Abstract
In this study, we developed a Phenotype MicroArray™ (PM) protocol to profile cellular phenotypes in Zymomonas mobilis, which included a standard set of nearly 2,000 assays for carbon, nitrogen, phosphorus and sulfur source utilization, nutrient stimulation, pH and osmotic stresses, and chemical sensitivities with 240 inhibitory chemicals. We observed two positive assays for C-source utilization (fructose and glucose) using the PM screen, which uses redox chemistry and cell respiration as a universal reporter to profile growth phenotypes in a high-throughput 96-well plate-based format. For nitrogen metabolism, the bacterium showed a positive test results for ammonia, aspartate, asparagine, glutamate, glutamine, and peptides. Z. mobilis appeared to use a diverse array of P-sources with two exceptions being pyrophosphate and tripolyphosphate. The assays suggested that Z. mobilis uses both inorganic and organic compounds as S-sources. No stimulation by nutrients was detected; however, there was evidence of partial inhibition by purines and pyrimidines, NAD, and deferoxamine. Z. mobilis was relatively resistant to acid pH, tolerating a pH down to about 4.0. It also tolerated phosphate, sulfate, and nitrate, but was rather sensitive to chloride and nitrite. Z. mobilis showed resistance to a large number of diverse chemicals that inhibit most bacteria. The information from PM analysis provides an overview of Z. mobilis physiology and a foundation for future comparisons of other wild-type and mutant Z. mobilis strains.
Keywords: Ethanol, Fermentation, Physiology, Systems biology, Global
Introduction
Biomass-based bioenergy processes are vital to make cellulosic ethanol cost-competitive with gasoline, and lignocellulosic materials represent an abundant feedstock for cellulosic biofuel production. Ethanol is presently sold nationwide mostly as a fuel oxygenate, and there is a reawakened interest in producing increasing amounts of ethanol in large-scale industrial fermentations to reduce fuel imports. The potential and challenges involved in supplanting a large amount of petroleum-derived transportation fuels by fuels from renewable resources like lignocellulosic ethanol has been the focus of several studies and reviews [1–5]. A core challenge to fermenting cellulosic material is accessing the sugars due to biomass recalcitrance, which is due in part to millions of years of plant evolution to resist microbial degradation [6]. Despite these challenges, a number of companies are presently attempting to produce biofuels from lignocellulosic feedstocks. DuPont and Broin have formed one such partnership, which will utilize recombinant Zymomonas mobilis strains for bioethanol fermentation from lignocellulosic residues such as corn stover [7].
Saccharomyces yeasts have been the preferred industrial biocatalysts in the current generation of fuel ethanol production processes as they are robust, efficient, and fermentative microorganisms with a long industrial history. A comparison of Z. mobilis and yeast has shown that they share many important attributes for ethanol production, although Z. mobilis can offer advantages in productivity [8]. Z. mobilis is a Gram-negative facultative anaerobic bacterium with a number of desirable industrial characteristics, such as unique anaerobic use of the Entner–Doudoroff pathway that results in low cell mass formation, high-specific productivity and ethanol yield, high ethanol tolerance (12%), broad pH range for ethanol production (pH 3.5–7.5), and designated status as generally regarded as safe [8–13]. Wild-type Z. mobilis strains are known to have a narrow substrate utilization range and utilize only C-6 sugars. However, recombinant strains have been developed that extend the substrate range to allow fermentation of pentose (C-5) sugars such as xylose and arabinose [14–16], and available genome sequence and genetic manipulation present opportunities for further strain developments [17, 18].
The availability of genome sequence, systems biology tools, and genetics presents new opportunities to elucidate the molecular basis of industrially relevant traits for strain development and process improvements and helps to provide fundamental insights into Z. mobilis physiology. However, systems biology tools such as transcriptomics, proteomics, and metabolomics have only recently begun to be applied for strain development, and there are few examples of their utility in industry. In a recent review, Park et al. [18] describe many of the tools that are available for genome-wide analyses for strain development, along with examples of successful systems metabolic engineering and future perspectives. The global analysis of bacterial phenotypes is another approach that can provide complementary data to other systems biology analyses (see [19] for a recent review). In this study, we developed a Phenotype MicroArray™ (PM) protocol to profile nearly 2,000 Z. mobilis cellular phenotypes and generated a baseline PM profile for the organism for future studies.
Materials and Methods
Bacterial Strains, Culture Conditions, and Chemicals
Z. mobilis ZM4 was obtained from the American Type Culture Collection (ATCC31821). PM inocula were prepared from ZM4 colonies incubated aerobically for 24 h and cultured on yeast extract peptone dextrose (YPD) agar plates (Sigma-Aldrich, St. Louis, MO) at 30 °C. Isolated colonies were removed from agar plates using a sterile swab and transferred into sterile inoculating fluids for inoculation (100 μL per well) into the PM panels.
PM Inoculation, Incubation, and Data Collection/Analysis
Z. mobilis cell suspensions were prepared and inoculated into each of the 20 PM plates for full metabolic profiling according to standard protocols recommended by Biolog for bacteria or for yeast. The PM plates were placed in an aerobic OmniLog incubator reader set at 30 °C using standard manufacturer operating conditions, which collected data every 15 min over a 72-h period. This was sufficient time for color to develop in the positive control wells, while the negative control wells remained colorless. Tests were conducted in duplicate, and the PMs were also examined visually at the end of each incubation for independent verification of results. The OmniLog® V. 1.5 comparison module and the average height parameter were used for data analysis with standard thresholds for detection.
Results and Discussion
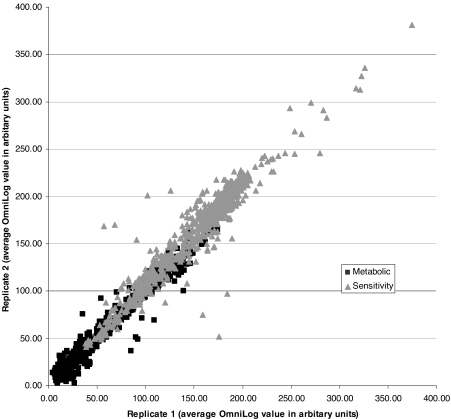
Z. mobilis is unlike most bacteria in that it produces high levels of ethanol and is very ethanol-tolerant. Z. mobilis and yeast share a number of important fermentation attributes, although productivity advantages have been reported for Z. mobilis [8]. It also behaved very differently from other bacteria and presented challenges when we attempted to assay it using protocols that we have used for PM testing of more than 1,000 diverse Gram-negative and Gram-positive species. Initial tests with a “universal” bacterial PM protocol failed to work for Z. mobilis (data not shown). After further preliminary PM testing, it was apparent that the properties of Z. mobilis more closely resembled Saccharomyces yeast than the bacteria assayed previously. We finally succeeded in testing its phenotypes when we employed an assay protocol used for testing the ethanol producing yeast, Saccharomyces. Like the yeast, Z. mobilis grows well at 30 °C on YPD agar. To assay Z. mobilis, we used a lower pH of 6.0 and, most importantly, a different tetrazolium redox dye, Dye Mix E, to measure energy production potential. We assayed nearly 2,000 cellular phenotypes in Z. mobilis, which included assays for carbon, nitrogen, phosphorus and sulfur source utilization, nutrient stimulation, pH and osmotic stresses, and chemical sensitivities with 240 inhibitory chemicals. The PM assays were conducted in duplicate, and correlation plots from the independent runs showed replicate profiles were reproducible (Fig. 1). All plates passed reproducibility analysis, and PM replicate experiments had r 2 values of 0.97 and 0.93 for the metabolic and sensitivity series, respectively.
Fig. 1.
Reproducibility of PM tests. Average OminLog value was calculated over a 72-h time course for each replicate and plotted as replicate 1 (x-axis) versus replicate 2 (y-axis). Identical values fall on a 45-degree line with few tests deviating by more than 50 units. Metabolic test values are represented by black squares, and sensitivity test values are represented by gray triangles
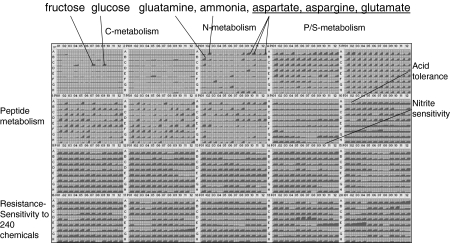
The PM analysis gave an overview of the Z. mobilis physiological characteristics (Fig. 2), such as the limited C-source utilization (d-fructose (PM1, C7) and d-glucose (PM1, C9)), which was consistent with literature reports [20]. The ability to hydrolyze sucrose has been reported to be strain-dependent [8], and our PM assays were negative for sucrose utilization even when repeated at higher cell densities. In subsequent growth studies, we confirmed that ZM4 grew in broth cultures and on plates where sucrose was supplied as the sole carbon source. Single ZM4 colonies from aerobic sucrose plates were used to inoculate aerobic broths of basal medium (i.e., without added carbon source) and medium containing either 50 g/L of glucose or sucrose prepared as described previously [21]. The initial culture turbidities were approximately OD600nm = 0.03 for each condition. After 18.5 h of static incubation at 30 °C, no growth was detected in the no-carbon source broth culture, which controlled for carryover of any carbon source from the plate culture and growth from components of the basal medium such as the 2.5 g/L of yeast extract. Broth cultures that contained either sucrose or glucose reached OD600nm values of approximately 1.5 and 2.1, respectively, during the same period. These results suggest that the ability to utilize indicate sucrose was subject to strong catabolite repression in our PM assays. With glucose as the carbon source, the nitrogen source panels (PM3, 6, 7, and 8) revealed that Z. mobilis can utilize ammonia, and for single amino acids, aspartate, asparagine, glutamate, and glutamine gave positive assay results. These findings are in accord with a report from a group at DuPont [22] which reported that they could only find evidence of transporters for aspartate and glutamate and that the growth rate was barely altered on rich versus minimal media. PM analysis expands this annotation by demonstrating potential utilization also of asparagine and glutamine, as it expanded and corrected Pseudomonas aeruginosa transporter annotations [23]. Z. mobilis utilized a large and somewhat diverse array of peptides in PM6, 7, and 8 and gave positive assay results for peptides with aspartate, asparagine, glutamate, glutamine, and glycine, although others were used more slowly. In addition to a variety of amino acids and dipeptides in PM3, the strain appeared to utilize ethanolamine (PM3, D9), glucuronamide (PM3, E6), adenosine (PM3, F3), and parabanic acid (PM3, G6). As P-sources, Z. mobilis appeared to use a diverse array of inorganic and organic phosphorus-containing compounds in PM4, rows A–E, with two exceptions being pyrophosphate and tripolyphosphate. Positive phosphorus wells were distinguished by their magnitude and kinetics exceeding the negative control. Both inorganic and organic sulfur sources were used, including sulfate (PM4, F2), thiosulfate (PM4, F3), tetrathionate (PM4, F4), thiophosphate (PM4, F5), dithiophosphate (PM4, F6), l-cysteine (PM4, F7), l-cysteine sulfinic acid (PM4, F12), glutathione (PM4, G5), and l-methionine (PM4, G7). No stimulation by nutrients (PM5) was detected; however, there was evidence of partial inhibition by purines and pyrimidines, NAD, and deferoxamine. Z. mobilis was relatively resistant to acid pH, tolerating a pH down to about 4.0, which is in agreement with published data [8]. It also tolerated phosphate, sulfate, and nitrate, but was rather sensitive to chloride and nitrite. Z. mobilis showed resistance to a large number of diverse chemicals that inhibit most bacteria, which is an unusual property shared with some Pseudomonas and few other bacteria assayed to date. These findings are again in accordance with those of the DuPont group [22]. As Z. mobilis is tolerant to a number of chemicals and antibiotics (~80% of those assayed), we report those to which it showed sensitivity under the conditions assayed (Table 1).
Fig. 2.
Graphical depiction of Z. mobilis metabolic and sensitivity phenotype profile. The figure is a 5 × 4 array of 96-well plate representations. Within each well representation, we plot time on the x-axis versus OmniLog value on the y-axis. Each plate contains a 12 × 8 array of well representations and is identified by a plate number in the upper left of the array that identifies the phenotypic tests for each array. PM1 and PM2 contain carbon tests and as a specific example, PM3; F3 contains adenosine as a nitrogen test. PM3 and PM6–8 contain nitrogen tests. PM4 contains phosphorus and sulfur tests. Negative control wells for metabolic substrate utilization tests are in the A1 position of PM1–4 and 6–8 and in the F1 position of PM4. PM5 is a minimal defined medium. A PM9–20 contain chemical sensitivity tests. A complete map to the content of each well can be found online at http://www.biolog.com/PM_Maps.html
Table 1.
Summary of chemicals Z. mobilis is sensitive to via PM profiling.
| PM | Chemical name | Mode of action |
|---|---|---|
| PM20 | Orphenadrine | Anti-cholinergic |
| PM20 | Pridinol | Anti-cholinergic |
| PM17 | 2-Hydroxybenzoic acid | Biofilm inhibitor, anti-capsule agent, chelator, prostaglandin synthetase inhibitor, mar inducer |
| PM13 | 2,2′-Dipyridyl | Chelator, lipophilic |
| PM20 | 8-Hydroxyquinoline | Chelator, lipophilic |
| PM12 | Novobiocin | DNA topoisomerase |
| PM16 | Chloroxylenol | Fungicide |
| PM16 | Dichlofluanid | Fungicide, phenylsulphamide |
| PM20 | Tolylfluanid | Fungicide, phenylsulphamide |
| PM17 | d-Serine | Inhibits 3PGA dehydrogenase (l-serine and pantothenate synthesis) |
| PM20 | Dodine | Membrane permeability, guanidine, fungicide |
| PM15 | Guanidine hydrochloride | Membrane, chaotropic agent |
| PM17 | Niaproof | Membrane, detergent, anionic |
| PM12 | Benzethonium chloride | Membrane, detergent, cationic |
| PM16 | Cetylpyridinium chloride | Membrane, detergent, cationic |
| PM12 | Dodecyltrimethyl ammonium bromide | Membrane, detergent, cationic |
| PM15 | Domiphen bromide | Membrane, detergent, cationic, fungicide |
| PM19 | Lauryl sulfobetaine | Membrane, detergent, zwitterionic |
| PM15 | Alexidine | Membrane, electron transport, biguanide |
| PM20 | Amitriptyline | Membrane, transport |
| PM20 | Captan | Multisite, carbamate, fungicide |
| PM14 | Iodoacetate | Oxidation, sulfhydryl |
| PM16 | 1-Chloro-2,4-dinitrobenzene | Oxidizes sulfhydryls, depletes glutathione |
| PM19 | d,l-Thioctic acid | Oxidizing agent |
| PM15 | 3,4-Dimethoxybenzyl alcohol | Oxidizing agent, free radical peroxidase substrate |
| PM17 | Chlorpromazine | Phenothiazine, anti-cholinergic, anti-psychotic, sedative |
| PM14 | Promethazine | Phenothiazine, anti-cholinergic, anti-psychotic, sedative |
| PM13 | Trifluoperazine | Phenothiazine, anti-cholinergic, anti-psychotic, sedative |
| PM19 | Phenyl-methyl-sulfonyl-fluoride (PMSF) | Protease inhibitor, serine |
| PM12 | Penimepicycline | Protein synthesis, 30S ribosomal subunit, tetracycline |
| PM11 | Chlortetracycline | Protein synthesis, 30S ribosomal subunit; tetracycline |
| PM11 | Demeclocyline | Protein synthesis, 30S ribosomal subunit; tetracycline |
| PM11 | Minocycline | Protein synthesis, 30S ribosomal subunit; tetracycline |
| PM19 | Josamycin | Protein synthesis, 50S ribosomal subunit, macrolide |
| PM15 | Oleandomycin | Protein synthesis, 50S ribosomal subunit, macrolide |
| PM15 | Fusidic acid | Protein synthesis, elongation factor |
| PM20 | 3,5-Dinitrobenzene | Respiration, ionophore, H+ |
| PM19 | Cinnamic acid | Respiration, ionophore, H+ |
| PM19 | Gallic acid | Respiration, ionophore, H+ |
| PM19 | Sodium caprylate | Respiration, ionophore, H+ |
| PM20 | Tetrazolium Violet | Respiration, uncoupler |
| PM20 | Thioridazine | Respiration, uncoupler |
| PM17 | Oxycarboxin | Respiratory enzymes, carboxamide, fungicide |
| PM14 | Boric acid | Toxic anion |
| PM13 | Potassium chromate | Toxic anion |
| PM18 | Sodium bromate | Toxic anion |
| PM18 | Sodium m-arsenite | Toxic anion |
| PM14 | Sodium nitrite | Toxic anion |
| PM16 | Sodium selenite | Toxic anion |
| PM18 | Sodium periodate | Toxic anion, oxidizing agent |
| PM14 | Sodium Dichromate | Toxic anion, SO4 analog |
| PM14 | Cadmium chloride | Toxic cation |
| PM13 | Cobalt chloride | Toxic cation |
| PM13 | Cupric chloride | Toxic cation |
| PM17 | Lithium chloride | Toxic cation |
| PM13 | Nickel chloride | Toxic cation |
| PM17 | Phenylarsine oxide | Tyrosine phosphatase inhibitor |
| PM19 | Phenethicillin | Wall, lactam |
Conclusions
This is the first report of PM profiles for the important ethanologenic bacterium Z. mobilis. Our results suggest that Z. mobilis differs fundamentally from most bacteria and resembles yeast not only in its ethanol metabolism, limited use of carbon sources, and high sugar tolerance but also in its mechanism of generating a redox potential and reducing tetrazolium dyes. Z. mobilis is known to have an unusual facultative anaerobic bacterial metabolism that generates one ATP/glucose via the Entner–Doudoroff pathway, whereas yeast and many other bacteria generate two net ATP molecules/glucose [8]. While the unusual physiology of Z. mobilis contributes to its overall success as an ethanologenic bacterium, many questions about its basic physiology remain unanswered [24]. The Z. mobilis protocol development and PM assay results provide an important baseline phenotypic profile for this bacterium. The protocol development and baseline data can be used to speed the optimization of Z. mobilis bioprocesses and to provide a means for detailed comparisons of other wild-type strains and mutant strains in future studies.
Acknowledgments
We thank Courtney Johnson for assistance in conducting sucrose growth tests. Research sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC for the US Department of Energy under contract no. DE-AC05-00OR22725. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Protocol development for Zymomonas was aided by funding provided to Barry Bochner and Biolog under STTR grant R42 GM073965 from NIH-NIGMS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Barry Bochner, Email: bbochner@biolog.com.
Steven D. Brown, Email: brownsd@ornl.gov
References
- 1.Farrell AE, Plevin RJTBT, Jones AD, O’Hare M, Kammen DM. Ethanol can contribute to energy and environmental goals. Science. 2006;311:506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 2.Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Koonin SE. Getting serious about biofuels. Science. 2006;311:435. doi: 10.1126/science.1124886. [DOI] [PubMed] [Google Scholar]
- 4.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert C, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 5.Service, R. F Cellulosic ethanol: Biofuel researchers prepare to reap a new harvest. Science. 2007;315:1488–1491. doi: 10.1126/science.315.5818.1488. [DOI] [PubMed] [Google Scholar]
- 6.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 7.Reisch M. Fuels of the future chemistry and agriculture join to make a new generation of renewable fuels. Chem. Eng. News. 2006;84:3. [Google Scholar]
- 8.Panesar PS, Marwaha SS, Kennedy JF. Zymomonas mobilis: An alternative ethanol producer. J. Chem. Technol. Biotechnol. 2006;81:623–635. doi: 10.1002/jctb.1448. [DOI] [Google Scholar]
- 9.Swings J, De Ley J. The biology of Zymomonas mobilis. Bacteriol. Rev. 1977;41:1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers PL, Goodman AE, Heyes RH. Zymomonas ethanol fermentations. Microbiol. Sci. 1984;1:133–136. [PubMed] [Google Scholar]
- 11.Gunasekaran P, Raj KC. Ethanol fermentation technology—Zymomonas mobilis. Current Science. 1999;77:56–68. [Google Scholar]
- 12.Dien BS, Cotta MA, Jeffries TW. Bacteria engineered for fuel ethanol production: Current status. Appl. Microbiol. Biotechnol. 2003;63:258–266. doi: 10.1007/s00253-003-1444-y. [DOI] [PubMed] [Google Scholar]
- 13.Rogers PL, Jeon YJ, Lee KJ, Lawford HG. Zymomonas mobilis for fuel ethanol and higher value products. Biofuels. 2007;108:263–288. doi: 10.1007/10_2007_060. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Eddy C, Deanda K, Finkestein M, Picataggio S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science. 1995;267:240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]
- 15.Deanda K, Zhang M, Eddy C, Picataggio S. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl. Environ. Microbiol. 1996;62:4465–4470. doi: 10.1128/aem.62.12.4465-4470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohagheghi A, Evans K, Chou YC, Zhang M. Cofermentation of glucose, xylose, and arabinose by genomic DNA-integrated xylose/arabinose fermenting strain of Zymomonas mobilis AX101. Appl. Biochem. Biotechnol. 2002;98–100:885–898. doi: 10.1385/ABAB:98-100:1-9:885. [DOI] [PubMed] [Google Scholar]
- 17.Seo JS, Chong HY, Park HS, Yoon KO, Jung C, Kim JJ, et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nature Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Pappas KM, Hauser LJ, Land ML, Chen G-L, Hurst GB, et al. Improved genome annotation for Zymomonas mobilis. Nat. Biotech. 2009;27:893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- 19.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker C, Peekhaus N, Zhang XH, Conway T. Kinetics of sugar transport and phosphorylation influence glucose and fructose cometabolism by Zymomonas mobilis. Appl. Environ. Microbiol. 1997;63:3519–3525. doi: 10.1128/aem.63.9.3519-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viikari L. Formation of levan and sorbitol from sucrose by Zymomonas mobilis. Appl. Microbiol. Biotechnol. 1984;19:252–255. doi: 10.1007/BF00251846. [DOI] [Google Scholar]
- 22.LaRossa, R. A., Smulski, D. R., Viitanen, P., & Jean-Francois Tomb, J.-F. (2007). Uncovering the unusual metabolic logic of Zymomonas mobilis, an ethanol producing microbe, by application of amino acid-centric genomic and genetic approaches. Invited talk, 15th Annual International Conference on Microbial Genomics, Maryland.
- 23.Johnson DA, Tetu SG, Phillippy K, Chen J, Ren QH, Paulsen IT. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genetics. 2008;4:e1000211. doi: 10.1371/journal.pgen.1000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalnenieks U. Physiology of Zymomonas mobilis: Some unanswered questions. Advances in Microbial Physiology. 2006;51:73–117. doi: 10.1016/S0065-2911(06)51002-1. [DOI] [PubMed] [Google Scholar]