Abstract
Mapping the vast quantities of short sequence fragments produced by next-generation sequencing platforms is a challenge. What programs are available and how do they work?
A new generation of DNA sequencers that can rapidly and inexpensively sequence billions of bases is transforming genomic science. These new machines are quickly becoming the technology of choice for whole-genome sequencing and for a variety of sequencing-based assays, including gene expression, DNA-protein interaction, human resequencing and RNA splicing studies1–3. For example, the RNA-Seq protocol, in which processed mRNA is converted to cDNA and sequenced, is enabling the identification of previously unknown genes and alternative splice variants; the ChIP-Seq approach, which sequences immunoprecipitated DNA fragments bound to proteins, is revealing networks of interactions between transcription factors and DNA regulatory elements4; and the whole-genome sequencing of tumor cells is uncovering previously unidentified cancer-initiating mutations5.
One of the challenges presented by the new sequencing technology is the so-called ‘read mapping’ problem. Sequencing machines made by Illumina of San Diego, Applied Biosystems (ABI) of Carlsbad, California, and Helicos of Cambridge, Massachusetts, produce short sequences of 25–100 base pairs (bp), called ‘reads’, which are sequence fragments read from a longer DNA molecule present in the sample that is fed into the machine. In contrast to whole-genome assembly, in which these reads are assembled together to reconstruct a previously unknown genome, many of the next-generation sequencing projects begin with a known, or so-called ‘reference’, genome. In this case, to make sense of the reads, their positions within the reference sequence must be determined. This process is known as aligning or ‘mapping’ the read to the reference. In one version of the mapping problem, reads must be aligned without allowing large gaps in the alignment (we describe this in more detail in the “Short-read mappers” section below). A more difficult version of the problem arises primarily in RNA-Seq, in which alignments are allowed to have large gaps corresponding to introns (discussed below in the “Spliced-read mappers” section).
These read mapping problems are certainly not new, and there are many programs that perform both spliced and unspliced alignment for the older Sanger-style capillary reads. Even so, these programs neither scale up to the much greater volumes of data produced by short-read sequencers nor scale down to the short read lengths. Aligning the reads from ChIP-Seq or RNA-Seq experiments can take hundreds or thousands of central processing unit (CPU) hours using conventional software tools such as BLAST or BLAT. Fortunately, new software packages designed to meet the computational challenges of short-read sequencing are quickly appearing. Before choosing one, it is essential to understand why the mapping problems are computationally difficult, which difficulties have been overcome and what challenges and opportunities remain.
Challenges of mapping short reads
The first challenge is a practical one: if the reference genome is very large, and if we have billions of reads, how quickly can we align the reads to the genome? Sequence alignment is a classic problem in bioinformatics, supported by a large body of literature describing different variants for both exact and inexact alignment. As a practical matter, the task of mapping billions of sequences to a mammalian-sized genome calls for extraordinarily efficient algorithms, in which every bit of memory is used optimally or near optimally.
The second challenge is strategic: if a read comes from a repetitive element in the reference, a program must pick which copy of the repeat the read belongs to. Because this may be impossible to decide with confidence, the program may choose to report multiple possible locations or to pick a location heuristically. Sequencing errors or variations between the sequenced chromosomes and the reference genome exacerbate this problem, because the alignment between the read and its true source in the genome may actually have more differences than the alignment between the read and some other copy of the repeat. The spliced mapping problem faces this same challenge but is further complicated by the possible presence of intron-sized gaps.
DNA sequencers from Illumina, ABI, Roche (of Basel, Switzerland), Helicos and other companies produce millions of reads per run. Complete assays may involve many runs, so an investigator may need to map millions or billions of reads to a genome. For example, the recent cancer genome sequencing project by Ley et al.5 generated nearly 8 billion reads from 132 sequencing runs. A large, expensive computer grid might map the reads from this experiment in a few days using traditional alignment algorithms such as BLAST or BLAT, but such grids are not accessible to everyone. To reduce the computing cost of analysis for sequencing-based assays and to make them available to all investigators, we and others have created a new generation of alignment programs capable of mapping hundreds of millions of short reads on a single desktop computer. Vendors of sequencing machines provide specialized mapping software, such as the ELAND program from Illumina, but in this article we focus on third-party packages, some of which are free and open source. These programs are built on algorithms that exploit features of short DNA sequencing reads to map millions of reads per hour while minimizing both processing time and memory requirements.
Short-read mappers
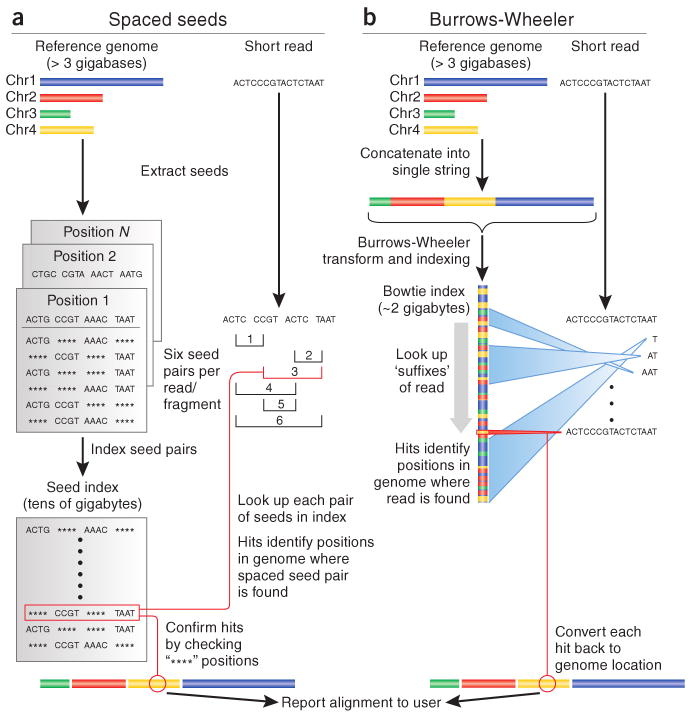
Such programs as Maq and Bowtie (Table 1) use a computational strategy known as ‘indexing’ to speed up their mapping algorithms. Like the index at the end of a book, an index of a large DNA sequence allows one to rapidly find shorter sequences embedded within it. Maq is based on a straightforward but effective strategy called spaced seed indexing6 (Fig. 1a). In this strategy, a read is divided into four segments of equal length, called the ‘seeds’. If the entire read aligns perfectly to the reference genome, then clearly all of the seeds will also align perfectly. If there is one mismatch, however, perhaps due to a single-nucleotide polymorphism (SNP), then it must fall within one of the four seeds, but the other three will still match perfectly. Using similar reasoning, two mismatches will fall in at most two seeds, leaving the other two to match perfectly. Thus, by aligning all possible pairs of seeds (six possible pairs) against the reference, it is possible to winnow the list of candidate locations within the reference where the full read may map, allowing at most two mismatches. Maq's spaced seed index enables it to perform this winnowing operation very efficiently. The resulting set of candidate reads is typically small enough that the rest of the read—that is, the other two seeds that might contain the mismatches—may be individually checked against the reference.
Table 1. A selection of short-read analysis software.
| Program | Website | Open source? | Handles ABI color space? | Maximum read length |
|---|---|---|---|---|
| Bowtie | http://bowtie.cbcb.umd.edu | Yes | No | None |
| BWA | http://maq.sourceforge.net/bwa-man.shtml | Yes | Yes | None |
| Maq | http://maq.sourceforge.net | Yes | Yes | 127 |
| Mosaik | http://bioinformatics.bc.edu/marthlab/Mosaik | No | Yes | None |
| Novoalign | http://www.novocraft.com | No | No | None |
| SOAP2 | http://soap.genomics.org.cn | No | No | 60 |
| ZOOM | http://www.bioinfor.com | No | Yes | 240 |
Figure 1.
Two recent algorithmic approaches for aligning short (20–200-bp) sequencing reads.
(a) Algorithms based on spaced-seed indexing, such as Maq, index the reads as follows: each position in the reference is cut into equal-sized pieces, called ‘seeds’ and these seeds are paired and stored in a lookup table. Each read is also cut up according to this scheme, and pairs of seeds are used as keys to look up matching positions in the reference. Because seed indices can be very large, some algorithms (including Maq) index the reads in batches and treat substrings of the reference as queries. (b) Algorithms based on the Burrows-Wheeler transform, such as Bowtie, store a memory-efficient representation of the reference genome. Reads are aligned character by character from right to left against the transformed string. With each new character, the algorithm updates an interval (indicated by blue ‘beams’) in the transformed string. When all characters in the read have been processed, alignments are represented by any positions within the interval. Burrows-Wheeler–based algorithms can run substantially faster than spaced seed approaches, primarily owing to the memory efficiency of the Burrows-Wheeler search. Chr., chromosome.
Bowtie takes an entirely different approach, borrowing a technique originally developed for compressing large files called the Burrows-Wheeler transform. Using this transform, the index for the entire human genome fits into less than two gigabytes of memory (an amount that is commonly available on today's desktop and even laptop computers)—in contrast to a spaced seed index, which may require over 50 gigabytes—and yet reads can still be aligned efficiently. Bowtie aligns a read one character at a time to the Burrows-Wheeler–transformed genome (Fig. 1b). Each successively aligned new character allows Bowtie to winnow the list of positions to which the read might map. If Bowtie cannot find a location where a read aligns perfectly, the algorithm backtracks to a previous character of the read, makes a substitution and resumes the search. In effect, the Burrows-Wheeler transform enables Bowtie to conquer the mapping problem by first solving a simple subproblem—align one character—and then building on that solution to solve a slightly harder problem—align two characters—and then continuing on to three characters, and so on, until the entire read has been aligned. Bowtie's alignment algorithm is substantially more complicated than Maq's, but Bowtie's alignment speed is more than 30-fold faster7.
Maq and Bowtie both report alignments with up to two mismatches when run in their default modes. In some alignment scenarios, a user may need to allow more mismatches. These two programs were originally designed for reads between 20 and 40 bp long, and both were optimized for human resequencing projects. Even so, Illumina sequencers can now produce reads longer than 100 bp. Additionally, some sequencing projects (such as bacterial or fungal genome sequencing) produce sequences that have many nucleotide-level differences with respect to the closest fully sequenced genome. Finally, the overall quality of reads produced by the new technologies is sensitive to factors such as library preparation, sequencing protocol and even the temperature of the room housing the sequencing machine. Thus, it is essential to know how to change the various default options for any short-read mapper and to be able to identify when those defaults are no longer appropriate.
Several of the new short-read mappers (Table 1) are open source, are simple to install and have good documentation and active user communities. The installation package for Bowtie includes a prebuilt index for Escherichia coli and a set of sample E. coli reads. To run the program on the sample data, just enter the following on the command line:
bowtie e_coli reads/e_coli_1000.fq
This command will produce a tabular report showing each matching read's identifier, the position(s) where it aligns to the reference sequence, and the number and location of mismatches. Maq reports this same information when you run it with the command:
maq.pl easyrun -d outdir
reference.fasta reads.fastq
For a given experiment, the fraction of reads that align to the genome depends on many factors. Assuming the sequenced DNA does not contain many mismatched nucleotides compared to the reference, and assuming the reads have passed rudimentary quality filters, most mapping software will find an alignment for 70–75% of the reads. This might seem surprisingly low, but the sequencing technology is still immature—and it's worth noting that Sanger sequencing had success rates of less than 80% until the late 1990s. Note that many reads will align to multiple positions in the genome. Most read mappers can be directed to report alignments only for reads that map to a unique location in the genome.
After aligning the reads, next one might want to call SNPs or view the alignments against the reference sequence. One package for this task is the SAM tools (http://samtools.sourceforge.net). SAM includes a consensus base caller and viewer that can be used either with Maq or with Bowtie.
Most read mapping software is designed with whole-genome resequencing in mind, but the programs can be configured for other assays. The manuals for Bowtie and Maq are quite detailed, and the array of choices a user can make can be daunting. Moreover, the list of programs capable of short-read mapping is rapidly growing (Table 1), and not every program is ideal or appropriate for every experiment. Fortunately, there are ways to get help. The SeqAnswers message board (http://www.seqanswers.com) is an excellent resource for novice and expert users, frequented by the developers of many short-read mapping programs. One of the most popular SeqAnswers threads contains a catalog of current software for primary analysis and visualization of short-read data.
Spliced-read mappers
The spliced alignment problem, in which cDNA (from processed mRNA) sequences are aligned back to genomic DNA, requires more specialized algorithms. Reads sampled from exon-exon junctions need to be mapped differently from reads that are contained entirely within exons (Fig. 2).
Figure 2.

RNA-Seq assays produce short reads sequenced from processed mRNAs. Aligning these reads to the genome with Bowtie or Maq will produce the alignments shown in black but will fail to align the blue reads. A spliced-read mapper such as TopHat or ERANGE will also report the (blue) alignments spanning intron boundaries.
To align cDNA reads from RNA-Seq1–3 experiments, packages such as ERANGE (http://woldlab.caltech.edu/rnaseq) use the positions of exons and introns within known genes as a guide. This allows ERANGE to construct the sequences spanning exon-exon junctions and use them as reference sequences, and then to invoke a standard read mapper such as Maq or Bowtie to align the spliced reads2. Because this approach will not discover entirely new splice junctions, some studies have used machine learning methods to predict possible junctions by training statistical models using available reference annotations8. In contrast, the TopHat spliced-read mapper (http://tophat.cbcb.umd.edu) does not rely on annotations. Instead, it uses Bowtie (in an initial alignment pass) to identify exons that fully contain some of the reads, and then aligns the remaining reads to junctions between those exons9. Another program, G-Mo.R-Se (http://www.genoscope.cns.fr/externe/gmorse), performs a similar spliced alignment while constructing gene models from RNA-Seq data10.
Limitations and open problems
The current solutions for short-read mapping all have limitations. Mapping programs such as Maq and Bowtie offer very limited support for aligning reads with insertions or deletions (indels). Some read mappers, such as SHRiMP (http://compbio.cs.toronto.edu/shrimp), support ABI's ‘color space’ sequence representation, but most do not. The spliced alignment programs suffer from these same problems and add a few of their own. Annotation-based methods are of course only as good as the annotations, and many organisms have annotations supported only by homology or computational predictions. Machine learning methods will perform poorly if they are trained on incorrect annotations, and they are prone to overtraining.
Many challenges and questions remain for developers of read mapping software. As all the sequencing machine vendors are trying to produce longer reads, will the short-read mapping programs scale well as the reads get longer? Maq, Bowtie and several other short-read packages support reads longer than 100 bp, but at some point, software designed for longer reads, such as BLAT, may be a better fit for downstream analysis. Furthermore, when mapping reads from an organism that has diverged significantly from its reference genome, how should a program's parameters be adjusted, and can that adjustment happen automatically? How useful is mapping quality in downstream analysis, and should it be computed while aligning reads, as Maq does, or later? The answers to each of these questions will depend on the type of assay and the scale of the analysis, and as long as the technology continues to change, the programs will have to change rapidly to keep up.
References
- 1.Nagalakshmi U, et al. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 3.Wang ET, et al. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DS, Mortazavi A, Myers RM, Wold B. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 5.Ley TJ, et al. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Ruan J, Durbin R. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 9.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009 March 16; doi: 10.1093/bioinformatics/btp120. pubished online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denoeud F, et al. Genome Biol. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]