisms (bacteria, viruses, plant…) or unauthorized GMO). Especially in the latter case, complex additional analyses might be required which are outside the scope or capacity of enforcement activities and will not be discussed here further (e.g. genome walking, DNA sequencing…).
Conclusions and future perspectives
CoSYPS represents a novel real-time qPCR GMP analysis approach based on SYBR®Green technology supported by a four-parameter DSS. CoSYPS is essentially a matrix-based approach aiming to identify the potential presence of GMP in a sample. CoSYPS combines for this purpose the analytical results (T m- and C t values) of a limited set of SYBR®Green qPCR methods with preferentially similar performance under the same conditions. While other technologies are also suitable to be applied in a matrix-approach, SYBR®Green detection offers the advantage of verification of the amplification products (based on the outcome of the melting analysis). Two decision thresholds, the LOD6 and the LOQ6, allow then to estimate the level of GMP present in a sample in a semi-quantitative way. The CoSYPS DSS can be developed in an Excel format wherein all concepts are integrated through simple logical and mathematical functions (own unpublished data).
As such, CoSYPS is considered a versatile, cost-effective approach in assessing the presence of GMP, particularly useful in routine analysis for enforcement. Further improvement of the CoSYPS may be (1) streamlining the SYBR®Green technology with other qPCR technologies (e.g., Taqman…), (2) adapting the DSS format to enable statistically valid analysis at low-level presence of GMP, and (3) developing a CoSYPS informatics tool which can be (easily) updated and electronically verified, and moreover made available to a broad range of stakeholders.
An integrated DSS for COSYPS screening and GM-event Taqman identification
To date, the confirmation on the final set of GMP identified to be possibly present in a sample by CoSYPS is obtained by identification analysis applying validated event-specific Taqman qPCR methods (http://gmo-crl.jrc.ec.europa.eu; CRL-GMFF). Recently, the so-called pre-spotted plates were developed allowing detecting all EU-regulated GMP using Taqman qPCR methods adapted from the official event-specific methods [21]. Such approach could be readily combined with a CoSYPS analysis, although the statistical model (including the acceptance criteria) for combining data obtained from Taqman qPCR and SYBR®Green qPCR analysis will have to be developed. Recently, a strategy for the interpretation of such analysis using a so-called “differential PCR approach” has been developed [22]. Such analysis is, to date, not yet available for CoSYPS and may require that the PCR amplification efficiencies for the different qPCR methods are well established and measurable in a single-well mode. Such determination is not (yet) available in the standard PCR programs of the PCR machines and requires considerable time investment using other alternative PCR analysis software [23].
GMP detection at low-level presence
A particular challenge for the EU legal system is the asynchronous commercialization of GMP on the global market. While segregation systems are put in place in the GMP exporting countries, greatly reducing the probability of escapes of non-EU authorized GMP in EU shipments, accidents or occasional failure seem to be inevitable (e.g. [3, 4]). Due to the zero-tolerance rules applied at the EU-level for non-authorized substances, these escapes represent illegal materials and are to be withdrawn from the EU market. In most cases, these non-authorized substances are present only as traces but are being observed in enforcement and reported as such in the European RASFF system (http://ec.europa.eu/food/food/rapidalert).
To monitor and manage the presence of substances at such low levels, new screening strategies may have to be implied based on analyzing more subsamples and more targets within a single sample analysis. For a more detailed discussion on some of the novel approaches that could be applied for this purpose, we refer to Querci at al. [7]. It remains, however, to be evaluated whether appropriate validation schemes according to the ISO-standards can be put into place for such low amounts of materials in a sample applying these complex technologies.
To date, a combinatory matrix-based approach based on simplex (eventually duplex or multiplex) qPCR methods in a 96-well (or larger) format could also be envisaged for such purposes as long as within the global GMP Universe, common elements (such as the p35S, the GM-traits…) are being applied in the transformation vectors. When GMP become however more specialized using only unique DNA sequences (as e.g., in the GM-maize LYO38 (1, 2, and 3)), the advantage of a combinatory approach may become a lesser efficient strategy.
An i-GMID platform: “twittering” technical tools and decision support on the web
Finally, given the complexity of the GMP analysis, it is preferred that a DSS such as that developed for CoSYPS, be maintained and updated centrally (e.g., by the European Commission or FAO) and made accessible through a web application. Such on-line data interpretation of GMP analytical results within a matrix-based DSS, should allow for designing customized GMP-screening modules. Within this set-up, access to DBases containing up-to-date information on validated qPCR methods and available certified reference materials should be included. Optimal combination of methods could be evaluated through “fuzzy logic” principles as outlined by [24]. In addition, algorithms such as the ‘GMOTrack” system recently developed by the National Institute of Biology that enable the choice of the better set of methods according to a number of sample parameters (presence of target elements, frequency of occurrence, and a cost function) could also be incorporated (http://kt.ijs.si/software/GMOtrack/). Applying all these decision-support principles within a web-based manner to a GMP detection system, such as CoSYPS, may assist in harmonizing and enabling transparency in GMP screening analysis for all stakeholders in the field of GMP production, processing, distribution, and control.
Acknowledgments
This work was supported by the Co-Extra EC-project (Contract No. 007158) and by a grant from the “Belgische Federale Overheidsdienst Volksgezondheid, Veiligheid van de Voedselketen en Leefmilieu” (GMODETEC project (RT-06/6): “Ontwikkeling van een algemene strategie voor detectie, identificatie en kwantificatie van genetisch gemodificeerd materiaal in voedingsproducten en veevoeder”)
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Marc Van den Bulcke, Email: gmolab@sbb.ihe.be, Email: marc.van-den-bulcke@ec.europa.eu.
Myriam Sneyers, Email: gmolab@sbb.ihe.be.
References
- 1.Regulation (EC) No 1829/2003 of the European Parliament and of the Council of 22 September 2003 on genetically modified food and feed. Off J Eur Union L 268:1-23
- 2.Regulation (EC) No 1830/2003 of the European Parliament and of the Council of 22 September 2003 concerning the traceability and labelling of genetically modified organisms and the traceability of food and feed products produced from GMOs and amending Directive 2001/18/EC. Off J Eur Union L 268:24-28
- 3.Commission Decision No 162/2008 of 26 February 2008 amending Decision 2006/601/EC on emergency measures regarding the non-authorised genetically modified organism ‘LL RICE 601’ in rice products. Off J Eur Union L 52/25
- 4.Commission Decision No 289/2008 of 3 April 2008 on emergency measures regarding the unauthorised genetically modified organism products. Off J Eur Union L 96/29
- 5.Commission Recommendation 2004/787/EC of 4 October 2004 on technical guidance for sampling and detection of genetically modified organisms and material produced from genetically modified organisms as or in products in the context of Regulation (EC) No 1830/2003. Off J Eur Union L 348:18-26
- 6.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′ 3′ exonuclease activity of Thermus aquaticus DNA polymerase. PNAS. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Querci M, Van den Bulcke M, Žel J, Van den Eede G, Broll H (2010) New approaches in GMO detection ABC in press [DOI] [PubMed]
- 8.Leimanis S, Hernández M, Fernández S, Boyer F, Burns M, Bruderer S, Glouden T, Harris N, Kaeppeli O, Philipp P, Pla M, Puigdomènech P, Vaitilingom M, Bertheau Y, Remacle J. A microarray-based detection system for genetically modified (GM) food ingredients. Plant Mol Biol. 2006;61:123–139. doi: 10.1007/s11103-005-6173-4. [DOI] [PubMed] [Google Scholar]
- 9.Leimanis S, Hamels S, Nazé F, Mbongolo Mbella G, Sneyers M, Hochegger R, Broll H, Roth L, Dallmann K, Micsinai A, La Paz JL, Pla M, Brünen-Nieweler C, Papazova N, Taverniers I, Hess N, Kirschneit B, Bertheau Y, Audeon C, Laval V, Busch U, Pecoraro S, Neumann K, Rösel S, van Dijk J, Kok E, Bellocchi G, Foti N, Mazzara M, Moens W, Remacle J, Van den Eede G. Validation of a GMO multiplex screening assay by the use of microarray. Eur Food Res Technol. 2008;227:1621–1632. doi: 10.1007/s00217-008-0886-y. [DOI] [Google Scholar]
- 10.Hamels S, Leimanis S, Mazzara M, Bellocchi G, Foti N, Moens W, Remacle J, Van den Eede G (2007) Microarray method for the screening of EU approved GMOs by identification of their genetic elements http://mbg.jrc.ec.europa.eu/home/docs.htm
- 11.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR: a perspective. J Mol Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 12.Directive 2001/18/EC of the European Parliament and of the Council of 17.4.2001 on the deliberate release into the environment of genetically modified organisms and repealing Council Directive 90/220/EEC. Off J Eur Union L 106/1
- 13.Marmiroli N, Maestri E, Gullì M, Malcevschi A, Peano C, Bordoni R, De Bellis G. Methods for detection of GMOs in food and feed. Anal Bioanal Chem. 2008;392:369–384. doi: 10.1007/s00216-008-2303-6. [DOI] [PubMed] [Google Scholar]
- 14.International Standard (ISO) 5725 . Accuracy (trueness and precision) of measurement methods and results . Genève, Switzerland: International Organization for Standardization; 1994. [Google Scholar]
- 15.Definition of minimum performance requirements for analytical methods of GMO testing, European Network of GMO Laboratories (ENGL) (2008) http://gmo-crl.jrc.ec.europa.eu/
- 16.Gödel K. Über formal unentscheidbare Sätze der Principia Mathematica und verwandter Systeme. Monatsheft für Math und Physik. 1931;38:173–198. doi: 10.1007/BF01700692. [DOI] [Google Scholar]
- 17.Barbau-Piednoir E, Lievens A, Mbongolo-Mbella G, Roosens N, Sneyers M, Leunda-Casi A, Van den Bulcke M (2010) SYBR(r)Green qPCR screening methods for the presence of "35S promoter" and "NOS terminator" elements in food and feed products. Eur Food Res Technol in press
- 18.Norme AFNOR XP V03-020-2 (04/2005): Produits alimentaires - Détection et quantification des organismes végétaux génétiquement modifiés et produits dérivés - Partie 2 : méthodes basées sur la réaction de polymérisation en chaîne
- 19.Thompson M, Ellison SLR, Wood R. Harmonized guidelines for single-laboratory validation of methods of analysis (IUPAC Technical Report) Pure Appl Chem. 2002;74:835–855. doi: 10.1351/pac200274050835. [DOI] [Google Scholar]
- 20.International Standard (ISO) 24276 . Foodstuffs—methods of analysis for the detection of genetically modified organisms and derived products—general requirements and definitions. Genève, Switzerland: International Organization for Standardization; 2006. [Google Scholar]
- 21.Querci M, Foti N, Bogni A, Kluga L, Broll H, Van den Eede G. Real-time PCR based ready-to-use multi-target analytical system for GMO detection. Food Analytical Methods. 2009 [Google Scholar]
- 22.Cankar K, Chauvensy-Ancel V, Fortabat MN, Gruden K, Kobilinsky A, Zel J, Bertheau Y. Detection of non-authorized genetically modified organisms using differential quantitative polymerase chain reaction: application to 35S in maize. Anal Biochem. 2008;376:189–199. doi: 10.1016/j.ab.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1045–1062. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellocchi G, Bertholet V, Hamels S, Moens W, Remacle J, van den Eede G. Fuzzy-logic based strategy for validation of multiplex methods: example with qualitative GMO assays. Transgenic Res. 2009 doi: 10.1007/s11248-009-9293-9. [DOI] [PubMed] [Google Scholar]