Abstract
BACKGROUND
A high copy number of CCL3L1, the most potent HIV-suppressive chemokine, associates with reduced HIV susceptibility. Whether CCL3L1 influences acquisition of multiple blood-borne infections (HCV, HIV-1, HBV) that occurs commonly among intravenous drug users (IDUs) is unknown.
METHODS
We determined CCL3L1 copy number by real-time PCR among 374 Caucasian IDUs from Estonia of whom 285 were HCV-positive, 208 HIV+, 177 HCV+/HIV+, and 57 HCV−/HIV−.
RESULTS
In univariate and multivariate analyses, HCV and HBV seropositivity, and duration of IDU each strongly predicted HIV seropositivity. A high CCL3L1 copy number (>2) associated with a 80% reduced risk of acquiring HIV, after adjusting for age, gender, HCV/HBV status, CCR5-Δ32 polymorphism and IDU duration (OR=0.20; 95% CI=0.09–0.45). By contrast, CCL3L1 gene dose did not influence HCV seropositivity. Among HCV+ IDUs, there was a 3.5-fold over- and 65% under-representation of a high CCL3L1 copy number among HCV+/HIV− and HCV+/HIV+ subjects, respectively.
CONCLUSION
Among IDUs exposed heavily to HCV/HIV, CCL3L1 copy number is a major determinant of HIV seropositivity, but not HCV seropositivity. The contrasting distribution of a protective high CCL3L1 copy number among HCV+/HIV− vs HCV+/HIV+ IDUs may reflect that HIV preferentially selects for subjects with a low CCL3L1 gene dose.
Keywords: chemokine copy number, HIV, HCV, IDU
INTRODUCTION
Complex interactions between three parameters -- environment, microbial agent, and host genotype --are thought to be a key determinant of the outcome to infectious challenge [1]. Conceptually, environmental factors may be parameters that are intrinsic to the host such as co-infections. Abundant data supports the relevance of investigating the factors that influence HIV pathogenesis in the context of microbial co-pathogens [2–8], especially HCV and HBV, which are frequent co-infections in HIV-infected subjects [9]. However, few studies have been undertaken to define the impact of these complex interactions on HIV-1 susceptibility.
With respect to host genotypes that may influence HIV susceptibility, significant attention has been focused on variations in CCR5, the primary co-receptor for the cell entry of HIV [10–12]. CC ligand 3 like-1 (CCL3L1), CCL3, CCL4, CCL4L1 and CCL5 are the main ligands of CCR5 and they inhibit entry of HIV into CD4+ T-cells in vitro [10, 11, 13–16]. In addition, CCR5 ligands may affect HIV pathogenesis by influencing several facets of the immune response, including cell-mediated immunity [17–22]. Notably, among the CCR5 chemokine ligands, CCL3L1 is the most potent and has maximal HIV-suppressive properties in vitro [14–16]. The region on chromosome 17q that encodes several CCR5 ligands is a hot-spot for segmental duplications such that individuals vary with respect to the number of CCL3L1-containing segmental duplications [16, 23–27]. Hence individuals differ with regards to the copy number of genes that are found in this segmental duplication, including chemokine genes such as CCL3L1 and CCL4L1 [16, 23–27]. Previous studies found that persons of European origin possess between one to six CCL3L1 gene-containing segmental duplications (henceforth designated as CCL3L1 copies) with an average of two copies [24–26, 28]. By contrast, those of African descent possess higher CCL3L1 copy numbers than those of European descent [24, 26, 29–31].
A low CCL3L1 copy number is associated with reduced chemokine levels and a higher proportion of CCR5 expressing CD4+ cells [24, 25]. Genetic association studies found that a low CCL3L1 copy number was associated with an increased risk of acquiring HIV infection in European-, African-, Hispanic-American adults infected primarily through the mucosal route [24]; hemophiliacs from Japan [32]; as well as Argentinean [24], South African [29, 30] and Ukrainian [28] children exposed perinatally to HIV.
Previous studies have found that a genetic factor may confer differential effects, depending on the route of infection. For example, Martin et al showed that a DC-SIGN promoter polymorphism was associated with increased risk for parenteral, but not mucosal, acquisition of HIV infection [33]. Although the results of the studies in hemophiliacs are suggestive [32], most of the prior studies which defined the association of a high CCL3L1 copy number with decreased HIV susceptibility were primarily in subjects infected via the mucosal or perinatal route [24, 28–30, 32]. To determine whether there was a relationship between CCL3L1 copy number and risk of acquiring infection following parenteral exposure of HIV, we examined intravenous drug users (IDUs).
We investigated IDU’s of European descent from Estonia, a geographic region in which the HIV epidemic is relatively new [34]. The outbreaks in Estonia were first detected among IDUs in 2000 and HIV infection is still concentrated among this population group [34]. Furthermore, infection with HIV among these IDUs is accompanied by a very high frequency of viral co-infections with HCV and HBV [35]. This provided the unique opportunity to examine whether there was an independent association of CCL3L1 copy number with these two viral infections. In addition, to the copy number of CCL3L1, we determined the association of the 32-bp deletion mutation in CCR5 (CCR5-Δ32) with HIV susceptibility because previous studies have demonstrated that homozygyosity for this polymorphism is associated with resistance to HIV infection [10–12]; the associations of CCR5-Δ32/Δ32 with HCV susceptibility vary between cohorts [36–39]. The results of our study confirm and extend significantly the notion that CCL3L1 plays a critical role in HIV susceptibility and also underscore the importance of accounting for complex interactions among HCV, HIV and CCL3L1 genotype when analyzing genotype-phenotype (HIV susceptibility) relationships.
METHODS
Subjects and sample collection
The study subjects were recruited in 2006 and 2007 from the syringe-exchange programs and three Estonian prisons; 374 ancestrally Caucasian IDUs (301 male; 55 female and gender unknown in 18; median age of 26 years; 95% antiretroviral therapy naïve) were enrolled. The data on the length of IV drug use (IVDU) was available for 249 subjects. All study subjects reported use of IV drugs for a minimum of one year, with most having longer periods of IVDU. From each subject two samples of 8 ml of venous blood were collected and transported immediately to the laboratory. The first blood sample was used for detection of HIV-1, HCV, and HBV serostatus and the second sample was used later for genotyping analyses.
HIV, HCV and HBV serostatus
HIV testing was performed in the Estonian Central HIV Reference Laboratory using a fourth generation enzyme-linked immunoassay (Vironistica HIV Uniform II Ag/Ab, BioMerieux, Marcy Etoile, France); all positive results were confirmed by a immunoblotting assay (INNO LIA HIV I/II Score Westernblot (Microgen Bioproducts Ltd, Surrey, UK). The presence of HCV antibodies was tested with the ETI-AB-HCVK-3 anti-HCV test (DiaSorin, Vercelli, Italy). HBV seropositivity was assessed by ETI-MAK-4 HBsAg (DiaSorin, Saluggia, Italy) and ETI-AB-COREK Plus (anti-HBc core) (DiaSorin, Saluggia, Italy).
Genotyping of CCR5 and CCL3L1 variations
Genomic DNA was extracted from peripheral blood mononuclear cells (courtesy of Professor Ismo Virtanen, University of Helsinki) using the Qiagen QIAamp DNA minikit (Qiagen, Hilden, Germany). CCR5-Δ32 mutation was assessed using PCR-RFLP as described previously [40]. CCL3L1 copy number was determined using quantitative real-time PCR with primers/probes and experimental conditions exactly as described previously [24].
Statistical analyses
Differences in gene copy numbers and other covariates were assessed by the Chi-square tests. Univariate and multivariate logistic regression models were used to determine the association of CCR5 or CCL3L1 genotype and other covariates with HIV or HCV status. Interactive multivariate logistic regression analyses were also conducted to assess the interactions between duration of IVDU and CCL3L1 copy number. STATA 6.0 (Stata Corporation, College Station, Texas, USA) was used for the statistical analyses. The study was approved by the Ethics Committee of Tallinn; all participants signed the informed consent.
RESULTS
HCV/HIV/HBV serostatus
Among the study subjects, 76%, 56% and 15% were infected with HCV, HIV and HBV, respectively (Table 1). Approximately one-third (133/374) were HIV+/HCV+, 1% (4/374) were HIV+/HBV+, 12% (44/374) were HIV+/HCV+/HBV+, and 7% (27/374) were HIV+ but HCV−/HBV− and 27% (100/374) were HCV+ but HIV−/HBV−. Of the remaining 66 subjects, 53 were HIV−/HCV−/HBV− or had other combinations of viral infection.
Table 1.
HIV, HCV and HBV serostatus in study subjects
One person had no HCV data collected
Four persons had no HBV data collected
44 subjects had HIV/HCV/HBV triple-infection
CCR5-Δ32 genotype and HIV infection
CCR5-Δ32 homozygotes were not detected among HIV+ subjects whereas four of 162 HIV− subjects had the CCR5-Δ32/Δ32 genotype. The prevalence of CCR5-Δ32 heterozygotes among HIV+ and HIV− subjects was similar (19% and 20%, respectively). The distribution of CCR5-Δ32 homozygosity or heterozygosity between HIV−/HCV+ vs HIV−/HCV− subjects was similar (3/105 vs 1/56 for Δ32/Δ32 and 24/108 vs 9/57 for Δ32 heterozygosity).
Distribution of CCL3L1 copy numbers
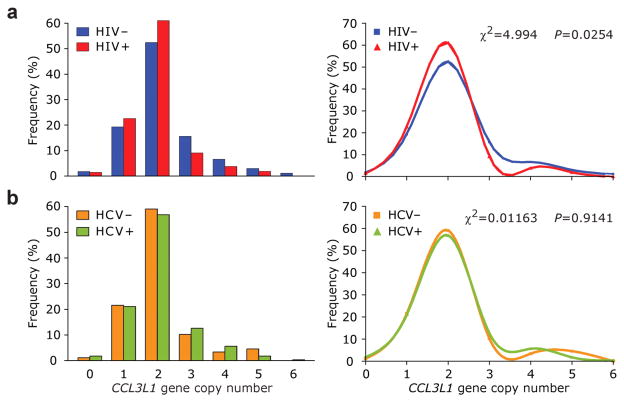
The median CCL3L1 copy number in the entire study population was two (range from 0 to 6). The distribution of the CCL3L1 copy numbers between HIV+ and HIV− subjects was significantly different (χ2 = 4.99; p < 0.05) such that HIV− subjects possessed higher gene copy numbers than HIV+ subjects (Figure 1a). By contrast, the distribution of CCL3L1 copy numbers in HCV+ vs HCV− subjects was similar (Figure 1b).
Figure 1. Distribution of CCL3L1 copy number in study subjects according to HIV or HCV serostatus.
Histograms and the cubic-spline smoothed frequency curves for the distribution of CCL3L1 copy number among HIV+ (a) and HCV+ (b) subjects.
Association of CCL3L1 copy number and HIV and HCV serostatus
We observed a clear shift in the frequency distributions of CCL3L1 copy number between 2 and 3 copies in HIV+ vs HIV− subjects (Figure 1a). Compared to those with ≤2 CCL3L1 copies, those with three copies had a nearly 50% lower risk of HIV seropositivity (odds ratio (OR) = 0.50; 95% confidence intervals (CI) = 0.26–0.96; P = 0.037). Similarly protective associations were observed for those with 4 (OR = 0.50; 95% CI = 0.19–1.29; P = 0.15) or 5–6 (OR = 0.39; 95% CI = 0.11–1.38; P = 0.14) copies, but these associations did not achieve statistical significance at p < 0.05.
Based on these results, and the distribution pattern of CCL3L1 copy number in HIV+ vs HIV− IDUs (Figure 1a), subjects were categorized into two groups - those with a CCL3L1 copy number between 0 to 2 and those with >2 (high CCL3L1 copy number). In univariate analyses, predictably, co-infection with HCV (OR = 3.01; 95% C I= 1.83–4.96) and HBV (OR = 5.10; 95% CI = 2.42–10.75), and longer duration of IVDU (OR = 1.08, 95% CI = 1.01–1.15) were each associated with a significantly increased likelihood of HIV seropositivity (Table 2). Whereas a copy number of CCL3L1 that was greater than two was associated with a 50% decreased likelihood of HIV seropositivity (OR = 0.49, 95% CI = 0.29–0.81; Table 2). By contrast, gender, age and CCR5-Δ32 genotype were not associated with HIV serostatus (Table 2). When examining the outcome of HCV serostatus, we found that co-infection with HIV and HBV, and longer duration of IVDU (OR = 1.24, 95% CI = 1.13–1.36) were also each associated with increased odds of HCV seropositivity, but >2 CCL3L1 copies was not associated with HCV (Table 2) or HBV serostatus (OR = 0.73; 95% CI, 0.34–1.56).
Table 2.
Associations by univariate logistic regression analyses of demographic factors, IV drug use duration, viral coinfection status, and CCL3L1 or CCR5 genotype with HIV or HCV serostatus
| Outcome: HIV serostatus | Outcome: HCV | |
|---|---|---|
| Covariate |
OR; 95% CI; P |
OR; 95% CI; P |
| Gender | ||
| Male* | 1.0 | 1.0 |
| Female | 1.08; 0.60–1.94; 0.797 | 0.52; 0.27–0.99; 0.0455 |
| Age | ||
| <26 years* | 1.0 | 1.0 |
| ≥26 years | 1.10, 0.70–1.72; 0.691 | 1.18; 0.69–2.02; 0.5378 |
| HCV status | ||
| HCV−* | 1.0 | |
| HCV+ | 3.01; 1.83–4.96; 1.45×10−5 | |
| HIV status | ||
| HIV−* | 1.0 | |
| HIV+ | 3.01; 1.83–4.96; 1.45×10−5 | |
| HBV status | ||
| HBV−* | 1.0 | 1.0 |
| HBV+ | 5.10; 2.42–10.75; 1.85×10−5 | 3.57; 1.38–9.25; 0.0088 |
| IV drug use duration | ||
| Yearsb | 1.08; 1.01–1.15; 0.025 | 1.24; 1.13–1.36; 1.10×10−5 |
| CCL3L1 copy number | ||
| 0–2* | 1.0 | 1.0 |
| 3–6 | 0.49; 0.29–0.81; 0.006 | 1.15; 0.62–2.12; 0.6558 |
| CCR5 genotype | ||
| wt/wt* | 1.0 | 1.0 |
| wt/Δ32 or | 0.81; 0.49–1.33; 0.398 | 1.47; 0.78–2.78; 0.2363 |
NOTE., reference group; OR, odds ratio; CI, confidence interval; P, significance value.
Years was categorized as a continuous variable in full-years, and data reflects an increase in OR with each additional year of IV drug use.
To assess the independent influence of the covariates analyzed in Table 2 on HIV and HCV seropositivity we considered the following. Because infection with HCV, HBV and HIV are all highly dependent on length of IVDU, and because these co-infections frequently occur concurrently (Table 1), we considered the possibility that the simultaneous inclusion of all these highly correlated variables in a single multivariate logistic regression model may confound the analyses. Hence, we conducted stepwise multivariate regression analyses in which we included these covariates sequentially (Tables 3 and 4).
Table 3.
Associations by multivariate logistic regression analyses of gender, IV drug use duration, viral coinfection status, and CCL3L1 copy number with HIV serostatus.
| Covariate | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 |
|---|---|---|---|---|---|
| OR; 95% CI | OR; 95% CI | OR; 95% CI | OR; 95% CI | OR; 95% CI | |
| P |
P |
P |
P |
P |
|
| Gender | |||||
| Male* | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Female | 1.29; 0.687–2.430 | 1.3; 0.519–3.255 | 1.297; 0.497–3.389 | 1.343; 0.706–2.554 | 1.031; 0.449–2.369 |
| 0.425 | 0.5751 | 0.5949 | 0.3690 | 0.9424 | |
| HCV status | |||||
| HCV−* | 1.0 | 1.0 | 1.0 | 1.0 | |
| HCV+ | 2.08; 1.211–3.572 | ** | ** | 2.138; 1.234–3.705 | |
| 0.008 | 0.9459 | 0.9441 | 0.0067 | ||
| HBV status | |||||
| HBV−* | 1.0 | 1.0 | 1.0 | 1.0 | |
| HBV+ | 4.13; 1.943–8.777 | 0.922; 0.221–3.851 | 0.657; 0.156–2.772 | 4.006; 1.875–8.56 | |
| 0.0002 | 0.9115 | 0.5674 | 0.0003 | ||
| IV drug use duration | |||||
| Yearsa | 1.016; 0.944–1.093 | 1.014; 0.938–1.095 | 1.08; 1.009–1.157 | ||
| 0.6665 | 0.7315 | 0.0276 | |||
| CCL3L1 copy number | |||||
| 0–2* | 1.0 | 1.0 | 1.0 | ||
| 3–6 | 0.204; 0.093–0.445 | 0.421; 0.244–0.727 | 0.267; 0.124–0.574 | ||
| <0.0001 | 0.0019 | 0.0007 | |||
NOTE. Models #1-5 depict multivariate logistic regression analyses with the indicated covariates for the outcome of HIV serostatus. For example, model 1 contains gender and HCV/HBV status whereas model 4 also includes CCL3L1 copy number. Although gender did not associate with HIV serostatus in univariate analyses (Table 2), it was included in the model to remain consistent with the models shown in Table 4.
reference group.
values reflected those of correlated variables (OR >999.99; 95% CI: <0.001–>999.99). OR, odds ratio; CI, confidence interval; P, significance value.
Years was categorized as a continuous variable in full-years, and data reflects an increase in OR with each additional year of IV drug use.
Table 4.
Associations by multivariate logistic regression analyses of gender, IV drug use duration, viral co-infection status, and CCL3L1 copy number with HCV serostatus
| Covariate | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 |
|---|---|---|---|---|---|
| OR; 95% CI | OR; 95% CI | OR; 95% CI | OR; 95% CI | OR; 95% CI | |
| P |
P |
P |
P |
P |
|
| Gender | |||||
| Male* | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Female | 0.493; 0.252–0.966 | 0.412; 0.122–1.39 | 0.288; 0.075–1.106 | 0.488; 0.249–0.958 | 0.512; 0.18–1.458 |
| 0.0394 | 0.1528 | 0.0698 | 0.0370 | 0.2100 | |
| HIV status | |||||
| HIV−* | 1.0 | 1.0 | 1.0 | 1.0 | |
| HIV+ | 2.088; 1.217–3.583 | ** | ** | 2.159; 1.246–3.738 | |
| 0.0075 | 0.9333 | 0.9282 | 0.0060 | ||
| HBV status | |||||
| HBV−* | 1.0 | 1.0 | 1.0 | 1.0 | |
| HBV+ | 2.535; 0.950–6.760 | 1.549; 0.159–15.113 | 2.549; 0.255–25.474 | 2.551; 0.956–6.808 | |
| 0.0631 | 0.7066 | 0.4257 | 0.0615 | ||
| IV drug use duration | |||||
| Yearsa | 1.272; 1.125–1.439 | 1.331; 1.158–1.531 | 1.351; 1.191–1.534 | ||
| 0.0001 | <0.0001 | <0.0001 | |||
| CCL3L1 copy number | |||||
| 0–2* | 1.0 | 1.0 | 1.0 | ||
| 3–6 | 7.64; 2.052–28.447 | 1.248; 0.641–2.429 | 2.995; 0.91–9.862 | ||
| 0.0024 | 0.5148 | 0.0712 | |||
NOTE. Models #1-5 depict multivariate logistic regression analyses with the indicated covariates with outcome of HCV serostatus.
reference group.
values reflected those of correlated variables (OR >999.99; 95% CI: <0.001–>999.99). OR, odds ratio; CI, confidence interval; P, significance value.
Years was categorized as a continuous variable in full-years, and data reflects an increase in OR with each additional year of IV drug use.
For the outcome of HIV seropositivity, we found that in models that did not include the duration of IV drug use, HCV and HBV infection status associated with 2- and 4-fold higher risk of HIV seropositivity, respectively (Table 3, models 1 and 4). Conversely, in the model that did not include HCV or HBV infection status, length of IVDU was associated with an increased risk of HIV seropositivity (Table 3, model 5). By contrast, and consistent with our prediction, inclusion of IVDU in the same model with HCV and HBV led to negative associations for each of these highly correlated parameters for the outcome of HIV seropositivity (Table 3, models 2 and 3). However, >2 CCL3L1 copies associated with a lower risk of HIV seropositivity, independent of length of IVDU and HCV/HBV co-infection status (Table 3, models 3–5).
Table 4 shows that HIV seropositivity is a strong predictor of HCV seropositivity but only in those models which do not include the length of IVDU (models 1 and 4); a similar trend is observed for co-infection with HBV infection. However, these positive associations of HIV and HBV infection status with HCV seropositivity are not observed after inclusion of length of IVDU into the model (compare models 1 and 4 vs models 2 and 3). In a model that accounts for the effects of gender, HIV and HBV serostatus, the CCL3L1 copy number was not associated with HCV seropositivity (Table 4, model 4). However, when length of IVDU was included in the model with or without HIV/HBV serostatus, a high CCL3L1 copy number was associated with an increased risk of HCV seropositivity (Table 4, models 3 and 5).
Selection of high copy number in HCV+/HIV− IDUs
Table 4 shows that the associations of CCL3L1 copy number with HCV seropositivity differs before and after accounting for the length of IVDU. The latter observations raised a conundrum with respect to whether CCL3L1 copy number is associated with HCV serostatus. We surmised that a high CCL3L1 copy number was not associated with an increased risk of HCV infection because when length of IVDU was excluded from the statistical models, a high CCL3L1 copy number did not associate with HCV serostatus (Table 4, model 4). Instead, the data in Table 3 and 4 may reflect three concurrent events that occur as the duration of exposure to both HIV and HCV increases. First, predictably, increasing durations of IVDU increases risk of HCV (Table 4, models 2, 3 and 5) and HIV (Table 3, model 5) infection. Second, however, those who resist acquiring HIV, are more likely to be those who also happen to have a high CCL3L1 copy number, and this might explain the increased odds ratios for a high CCL3L1 copy number when both it and duration of IVDU are placed concurrently in the same model (Table 4, models 3 and 5). Third, those subjects who acquire HCV and are also susceptible to HIV are less likely to possess a high CCL3L1 copy number (Table 3, models 2–5). In this scenario, there should be an overrepresentation of subjects with a high CCL3L1 copy number among HCV+ IDUs who resist acquiring HIV infection, and conversely an underrepresentation of high CCL3L1 copy numbers in those who are both HCV+ and HIV+.
To test this premise, we conducted two analyses. First, to determine whether there is an overrepresentation and underrepresentation of high CCL3L1 copy number among HCV+/HIV− vs HCV+/HIV+ subjects, respectively, we stratified HCV+ subjects according to their HIV status, and conducted the logistic regression analyses. The results showed (Table 5) that, consistent with our hypothesis, there was a 3.5-fold overrepresentation of a high CCL3L1 copy number among HCV+/HIV− subjects (OR = 3.57; 95% CI = 1.47–8.67), whereas there was a 65% lower representation of a high CCL3L1 copy number among HCV+/HIV+ subjects (OR = 0.35; 95% CI = 0.14–0.85).
Table 5.
Likelihood of high CCL3L1 copy number among HCV+ subjects according to HIV infection status.
| n |
OR |
95% CI |
P |
|
|---|---|---|---|---|
| HIV− | 166 | |||
| CCL3L1 0–2 copies | 122 | 1.0 | ||
| CCL3L1 3–6 copies | 44 | 3.57 | 1.47–8.67 | 0.005 |
| HIV+ | 208 | |||
| CCL3L1 0–2 copies | 177 | 1.0 | ||
| CCL3L1 3–6 copies | 31 | 0.35 | 0.14–0.85 | 0.020 |
NOTE. OR, odds ratio; CI, confidence interval; P, significance value
Second, to affirm further that CCL3L1 copy number does not influence risk of HCV infection, despite increased durations of IVDU, we conducted the analyses shown in Table 6. The median duration of IVDU was 8 years among the study subjects. Relative to those with IVDU of ≤8 years, those with a history of >8 years had a 3-fold greater risk of HCV seropositivity (Table 6, model 1). We next determined the association of CCL3L1 with HCV serostatus, but for the reasons discussed in the preceding paragraphs, in this model we also accounted for the possible interaction between duration of IVDU and CCL3L1 copy number (Table 6, model 2). These data indicated that length of IVDU was a strong predictor of HCV seropositivity, but CCL3L1 copy number was not (Table 6, model 2).
Table 6.
Association of IV drug use length and CCL3L1 copy number with HCV infection
| OR | 95% CI | P | |
|---|---|---|---|
| Model 1: IVDU duration with outcome of HCV | |||
| IVDU ≤ 8 years* | 1 | ||
| IVDU >8 year | 3.321 | 1.654–6.666 | 0.0007 |
| Model 2: IVDU x CCL3L1 for outcome of HCV | |||
| IVDU* | 3.010 | 1.46–6.20 | 0.0028 |
| CCL3L1** | 2.174 | 0.68–6.91 | 0.1879 |
| IDUxCCL3L1 | § | § | 0.9717 |
NOTE. Model 1 is a univariate analysis of the association of duration of IV drug use (IVDU) with HCV status. Model 2 is an interactive multivariate logistic regression model with the indicated covariates.
IVDU was categorized according to median duration of IV drug use.
CCL3L1 copy number is categorized as 0–2 vs >2 copies. OR, odds ratio; CI, confidence interval; P, significance value. X, interactio
DISCUSSION
In this study, by examining an IDU population of Caucasian ancestry, we demonstrate that a high CCL3L1 copy number is associated with strong resistance to acquiring HIV infection. Notably, this association of CCL3L1 copy number with HIV serostatus is independent of the length of IVDU and co-infection with HCV and HBV. The specificity of this association is highlighted by the observation that by contrast to its impact on HIV susceptibility, CCL3L1 copy number does not influence risk of acquiring HCV infection.
The study was conducted in Estonia, a geographic region in which HIV is thought to be recent, with HIV incidence peaking in 2001 [34, 35, 41]. Thus, infection with HCV and HBV antedated HIV infection among Estonian IDU’s [35, 42]. This is reflected by both the higher prevalence of HCV (76%) than HIV (56%) infection in our study subjects and that the duration of IVDU was a stronger predictor of HCV than HIV seropositivity, with each additional year of IVDU increasing the risk of HCV and HIV by 27–35% and 8%, respectively. Predictably then, concordant with studies conducted in other countries [9, 41, 43], we found that among Estonian IDUs, HCV status was a strong predictor of HIV seropositivity, and conversely, HIV seropositivity associated strongly with HCV serostatus. Hence, given the very heavy exposure to multiple viral infections, and the relatively recent nature of the HIV epidemic in Estonia, this IDU study group was ideal to examine the host genetic factors that confer resistance to infection of HIV and/or HCV infection.
Another strength of this study is the relative homogeneity of both the viral and host population [34]. The HIV epidemic in Estonia commenced in 2000 and was characterized by concentrated one-source infections introduced into the IDU population during the early years of the epidemic [34]. Recent epidemiological studies have confirmed the low sequence heterogeneity of this mainly monophyletic population of recombinant circulating form HIV-1 viruses CRF06_cpx [44–46]. This provides an advantage as it allows the study of the influence of host genetic factors on HIV susceptibility in the context of minimal viral genetic heterogeneity. Another strength of this study was its homogeneous study population consisting mainly of young male IDUs of Caucasian descent. In contrast to some previous studies where distribution of CCL3L1 copy numbers in HIV+ adults was compared with the distribution in a HIV-negative reference population, in this study all subjects had documented exposure risk factors (e.g. IVDU) [24].
Our results not only affirm the previously reported strong association of a high CCL3L1 copy number with a reduced likelihood of HIV seropositivity, but also extend them significantly in three notable ways. First, our study provides the first evidence of this association among IDUs with heavy parenteral exposure to both HCV and HIV, a risk-group that is highly understudied with respect to the host genetic factors that influence susceptibility to viral infections. Most of the prior studies that have examined the association of CCL3L1 with HIV susceptibility have focused on either men with risk of mucosal transmission [24] or children exposed perinatally to HIV [24, 28–30]. Our results are consistent with those of Nakajima et al who found that a high CCL3L1 copy number was associated with a lower risk of acquiring HIV infection among Japanese hemophiliacs [32].
Second, the strength and magnitude of the association was high, and depending on the statistical model, a high CCL3L1 copy number afforded a nearly 60–80% lower odds of HIV seropositivity. One possible mechanism for this strong association is the extensive in vitro data showing that among the chemokines that bind to CCR5, CCL3L1 displays the most prominent HIV suppressive activity in vitro [14–16]. However, other mechanisms are possible. Chemokines influence immune responses [18–22, 47], and Dolan et al showed that subjects who lacked or had a low CCL3L1 copy number had reduced cell-mediated immune responses, as estimated by delayed type hypersensitivity skin test reactions to the neoantigen KLH [17]. Thus, it also possible that a low CCL3L1 copy may associate with increased HIV susceptibility because of both low chemokine production and impaired cell mediated immune responses.
Third, we provide experimental evidence for the previous suggestion [24] that among population groups that are under continuous and high exposure to HIV infection, such as among IDUs, there will be a shift in the distribution of CCL3L1 copy number over time, such that subjects who resist HIV infection will be enriched for a high CCL3L1 copy number. Given the heavy exposure to both HCV and HIV among IDUs, HCV+/HIV− subjects reflect subjects traditionally categorized as highly exposed, HIV-uninfected. When we examined HCV+ subjects according to their HIV infection status, we found that there was a 3-fold overrepresentation of subjects with a high CCL3L1 copy number among exposed HIV-uninfected HCV+ subjects and conversely, an underrepresentation of IDUs with a high CCL3L1 copy number among HCV+/HIV+ subjects. That is, the increased frequency of a higher CCL3L1 gene dose among HCV+/HIV− subjects resulted from resistance to infection with HIV among subjects who also happen to be at high risk for acquiring HCV infection. Thus, the overrepresentation of CCL3L1 among HCV+/HIV− subjects does not reflect increased susceptibility to HCV. This scenario is similar to studies of Zhang et al, who suggested that the increased frequency of the protective CCR5-Δ32/Δ32 genotype among HCV+/HIV− hemophiliacs resulted from resistance to infection with HIV, and not increased susceptibility to HCV [39]. In this respect it is noteworthy that consistent with prior reports [12, 39], subjects with the CCR5-Δ32/Δ32 genotype were not found among HIV+ subjects and that among the four HIV-negative CCR5-Δ32/Δ32-bearing subjects, three were HCV+/HIV−.
In conclusion, these data suggest that among individuals at high risk for infection with both HCV and HIV, those with a low CCL3L1 copy number are preferentially infected with HIV, and conversely, those with a high CCL3L1 copy number resist infection with HIV. However, a high CCL3L1 copy number does not afford protection against acquiring infection with HCV. In turn, this differential impact of CCL3L1 copy number on risk of HIV and HCV seropositivity shifts the distribution of CCL3L1 copy number wherein HCV+/HIV− subjects have a higher CCL3L1 copy number whereas HCV+/HIV+ subjects are underrepresented for subjects with a high CCL3L1 copy number. Together, these findings suggest a possible selection by HIV for subjects with a low CCL3L1 copy number, underscore the pivotal importance of CCL3L1, and by extension, its cognate receptor CCR5 in HIV susceptibility. Additionally, our results highlight the importance of investigating the genotype-phenotype relationships for HIV serostatus in the context of microbial co-pathogens.
Acknowledgments
The study was supported by European Union through the European Regional Development Fund and by the Archimedes Foundation and Norwegian Financial Mechanism/EEA(grant EE0016). The authors are grateful to Inge Ringmets and Heti Pisarev for statistical advice and the participants and teams from the Tartu Prison, from NGOs “Convictus” and “Me aitame sind”.
Financial support: Basic Financing and the Target Financing of Estonian Ministry of Education and Research (SF0182726s06); European Commission funded project Expanding Network for Comprehensive and Coordinated Action on HIV/AIDS prevention among IDUs and Bridging Population Nr 2005305 (ENCAP); Global Fund to Fight HIV, Tuberculosis and Malaria Program “Scaling up the response to HIV in Estonia” for 2003–2007. National HIV/AIDS Strategy for 2006–2015; US Civilian Research Development Foundation grant (ESX0-2722-TA-06); US National Institutes of Health, National Institute on Drug Abuse (grant R01DA03574); European Regional Development Fund (project SFOS, WP1-NeuroAIDS); Estonian Science Foundation grant (8004).
Footnotes
Potential conflicts of interest: The authors do not have a commercial or other association that might pose a conflict of interest.
Presented in part: Conference of Retroviruses and Opportunistic Infections 2008, Massachusetts, Boston, 3–6 February 2008 (poster 296).
References
- 1.Clementi M, Di Gianantonio E. Genetic susceptibility to infectious diseases. Reprod Toxicol. 2006;21:345–9. doi: 10.1016/j.reprotox.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Shankar EM, Solomon SS, Vignesh R, et al. GB virus infection: a silent anti-HIV panacea within? Trans R Soc Trop Med Hyg. 2008;102:1176–80. doi: 10.1016/j.trstmh.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 3.Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Med. 2006;7:173–80. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams CF, Klinzman D, Yamashita TE, et al. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med. 2004;350:981–90. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 5.Stapleton JT, Balfour HH., Jr Coinfection alters the playing field: herpesviruses induce acyclovir to inhibit HIV. Cell Host Microbe. 2008;4:194–5. doi: 10.1016/j.chom.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Pilotti E, Elviri L, Vicenzi E, et al. Postgenomic up-regulation of CCL3L1 expression in HTLV-2-infected persons curtails HIV-1 replication. Blood. 2007;109:1850–6. doi: 10.1182/blood-2006-07-036046. [DOI] [PubMed] [Google Scholar]
- 7.Grivel JC, Ito Y, Faga G, et al. Suppression of CCR5- but not CXCR4-tropic HIV-1 in lymphoid tissue by human herpesvirus 6. Nat Med. 2001;7:1232–5. doi: 10.1038/nm1101-1232. [DOI] [PubMed] [Google Scholar]
- 8.Berzsenyi MD, Bowden DS, Kelly HA, et al. Reduction in hepatitis C-related liver disease associated with GB virus C in human immunodeficiency virus coinfection. Gastroenterology. 2007;133:1821–30. doi: 10.1053/j.gastro.2007.08.076. [DOI] [PubMed] [Google Scholar]
- 9.Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44:S6–9. doi: 10.1016/j.jhep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 11.Lederman MM, Sieg SF. CCR5 and its ligands: a new axis of evil? Nat Immunol. 2007;8:1283–5. doi: 10.1038/ni1207-1283. [DOI] [PubMed] [Google Scholar]
- 12.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191 (Suppl 1):S68–77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–5. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Nibbs RJ, Yang J, Landau NR, Mao JH, Graham GJ. LD78beta, a non-allelic variant of human MIP-1alpha (LD78alpha), has enhanced receptor interactions and potent HIV suppressive activity. J Biol Chem. 1999;274:17478–83. doi: 10.1074/jbc.274.25.17478. [DOI] [PubMed] [Google Scholar]
- 15.Menten P, Struyf S, Schutyser E, et al. The LD78beta isoform of MIP-1alpha is the most potent CCR5 agonist and HIV-1-inhibiting chemokine. J Clin Invest. 1999;104:R1–5. doi: 10.1172/JCI7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 17.Dolan MJ, Kulkarni H, Camargo JF, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 18.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–5. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 19.Karpus WJ, Lukacs NW, Kennedy KJ, Smith WS, Hurst SD, Barrett TA. Differential CC chemokine-induced enhancement of T helper cell cytokine production. J Immunol. 1997;158:4129–36. [PubMed] [Google Scholar]
- 20.Lillard JW, Jr, Singh UP, Boyaka PN, Singh S, Taub DD, McGhee JR. MIP-1alpha and MIP-1beta differentially mediate mucosal and systemic adaptive immunity. Blood. 2003;101:807–14. doi: 10.1182/blood-2002-07-2305. [DOI] [PubMed] [Google Scholar]
- 21.Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–8. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- 22.Pinto LA, Williams MS, Dolan MJ, Henkart PA, Shearer GM. Beta-chemokines inhibit activation-induced death of lymphocytes from HIV-infected individuals. Eur J Immunol. 2000;30:2048–55. doi: 10.1002/1521-4141(200007)30:7<2048::AID-IMMU2048>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 23.Modi WS. CCL3L1 and CCL4L1 chemokine genes are located in a segmental duplication at chromosome 17q12. Genomics. 2004;83:735–8. doi: 10.1016/j.ygeno.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 25.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–26. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 26.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gornalusse G, Mummidi S, He W, Silvestri G, Bamshad M, Ahuja SK. CCL3L Copy number variation and the co-evolution of primate and viral genomes. PLoS Genet. 2009;5:e1000359. doi: 10.1371/journal.pgen.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shostakovich-Koretskaya L, Catano G, Chykarenko ZA, et al. Combinatorial content of CCL3L and CCL4L gene copy numbers influence HIV-AIDS susceptibility in Ukrainian children. Aids. 2009;23:679–88. doi: 10.1097/QAD.0b013e3283270b3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meddows-Taylor S, Donninger SL, Paximadis M, et al. Reduced ability of newborns to produce CCL3 is associated with increased susceptibility to perinatal human immunodeficiency virus 1 transmission. J Gen Virol. 2006;87:2055–65. doi: 10.1099/vir.0.81709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn L, Schramm DB, Donninger S, et al. African infants’ CCL3 gene copies influence perinatal HIV transmission in the absence of maternal nevirapine. Aids. 2007;21:1753–61. doi: 10.1097/QAD.0b013e3282ba553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalekoff S, Meddows-Taylor S, Schramm DB, et al. Host CCL3L1 gene copy number in relation to HIV-1-specific CD4+ and CD8+ T-cell responses and viral load in South African women. J Acquir Immune Defic Syndr. 2008;48:245–54. doi: 10.1097/QAI.0b013e31816fdc77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima T, Ohtani H, Naruse T, et al. Copy number variations of CCL3L1 and long-term prognosis of HIV-1 infection in asymptomatic HIV-infected Japanese with hemophilia. Immunogenetics. 2007;59:793–8. doi: 10.1007/s00251-007-0252-4. [DOI] [PubMed] [Google Scholar]
- 33.Martin MP, Lederman MM, Hutcheson HB, et al. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol. 2004;78:14053–6. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruutel K, Uuskula A. HIV epidemic in Estonia in the third decade of the AIDS era. Scand J Infect Dis. 2006;38:181–6. doi: 10.1080/00365540500388743. [DOI] [PubMed] [Google Scholar]
- 35.Uuskula A, McNutt LA, Dehovitz J, Fischer K, Heimer R. High prevalence of blood-borne virus infections and high-risk behaviour among injecting drug users in Tallinn, Estonia. Int J STD AIDS. 2007;18:41–6. doi: 10.1258/095646207779949907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woitas RP, Ahlenstiel G, Iwan A, et al. Frequency of the HIV-protective CC chemokine receptor 5-Delta32/Delta32 genotype is increased in hepatitis C. Gastroenterology. 2002;122:1721–8. doi: 10.1053/gast.2002.33660. [DOI] [PubMed] [Google Scholar]
- 37.Mangia A, Santoro R, D’Agruma L, Andrilli A. HCV Chronic Infection and CCR5-Δ32/Δ32. Gastroenterology. 2003;124:868–869. doi: 10.1053/gast.2003.50134. [DOI] [PubMed] [Google Scholar]
- 38.Poljk M, Seme K, Marin I, Babic D, Maticic M, Meglic J. Frequency of the 32-Base Pair Deletion in the Chemokine Receptor CCR5 Gene Is Not Increased in Hepatitis C Patients. Gastroenterology. 2003;124:1558–1559. doi: 10.1016/s0016-5085(03)00349-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Goedert J, O’Brien T. High Frequency of CCR5-Δ32 Homozygosity in HCV-Infected, HIV-1-Uninfected Hemophiliacs Results From Resistance to HIV-1. Gastroenterology. 2003;124:867–868. doi: 10.1053/gast.2003.50132. [DOI] [PubMed] [Google Scholar]
- 40.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 41.Aceijas C, Stimson GV, Hickman M, Rhodes T. Global overview of injecting drug use and HIV infection among injecting drug users. Aids. 2004;18:2295–303. doi: 10.1097/00002030-200411190-00010. [DOI] [PubMed] [Google Scholar]
- 42.Platt L, Bobrova N, Rhodes T, et al. High HIV prevalence among injecting drug users in Estonia: implications for understanding the risk environment. Aids. 2006;20:2120–3. doi: 10.1097/01.aids.0000247586.23696.20. [DOI] [PubMed] [Google Scholar]
- 43.Aceijas C, Rhodes T. Global estimates of prevalence of HCV infection among injecting drug users. Int J Drug Policy. 2007;18:352–8. doi: 10.1016/j.drugpo.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Adojaan M, Kivisild T, Mannik A, et al. Predominance of a rare type of HIV-1 in Estonia. J Acquir Immune Defic Syndr. 2005;39:598–605. [PubMed] [Google Scholar]
- 45.Avi R, Huik K, Sadam M, et al. Absence of genotypic drug resistance and presence of several naturally occurring polymorphisms of human immunodeficiency virus-1 CRF06_cpx in treatment-naive patients in Estonia. J Med Virol. 2009;81:953–8. doi: 10.1002/jmv.21482. [DOI] [PubMed] [Google Scholar]
- 46.Zetterberg V, Ustina V, Liitsola K, et al. Two viral strains and a possible novel recombinant are responsible for the explosive injecting drug use-associated HIV type 1 epidemic in Estonia. AIDS Res Hum Retroviruses. 2004;20:1148–56. doi: 10.1089/aid.2004.20.1148. [DOI] [PubMed] [Google Scholar]
- 47.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]