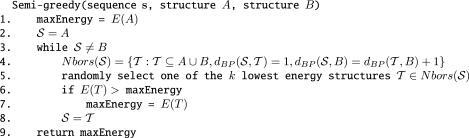Abstract
Given an RNA sequence and two designated secondary structures A, B, we describe a new algorithm that computes a nearly optimal folding pathway from A to B. The algorithm, RNAtabupath, employs a tabu semi-greedy heuristic, known to be an effective search strategy in combinatorial optimization. Folding pathways, sometimes called routes or trajectories, are computed by RNAtabupath in a fraction of the time required by the barriers program of Vienna RNA Package. We benchmark RNAtabupath with other algorithms to compute low energy folding pathways between experimentally known structures of several conformational switches. The RNApathfinder web server, source code for algorithms to compute and analyze pathways and supplementary data are available at http://bioinformatics.bc.edu/clotelab/RNApathfinder.
INTRODUCTION
In this article, we describe a new computational tool to determine nearly optimal folding pathways between two given secondary structures of an RNA sequence. Our tool, RNAtabupath, and related web server, RNApathfinder, have potential applications in synthetic biology; in particular, our work can be used to help engineer bistable conformational switches with reasonable folding kinetics [see Abfalter et al. (1) and Flamm et al. (2) for methods to computationally design bistable switches.). Folding pathways play an important role in various biological processes, including the hok/sok (host killing/suppression of killing] system (3) and transition between two metastable structures, as in the conformational switch in spliced leader (SL) RNA from Leptomonas collosoma (4).
In the hok/sok system, the hok gene of Escherichia coli codes a small (52 amino acids) toxin causing irreversible damage to the cell membrane. While the very stable hok-mRNA is constitutively expressed from a weak promoter, the highly unstable (rapidly degraded) sok-RNA is constitutively expressed from a strong promoter. The hok-mRNA is initially inactive, since a foldback sequesters the Shine–Dalgarno sequence; however, slow exonucleolytic processing digests the last ∼40 nt of the 3′-end of hok-mRNA, thus transforming the molecule into its active form in which the Shine–Dalgarno sequence is no longer sequestered. If R1 plasmids of E. coli are present in sufficient copy number, then a portion of the 64 nt sok-RNA, which is complementary to hok-mRNA leader region, binds to the active conformation of hok-mRNA, thus causing degradation of the complex by RNaseIII (3). If plasmids are not present in sufficient copy number, then the cell is killed by hok toxin. In this fashion, efficient plasmid stabilization is ensured in the population. [See (3) for a review of the hok/sok system.]
In the case of SL RNA from certain trypanosomes and nematodes, a portion of the 5′ exon is donated to another mRNA by trans splicing. Intermediate structures may be important for the process of splicing, as shown by LeCuyer and Crothers (4), who performed stopped-flow rapid-mixing and temperature-jump measurements of the kinetics for the structural transition between two low energy structures of SL RNA from L. collosoma. Conformational switches are thought not only to play a role in such trans splicing but also in transcriptional and translational regulation, protein synthesis and mRNA splicing.
As indicated by the examples of hok/sok and SL RNA, it is biologically important to determine low energy RNA folding pathways. For that reason, this problem has been considered by a number of authors, both in the context of RNA secondary and tertiary structure. Mathews and Case (5) implemented the ‘Nudge Elastic Band’ (NEB) method in amber to sample low energy paths for RNA conformational changes at the three-dimensional atomic scale. They used NEB to study RNA cis Watson–Crick/Hoogsteen GG non-canonical pairs, where one G is syn around the glycosidic bond while the other G is anti. Since prior NMR-constrained modeling had demonstrated that the GG pairs change from (syn)G-(anti)G to (anti)G-(syn)G on the millisecond timescale, such atomic-level simulations using amber were feasible.
Due to large structural transitions between metastable structures in conformational switches, it seems clear that three-dimensional atomic scale simulations using molecular dynamics cannot adequately address the general problem posed in this article. For that reason, it is important to develop efficient algorithms to determine optimal and suboptimal folding pathways between RNA secondary structures. Intermediate structures from such low energy pathways can then be further investigated using atomic scale methods such as NEB.
Morgan and Higgs (6) appear to be among the first to have considered the problem of determining an optimal ‘folding pathway’ between two given secondary structures A, B of a given RNA sequence. If A, B are secondary structures for a given RNA sequence s, then a ‘folding pathway’ from A to B is a sequence A=𝒮0, 𝒮1, … , 𝒮n = B such that each intermediate structure 𝒮i differs from the next structure 𝒮i+1 by exactly one base pair. A folding pathway is ‘direct’ if every intermediate structure 𝒮i+1 is obtained from the preceding structure 𝒮i by either adding a base pair that belongs to B but not A or by removing a base pair that belongs to A but not B. If a pathway is not direct, then it is ‘indirect’. The ‘saddle point’ in a pathway A = 𝒮0, 𝒮1, … , 𝒮n = B is the intermediate structure 𝒮i of highest energy (In case there is more than one intermediate structure having maximum energy along the path, we define the saddle point to be the first such structure, having smallest index.). The ‘barrier energy’ of a pathway from A to B is the energy difference E(𝒮) − E(A), where 𝒮 is the saddle point of the pathway. Clearly, the barrier energy is of fundamental importance in folding kinetics.
Morgan and Higgs describe both a ‘greedy’ algorithm to construct a direct pathway, as described in the ‘Materials and Methods’ section, as well as an algorithm to construct an indirect pathway by gluing together greedy direct pathways between low energy structures sampled from the partition function (In (6), Morgan and Higgs compute the partition function Z = ∑S exp(−E(S)/RT), where the sum is over all secondary structures of a given RNA sequence, and E(S) is the Nussinov energy model (7). Since the partition function is inductively computed, it is simple to the stochastically sampled structures from the low energy Boltzmann ensemble. Later, Ding and Lawrence (8) describe the same stochastic sampling algorithm, Sfold, with the exception that Turner energy model (9) is used in the place of the Nussinov energy model.). While Morgan and Higgs had worked with the Nussinov energy model (7), which ascribes −1 per base pair, with no energetic contribution due to base stacking or loop entropies, our implementation of the direct and indirect pathway algorithms of Morgan and Higgs uses the Turner nearest neighbor energy model (9–14), whose parameters have been obtained by UV absorption (optical melting) experiments. Since the pioneering work of Morgan and Higgs, other groups have developed methods to compute folding pathways between secondary structures. Flamm et al. (2,15) describe an exact algorithm, barriers (barriers is available at http://www.tbi.univie.ac.at/∼ivo/RNA/Barriers/.), that computes optimal (possibly indirect) folding pathways between any two locally optimal secondary structures (A locally optimal secondary structure is one in which the energy is not lowered if a single base pair is either added or removed. Sometimes such structures are called ‘metastable’). While most biologically important examples of pathway computation concern metastable or locally optimal structures, there are nevertheless important exceptions, such as conformational switches (incompletely) determined by experimental methods; the adenine riboswitch from Vibrio vulnificus (rb2) (16) is indeed one such example. barriers relies on the Vienna RNA Package program RNAsubopt (17) that exhaustively generates all secondary structures within a user-specified energy upper bound. For this reason, although barriers is the only exact algorithm, it is generally limited to relatively small RNA sequences or those for which the energy of the saddle point between A and B is not too large. In (18), Flamm et al. describe a breadth-first search algorithm with bounded look-ahead, to compute nearly optimal ‘direct’ pathways. The algorithm is implemented in the program findpath.c, now part of the Vienna RNA Package. Finally, as part of the method paRNAss, Voss et al. (19) describe a straightforward, greedy method to construct ‘direct’ pathways.
Our new algorithm, RNAtabupath, produces (possibly indirect) almost optimal folding pathways by using a heuristic from combinatorial optimization theory known as ‘tabu search’. Tabu search, described in the text by F. W. Glover and M. Laguna (20), is a meta-heuristic to avoid being trapped in local optima in local search algorithms. One of its key components, which we use in this article, is a short memory, often called the ‘tabu list’, that prevents the local search from returning to configurations visited recently. Tabu search then selects the best configuration in the neighborhood which is not in the tabu list. This neighbor may in fact degrade the value of the objective function. Tabu search has been a very effective technique in combinatorial optimization for a wide variety of problems and is an integral part of the repertoire of optimization techniques.
To fix ideas, Figure 1 depicts three folding pathways for a toy 12 nt RNA sequence GGGGGGCCCCCC, with structures A = .((.....)).. having free energy of −1.40 kcal/mol and B = ..(((...))). having free energy of −1.70 kcal/mol. Structure B is not locally optimal, since by adding the base pair (1, 11) to A and by adding base pair (2, 12), one obtains structures (((.....))). and .((((...)))) having free energies −4.70 and −4.20 kcal/mol, respectively. It follows that barriers cannot be applied (In such cases, following the suggestion of an anonymous referee, one could first determine locally optimal structures 𝒮, 𝒯 that, respectively, contain A, B, apply barriers to find an optimal path between 𝒮, 𝒯. This yields a near-optimal path between A, B.). The left, middle and right panels displays the path computed by our implementation of the Morgan–Higgs direct algorithm, Morgan–Higgs indirect path algorithm and RNAtabupath, respectively.
Figure 1.
Three folding pathways for the (toy) RNA sequence S = GGGGGGCCCCCC, between the secondary structure A = .((.....)).. with free energy −1.40 kcal/mol and the structure B = ..(((...))). with −1.70 kcal/mol. The left panel of this figure depicts a (direct) folding pathway from A to B produced by our implementation of the Morgan–Higgs algorithm (6) to produce a (greedy) direct path. The middle panel depicts the indirect folding pathway produced by our implementation of the extension of Morgan–Higgs indirect algorithm to the Turner energy model. Note that the structure .(.........) contains the base pair (2, 12) which is present in neither A nor B. The right panel depicts a folding pathway from A to B produced by our RNAtabupath algorithm. Although RNAtabupath often yields indirect pathways, in this case, the pathway returned by RNAtabupath is direct. Note that the last three structures proposed by RNAtabupath are ...(.....).., ..((.....))., ..(((...)))., respectively, having free energy of 1.90, −1.40 −1.70 kcal/mol. This nucleation and zipping of the stem–loop is energetically more favorable than the alternative (not proposed by RNAtabupath), given by structures ....(...)..., ...((...)).., ..(((...)))., respectively, having free energy of 4.90, 1.60 and −1.70 kcal/mol. Secondary structures are indicated in the familiar (Vienna) dot bracket notation, while free energy in kcal/mol appears to the right of each structure. Free energies are determined by the program RNAeval from the Vienna RNA Package (27).
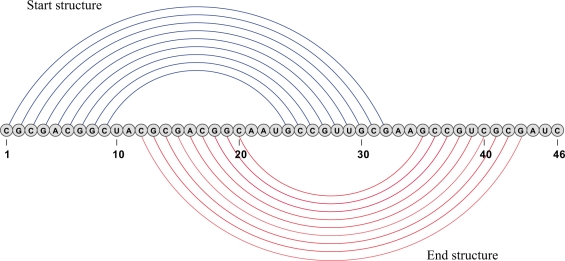
Figures 2–4 depict examples where indirect pathways may have (provably) lower barrier energies than every direct pathway, while Figure 5 displays the two meta-stable structures of host killing (hok) RNA. Type-H pseudoknots, described in the data base ‘PseudoBase’ (21), furnish canonical examples where direct pathways are likely to have greater barrier energies than even naive indirect pathways. Type-H pseudoknots admit a planar representation where certain base pairs are depicted above the horizontal line corresponding to the RNA sequence, while others are depicted below the line—see Figure 2 for illustration. Define structure A [respectively B] to consist of those base pairs above [respectively below] the line. Clearly any direct path from A to B must proceed by removal of all base pairs from A, resulting in the empty structures, followed by addition of all base pairs from B. It follows that E(A) is a lower bound for the barrier energy of every direct path from A to B, where A, B are indicated in Figure 2.
Figure 2.
Consider the 46 nt RNA sequence CGCGACGGCU ACGCGACGGC AAUGCCGUUG CGAAGCCGUC GCGAUC, with secondary structures A = (((((((((..............))))))))).............. having free energy −16.04 kcal/mol and B = ...........(((((((((..............)))))))))... having free energy −18.14 kcal/mol. The structure A consists of the base pairs lying above the line in this figure, while the structure B consists of the base pairs lying below the line. Program barriers cannot be used, since neither A nor B is locally optimal.
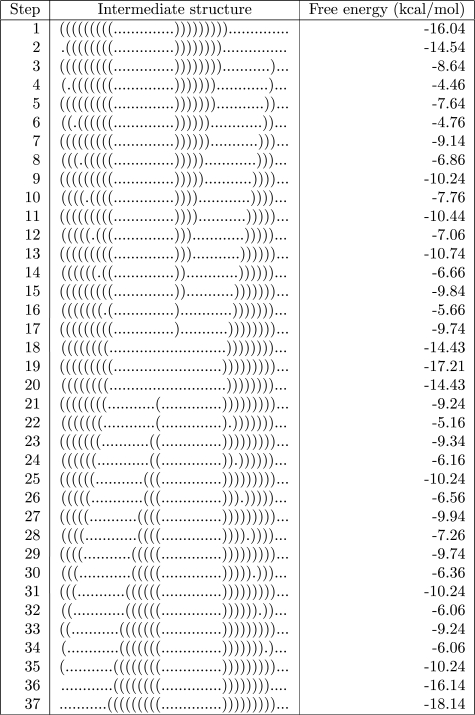
Figure 3.
A manually designed indirect folding pathway for the 46 nt RNA sequence CGCGACGGCU ACGCGACGGC AAUGCCGUUG CGAAGCCGUC GCGAUC, proceeding from locally optimal secondary structure A = (((((((((..............))))))))).............. having free energy −16.04 kcal/mol to locally optimal structure B = ...........(((((((((..............)))))))))... having free energy −18.14 kcal/mol. Intuitively, this pathway can be visualized as repeatedly moving the remaining rightmost right-parenthesis to the right, then repeatedly moving the rightmost left-parenthesis to the right. In this manner, all intermediate structures have negative free energy. The barrier energy of this indirect path is 13.68 kcal/mol, while every direct path must have a barrier energy of at least 16.04 kcal/mol, since the empty structure must be an intermediate structure in every direct path in this example. Indeed, due to nucleation energy required to start a hairpin in the empty structure, the barrier energy of every direct path must properly exceed 16.04. In this case, Vienna Package program findpath.c with look-ahead 100 returns a barrier energy of 18.27, while RNAtabupath returns a barrier energy of 16.84.
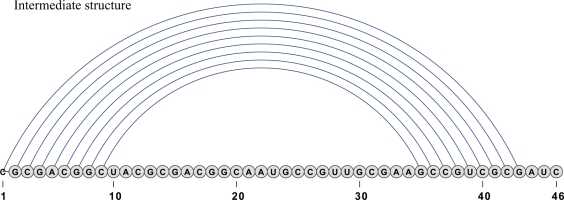
Figure 4.
This figure depicts a folding intermediate in a low energy ‘indirect’ path from A to B (unexplained notation taken from Figure 2). Clearly, every direct path from A to B must have the empty structure as an intermediate structure, hence the lowest barrier energy of a direct folding pathway must be at least 16.04 kcal/mol (in fact even larger due to nucleation energy). However the indirect folding pathway depicted in Table 3 has a barrier energy of 13.68 kcal/mol.
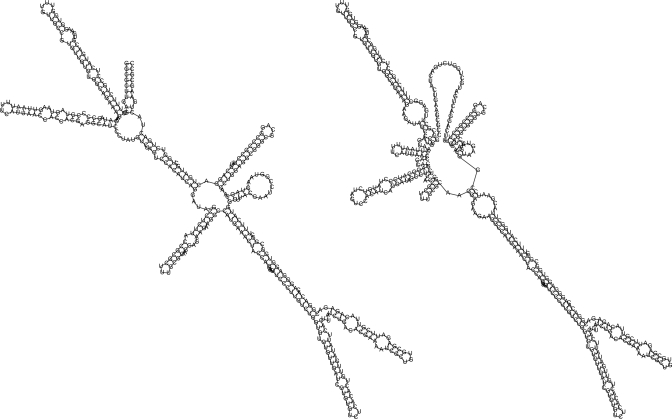
Figure 5.
Two secondary structures of host killing (hok) RNA, taken from Figure 8 of Shapiro et al. (28). The left panel depicts the secondary structure of 396 nt hok-RNA, presumably based on Figure 1B of Franch et al. (29), which latter was obtained by chemical probing experiments. The right panel depicts the secondary structure of the 361 nt truncated hok-RNA, after 3′ processing. Since RNAtabupath requires two structures A, B, of the same length for a given RNA sequence, we have extended the secondary structure of truncated hok-RNA to consist of unpaired nucleotides. Free energy of tructure A is −186.1 kcal/mol, while that of structure B is −142.8 kcal/mol. In this case, findpath (18) obtained the best barrier energy.
Given the combinatorial difficulty of determining optimal pathways for the Turner energy model and the inherent exponential time complexity of the program barriers, it is perhaps not surprising that the problem of computing the minimum energy path between two given RNA structures has recently been announced to be NP-complete. The NP-completeness of computing an optimal pathway is proven in the pre-print, Manuch,J., Thachuk,C., Stacho,L. and Condon,A. (2009) ‘NP-completeness of the direct energy barrier problem without pseudoknots’, in 15th International Meeting on DNA Computing and Molecular Programming, June 8–11, Fayetteville, Arkansas.
MATERIALS AND METHODS
In this section, we survey several known heuristics for determining folding pathways between two secondary structures, as well as present our novel semi-greedy and RNAtabupath methods.
Morgan–Higgs
To explain the Morgan–Higgs greedy direct pathway algorithm, we first define the notion of a base pair ‘clashing’ with another base pair—base pair (i, j) is said to clash with base pair (x, y) if either x ≤ i ≤ y ≤ j or i ≤ x ≤ j ≤ y. More generally, a base pair (i, j) clashes with a secondary structure A if there exists (x, y) ∈ A such that (i, j) clashes with (x, y). The set of base pairs (x, y) ∈ A such that (i, j) clashes with (x, y) is denoted Clash(i, j, A); i.e.
With this definition, the Morgan–Higgs greedy algorithm repeatedly performs the following steps: (i) determine the base pair (i, j) belonging to B but not A which has minimum size clash set C, (ii) remove base pairs from C, and (iii) add base pairs in B that do not induce any new clashes. Pseudocode for this algorithm is described in Figure 6.
Figure 6.
Morgan–Higgs Greedy Algorithm (6) to construct a greedy direct pathway from secondary structure A to B.
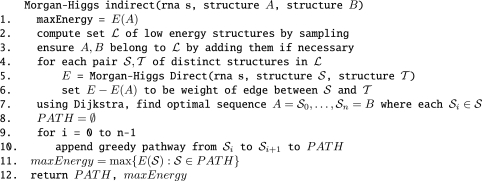
The Morgan–Higgs algorithm to compute a nearly optimal (possibly) indirect pathway between secondary structures A, B proceeds as follows. By sampling, create a set 𝒮 of low energy secondary structures. If either A or B does not belong to 𝒮, then add the missing structure to 𝒮. Define a complete, weighted, undirected graph G = (V, E), where the set V of vertices consists of all structures in 𝒮, and the edge weight between any two structures 𝒮, 𝒯 is defined to be energy barrier max{E(𝒮i) − E(𝒮): 1 ≤ i ≤ n}, where 𝒮 = 𝒮0, … , 𝒮n = 𝒯 is the greedy direct pathway from 𝒮 to 𝒯, as determined by the Morgan–Higgs direct algorithm described in Figure 6. Morgan and Higgs then apply ‘single link cluster’ (SLC) algorithm, as described in (22), in order to determine an optimal pathway, starting from structure A, proceeding by hopping from one low energy structure in 𝒮 to another via a greedy direct pathway, and terminating by the structure B.
In our implementation of the Morgan–Higgs indirect algorithm, we sample low energy structures with respect to the Turner energy model by applying the Ding–Lawrence algorithm (8), as implemented in RNAsubopt-p from the Vienna RNA Package (Step 2 of Figure 7). In place of the SLC algorithm, we apply a modified form of Dijkstra's single source shortest path algorithm (23), in order to determine a sequence A=𝒮0, 𝒮1, … , 𝒮n = B of structures, where each 𝒮i ∈ 𝒮, then concatenate the direct pathways between successive 𝒮i to 𝒮i+1, as determined by the Morgan–Higgs direct pathway algorithm. Figure 7 depicts the pseudocode for this algorithm.
Figure 7.
Morgan–Higgs algorithm (6) to construct an indirect pathway from secondary structure A to B. In line 2 of this algorithm, we use stochastic sampling of Ding and Lawrence (8), as implemented in RNAsubopt-p, and applied a modified version of Dijkstra's single source shortest path algorithm to determine low energy structures, whose greedy direct paths can be glued together for a pathway from A to B.
Greedy direct algorithm of Voss et al.
Perhaps the simplest possible algorithm to find a nearly optimal (direct) pathway between A and B is to apply a greedy approach, where at each step we choose to remove a base pair belonging to A but not B, or add a base pair belonging to B but not A, where the choice of base pair to be removed or added is made so as to ensure the lowest energy next structure. Pseudocode for this method, described by Voss et al. (19), is depicted in Figure 8.
Figure 8.
Greedy method to determine direct pathway between A and B, as described by Voss et al. (19). Secondary structures A, B can be considered to be sets of base pairs, so the requirement that 𝒯 ⊆ A ∪ B means that every base pair of 𝒯 belongs to either A or B. This condition ensures that the pathway produced is direct. The notation dbp(𝒮, 𝒯) = 1 means that the base pair distance (30) between 𝒮, 𝒯 is 1; i.e. 𝒮, 𝒯 differ by one base pair. Moreover, since dBP(𝒮, B) = dBP(𝒯, B) + 1, each iteration in the while loop ensures advancement by one base pair to the target structure B. It follows that the while loop involves dBP(A, B) iterations.
Searching exhaustively all the possible direct routes is impractical. However, we can benefit from a more randomized approach in which we randomly add or remove a valid base pair that yields a structure that is among the k lowest energy structures. This semi-greedy approach is depicted in Figure 9. Since the result is clearly dependent on a parameter k we can iterate the same approach for several values of k and return the route with the lowest energy barrier.
Figure 9.
Semi-greedy method to determine direct pathway between A and B. The only difference between the greedy and semi-greedy method is that the latter randomly selects one of the k lowest energy neighbors (Step 5) rather than the minimum energy neighbor. Benchmarking indicates that the semi-greedy method generally outperforms the greedy method when determining low energy pathways between conformers of a riboswitch.
Semi-greedy and tabu semi-greedy methods
Indirect routes present more opportunities and challenges, since the space of possible routes increases considerably. Also, a purely greedy approach is not possible since the algorithm would not be able to escape from cycles. Indeed, suppose that the structure A is the minimum free energy structure for the given RNA sequence; then the first step would add or remove a base pair, yielding a structure that is no longer the minimum free energy structure. In the next step, the added (respectively removed) base pair would then be removed (respectively added), in order to return to the minimum free energy structure. For that reason, it makes sense to exclude certain moves at certain times during the search. Tabu search (20) is a well-studied combinatorial optimization method that entails a greedy strategy where a list of recently taken moves is placed temporarily on a ‘tabu list’, and cannot be applied until removed from the tabu list.
In Figure 10, we present pseudocode for a TABU semi-greedy algorithm (RNAtabupath) to find nearly optimal, possibly indirect pathways between designated secondary structures A and B. The algorithm starts with the initial structure. At each successive step in the execution of the algorithm, we choose to add or remove that base pair resulting in the lowest energy (greedy), after which the base pair is placed in the tabu list, hence cannot be added or removed for a certain number of steps. The algorithm iterates this strategy until the target structure is reached.
Figure 10.
TABU semi-greedy algorithm to compute near-optimal folding pathway between two designated structures A, B for a given RNA sequence. In line 11, we assume that 𝒯 is obtained from 𝒮 without using a base pair in the tabu list. The tabu list contains base pairs that were recently added or removed from an intermediate structure. When added to the tabu list, a base pair is given a time stamp. It is removed from the tabu list after a system dependent waiting time. Fitness F of a structure is defined by F = E + w · BP, where E is energy of current structures, w is weight defined in line 2 and BP is ‘incremental‘ distance toward the target, i.e. ±1.
As in every optimization algorithm we need to define the fitness function, F. The fitness function is a measure of quality of each state. In the case at hand, a state is a secondary structure 𝒮, and the fitness function must account for the free energy E(𝒮) as well as the distance dBP(𝒮, B) from 𝒮 to the target structure B. Hence, the fitness F(𝒮) of secondary structure 𝒮 is defined by
where w represents a weight that regulates the importance of reaching the target structure versus choosing a low energy structure. A low weight has the potential of driving the algorithm to structures that are too far away from the target, B, while a higher weight can quickly converge to the target structure at the expense of including higher energy intermediate structures in the path produced. An intermediate value for weight w will tend to cause the algorithm to behave in a manner similar to that of the greedy algorithm for direct pathways. In order to avoid the latter, we have developed a ‘weight oscillation’ strategy that can be explained in the following steps:
Start with a given initial weight w0.
Increase the value of w when the distance to the target has not been improved for a number of iterations and restart from the structure found to be closest to the target.
Decrease the weight when the distance to the target is improved.
If the weight reaches a certain value wMax, increase the value of w0 and restart the search (with w = w0).
Our TABU strategy starts with the initial structure A, and in each step either adds or removes the base pair that minimizes the fitness function F. The base pair that has just been added or removed will be kept in a tabu list for a certain number of steps during which time it cannot be added or removed to any structure in the pathway being constructed. The fitness function F is adaptive, since it depends on the weight oscillation scheme. The algorithm terminates when the target structure is reached. Additionally, the algorithm introduces an aspiration criterion for which a base pair can be changed (even if it is tabu) when the resulting structure reduces the best distance to the target found so far, provided that its free energy does not exceed that of the maximum energy of a structure in the pathway constructed so far. Additionally, we introduce two stochastic aspects to the TABU algorithm: the time a base pair remains on the tabu list, and the way to break ties when choosing the best base pair. See Figure 10 for pseudocode of the resulting TABU algorithm. Note that the algorithm depends on parameter w0. Consequently, we can start with a given value and iterate the algorithm using different values while maintaining the best pathway so far found. Note that we assume that in line 10 of the pseudocode of Figure 10, we assume that 𝒯 is obtained from 𝒮 without using a base pair in the tabu list unless the aspiration criterion just mentioned has been applied, and that the tabu list is updated.
Our initial implementation of TABU method used a greedy search strategy. Upon subsequent testing, we found that by adding a semi-greedy component to TABU search, the resulting algorithm was substantially improved. Similarly, we found that the greedy algorithm of Voss et al. (19), described in Figure 8, is improved by adding a semi-greedy component for the search. The resulting pseudocode is given in Figure 8. Clearly, one could apply Monte Carlo and simulated annealing strategies to sample low energy folding pathways, as well as envision a genetic algorithm, that permits the crossover between folding pathways having a common source A and target B. Nevertheless, the TABU semi-global approach of RNAtabupath appears to be a very fast method to quickly determine near-optimal folding pathways. The web site for RNApathfinder includes additional tools to determine the frequency of occurrence of secondary startures in (say) 1000 low energy folding pathways, and to determine the similarity between two pathways.
In this section, we survey several known heuristics for determining folding pathways between two secondary structures, as well as present our novel semi-greedy and RNAtabupath methods.
RESULTS
In this section, we present summary results on folding pathways and energy barriers computed for each of the algorithms: greedy (19), semi-greedy, RNAtabupath, Morgan–Higgs direct (6), Morgan–Higgs indirect (6), findpath (18) and barriers (2,15). Due to the stochastic nature of the semi-greedy method and RNAtabupath, we report the best results found over 1000 runs. In the case of RNAtabupath, fitness of the current structure is defined by F = E + w · BP, where E is the free energy of the current structure, and BP is the ‘incremental’ distance toward the target (i.e. ±1). RNAtabupath allows the user to input parameters wMin, wMax that confine the weight w ∈ [wMin, wMax]. Reported values were for wMin = 1 and wMax = 7, which are the default values on the web server.
Table 1 presents the ‘energy barrier’ in the pathway A=𝒮0, 𝒮1, … , 𝒮n = B between low energy structures A and B of known conformational switches, where energy barrier is defined to be max{E(𝒮i) − E(A) : i = 1, … , n}. Structures A and B are two metastable states of five riboswitches, guanine riboswitch from Bacillus subtilis (rb1), adenine riboswitch from V. vulnificus (rb2), S-adenylmethionine riboswitch from Thermoanaerobacter tecongensis (rb3), thymine pyrophosphate riboswitch from T. tecongensis (rb4), and xpt-pbuX riboswitch from B. subtilis (rb5), whose metastable secondary structures were found by in-line probing experiments of various groups. See references from Wakeman et al. (16) for riboswitches rb1–rb4 and Mandal et al. (24) for rb5. Table 1 also contains results for some conformational switches found on the paRNAss web site, http://bibiserv.techfak.uni-bielefeld.de/parnass/examples.html; however, since the metastable structures for the latter conformational switches have not been experimentally determined, we ran the software RNAbor, which determines for each integer value of δ, the minimum free energy structure MFE(δ) and partition function Z(δ) over all δ-neighbors of the minimum free energy structure. Here a structure 𝒯 is said to be a δ-neighbor of structure 𝒮 if base pair distance between 𝒮, 𝒯 is δ. [See (25) for details on RNAbor.] For the conformational switches taken from the paRNAss web site, we defined A to be the minimum free energy structure and B to be that structure which is the minimum free energy structure over all δ-neighbors of A, where 10 < δ and the output of RNAbor indicated a second peak at the value δ.
Table 1.
Algorithm benchmarks for computing folding pathways between two low energy secondary structures
| Instance | Greedy | Semi-greedy | RNAtabupath | GreedyMH | IndirectMH | Findpath | barriers |
|---|---|---|---|---|---|---|---|
| rb1 | 32.80 | 25.24 | 24.04 | 26.24 | 23.99 | 24.04 | † |
| rb2 | 14.64 | 9.20 | 7.25 | 10.00 | 10.00 | 8.20 | * |
| rb3 | 24.80 | 22.70 | 17.90 | 28.40 | 20.00 | 22.40 | † |
| rb4 | 16.90 | 16.90 | 16.90 | 16.90 | 16.90 | 16.90 | † |
| rb5 | 33.30 | 25.67 | 24.54 | 26.74 | 26.74 | 24.54 | † |
| hok | 36.37 | 33.70 | 29.66 | 36.30 | 36.30 | 28.5 | † |
| SL | 14.09 | 14.09 | 12.90 | 18.20 | 16.20 | 13.00 | 11.80 |
| attenuator | 11.50 | 9.00 | 8.60 | 12.60 | 14.70 | 8.70 | 8.30 |
| s15 | 7.10 | 7.10 | 6.60 | 9.70 | 9.70 | 7.10 | 6.60 |
| s-box leader | 7.10 | 5.30 | 5.20 | 10.20 | 9.30 | 5.20 | * |
| thiM leader | 21.44 | 16.67 | 14.84 | 20.57 | 31.00 | 16.13 | † |
| ms2 | 8.30 | 6.60 | 6.60 | 11.70 | 11.70 | 6.60 | * |
| HDV | 25.50 | 21.70 | 17.00 | 23.53 | 22.50 | 17.4 | † |
| dsrA | 8.30 | 8.30 | 8.20 | 14.60 | 10.77 | 8.30 | 8.00 |
| ribD leader | 13.84 | 11.70 | 9.50 | 18.11 | 16.90 | 10.71 | † |
| amv | 10.00 | 6.40 | 5.80 | 15.6 | 10.4 | 5.80 | * |
| alpha operon | 6.50 | 6.50 | 6.50 | 9.90 | 6.50 | 6.50 | * |
| HIV-1 leader | 14.28 | 13.49 | 11.30 | 17.90 | 18.50 | 9.30 | † |
‘Greedy’ refers to our implementation of Voss et al. (19), where a direct path is constructed by choosing the lowest energy base pair to remove or add at each step; ‘Semi-greedy’ refers to to our modification of Voss et al. (19), where a direct path is constructed by choosing one of the k lowest energy base pairs to remove or add at each step; RNAtabupath refers to our semi-greedy tabu search method described in the text; ‘GreedyMH’ refers to Morgan–Higgs greedy method (6) to produce a direct path; ‘IndirectMH’ refers to our implementation of Morgan–Higgs method (6) to produce a possibly indirect path; ‘Findpath’ refers to Vienna RNA package findpath.c method described in Flamm et al. (18) with look-ahead parameter k = 10; barriers refers to the exact method of Flamm et al. (2,15), that relies on RNAsubopt. In each case, near-optimal low energy pathways between two low energy secondary structures of five different riboswitches: guanine riboswitch from B. subtilis (rb1), adenine riboswitch from V. vulnificus (rb2), S-adenylmethionine riboswitch from T. tecongensis (rb3), thymine pyrophosphate riboswitch from T. tecongensis (rb4) and xpt-pbuX riboswitch from B. subtilis (rb5). Secondary structures for rb1–rb5 were experimentally determined; see Wakeman et al. (16) for rb1–rb4 and Mandal et al. (24) for rb5. Sequences of additional conformational switches were taken from the paRNAss web site http://bibiserv.techfak.uni-bielefeld.de/parnass/examples.html, courtesy of the Giegerich Lab. For the latter, the two low energy structures were taken to be the minimum free energy structure A and the structure B determined by RNAbor (25) to be the minimum free energy structure over all structures having base pair distance k with A, where 10 ≤ k and a second peak was found at position k in the output of RNAbor. Energy barrier in the pathway A = 𝒮0, 𝒮1, … , 𝒮n = B from A to B is here defined to be max{E(𝒮i) − E(A)} : i = 1, … , n}, where free energy is measured in kcal/mol, as computed by RNAeval from Vienna RNA Package. Notation used in last column given as follows: †means barriers could not converge; *means that either structure A or B is not locally optimal, hence barriers could not be directly applied. However, one could apply barriers in the following manner, as suggested by an anonymous referee. Given non-locally optimal structures A, B, one can first determine locally optimal structures 𝒮, 𝒯 that, respectively, contain A, B, then apply barriers to find an optimal path between 𝒮, 𝒯. This will yield a near-optimal path from A to B. Boldface numbers indicate the minimal barrier energy found by one of the heuristic algorithms, while underlined numbers indicate the minimum barrier energy found by the exact algorithm, BARRIERS.
For technical reasons having to do with computation of the partition function, the treatment of dangles in RNAbor is identical to that of Vienna RNA Package RNAfold with option −d2. In some instances, the metastable structure we chose using RNAbor was no longer locally optimal under the −d1 treatment of dangle, which latter is used in all the algorithms appearing in Table 1. In particular, we should mention that one must explicitly use −d1 option with RNAsubopt, to ensure that RNAfold, RNAeval and barriers all use the same treatment of dangles. Due to the energy model differences (−d2 versus −d1) in using RNAbor to choose one of the metastable structures, barriers could not be used in some instances—rb2, s-box leader, ms2, amv and alpha operon.
We see that the greedy approach is simple, but yields considerably poorer results than other methods tested. However, a small change such as a semi-greedy component yields great improvements. Tabu search for indirect routes outperforms both greedy and semi-greedy approaches (data for the tabu greedy method is not shown). In the semi-greedy algorithm and RNAtabupath, we experimented with different choices of the value k, where randomly one of the best k neighbors is chosen. After computational experiments over the range of lengths typical for conformational switches, we fixed the value k = 8 for semi-greedy algorithm and k = 5 for RNAtabupath. The initial weight w0 in RNAtabupath ranges from 1 to 7, the default setting for the web server, although best results for this range depend on the input sequence. In general, w0 ∈ [4, 7] works better for larger sequences. Morgan–Higgs direct and indirect algorithms did not perform well in all but one instance; Morgan–Higgs indirect algorithm curiously outperformed all algorithms for rb1. In general, findpath is a very fast algorithm that produces excellent quality direct pathways, with barrier energies often equal or close to those of RNAtabupath. In the case of hok-RNA and HIV-1 leader, findpath outperformed all other approaches. In our benchmarking, we set the look-ahead of findpath to be 10; often increasing the look-ahead to 100 did not change the results. However, in the 396 nt hok-RNA, findpath improved dramatically with increased look-ahead k: barrier energy of 28.5 for k = 10, 28.17 for k = 20, 23.5 for k = 100, 22.7 for k = 200, 21.4 for k = 500 and k = 1000.
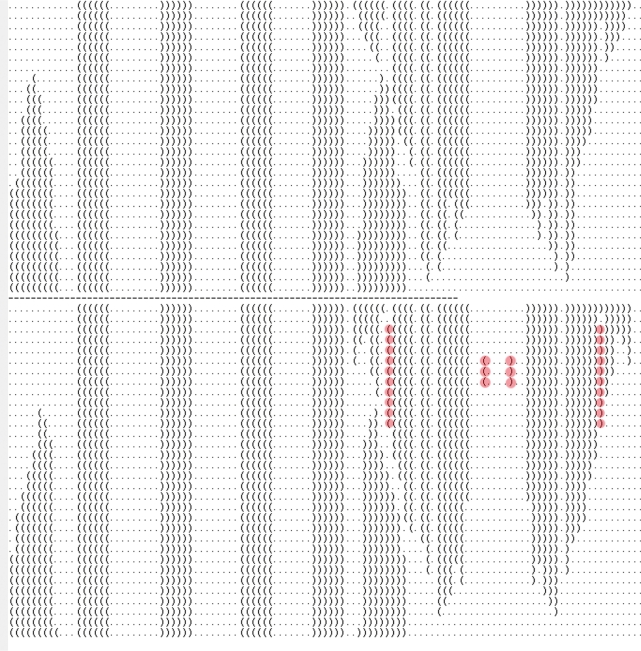
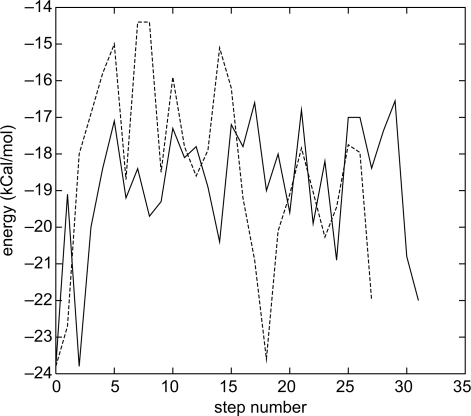
Figures 2 and 3 demonstrate cases where a well-chosen indirect pathway necessarily has lower barrier energy than that of any direct pathway. Applying this principle to 304 examples derived from pseudoknotted structures in Pseudobase (21), we found that in roughly half the examples, RNAtabupath and findpath produced the same barrier energy, while in all other instances, RNAtabupath produced a lower barrier energy barrier than did findpath; indeed, the maximum difference in barrier energy was 6.51, while the average was 1.93 kcal/mol with standard deviation of 1.45. Figure 11 depicts a folding pathway computed by RNAtabupath between the two meta-stable secondary structures of the adenine riboswitch from V. vulnificus (rb2) (16). Figure 12 depicts the free energy of intermediate structures in this pathway as a function of step number.
Figure 11.
Comparison of best direct pathway (above) and best indirect pathway (below), as found by RNAtabupath between the two metastable secondary structures of the adenine riboswitch from V. vulnificus (rb2), as reviewed in Wakeman et al. (16). Base pairs not belonging to either start and target structure are highlighted.
Figure 12.
Graph of free energy of intermediate structure as a function of step number or index in the RNAtabupath folding pathway between two metastable secondary structures of the adenine riboswitch from V. vulnificus (rb2), corresponding to data from Figure 11. Dotted lines depict a similar graph for a folding pathway computed by the semi-global method.
One useful application of RNAtabupath is to provide an energy upper bound for subsequent application of barriers, an observation pointed out by an anonymous referee. Specifically, given RNA sequence s and two metastable structures A, B, let E0 denote the minimum free energy of s and let E(A) denote the free energy of structure A. If E is the barrier energy computed by RNAtabupath (or another method) for a folding pathway from A to B, then barriers with bound E + (E(A) − E0) will compute an optimal pathway, provided it converges.
The barrier energies obtained by barriers in Table 1 were computed in this fashion. Since barriers is the only exact algorithm, when it converges, a provably optimal pathway is produced. In the cases of rb1, rb3, rb4, rb5, thiM leader, ribD leader and HIV-1 leader, barriers did not converge, even when started with the energy bound obtained by RNAtabupath.
DISCUSSION
Molecular folding pathways are low energy routes taken along an energy surface. As previously noticed by Morgan and Higgs (6), indirect pathways in general involve lower energy structures than do direct pathways. This is clear from the toy example presented in Figures 2 and 3. In other data, we see how the creation of a base pair in a region with no base pairs leads to the stabilization of other secondary structures along the folding pathway.
Since barriers is an exact algorithm, it should be used whenever possible; i.e. one should first apply findpath or RNAtabupath to obtain an energy upper bound for subsequent application of barriers. In other cases, findpath and RNAtabupath appear to produce energy barriers of roughly the same quality. If large type-H pseudoknots appear in the structure obtained by adjoining two metastable structures, then RNAtabupath is likely to be the best algorithm, since indirect pathways will have lower barrier energy in this case.
To assist those interested in computing near-optimal folding pathways, we have created the web server RNApathfinder, located at http://bioinformatics.bc.edu/clotelab/RNApathfinder. In addition to supporting RNAtabupath computations, source code can be downloaded for several algorithms discussed in this article.
FUNDING
Fundacion Caja Madrid (to I.D.); National Science Foundation (grants DBI-0543506 and DMS-0817971 to P.C. and W.A.L.); RNA Ontology Consortium (to I.D., P.C. and W.A.L.); National Science Foundation (grant DMI-0600384 to I.D. and P.VH.); Deutscher Akademischer Austauschdienst (to P.C. for funding a visit to Martin Vingron's; group in the Max Planck Institute of Molecular Genetics); Digiteo Foundation (to P.C.). Funding for open access charge: National Science Foundation (grant DBI-0543506).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Robert Giegerich for use of his RNAmovies software that converted the output of our RNAtabupath pathways into movie format, subsequently converted to FLASH format by Irina Bariakhtar for display on the web server. We would like to thank three anonymous referees for their helpful comments. We would also like to thank Christian Forst, for permission to use the name, ‘RNApathfinder’, which appears in the title of paper (26) by U. Göbel and Ch. V. Forst on the neutral network theory of P. Schuster and co-workers.
REFERENCES
- 1. Abfalter,I., Flamm,C. and Stadler,P. Design of multi-stable nucleid acid sequences. German Conference on Bioinformatics 2003: 1–7.
- 2.Fontana W, Hofacker I, Schuster P. RNA folding at elementary step resolution. RNA. 2000;6:325–338. doi: 10.1017/s1355838200992161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes K, Gultyaev AP, Franch T, Pedersen K, Mikkelsen ND. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Harris K, Crothers D. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993;32:5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- 5.Mathews DH, Case DA. Nudged elastic band calculation of minimal energy paths for the conformational change of a GG non-canonical pair. J. Mol. Biol. 2006;357:1683–1693. doi: 10.1016/j.jmb.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 6.Morgan S, Higgs P. Barrier heights between ground states in a model of RNA secondary structure. J. Phys. A: Math. Gen. 1998;31:3153–3170. [Google Scholar]
- 7.Nussinov R, Jacobson AB. Fast algorithm for predicting the secondary structure of single stranded RNA. Proc. Nat. Acad. Sci. USA. 1980;77:6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia T, J. SantaLucia J, Burkard M, Kierzek R, Schroeder S, Jiao X, Cox C, Turner D. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1999;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 10.Turner DH, Sugimoto N, Freier SM. RNA structure prediction. Annu. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 11.Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Kierzek R, SantaLucia J, Jr, Walter AE, Turner DH. Nearest-neighbor parameters for G.U mismatches: [formula; see text] is destabilizing in the contexts [formula; see text] and [formula; see text] but stabilizing in [formula; see text] Biochemistry. 1991;30:11124–11132. doi: 10.1021/bi00110a015. [DOI] [PubMed] [Google Scholar]
- 13.Peritz AE, Kierzek R, Sugimoto N, Turner DH. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991;30:6428–6436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- 14.Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flamm C, Hofacker I, Stadler P, Wolfinger M. Barrier trees of degenerate landscapes. Z. Phys. Chem. 2002;216:155–173. [Google Scholar]
- 16.Wakeman CA, Winkler WC, CE D. Structural features of metabolite-sensing riboswitches. Trends Biochem. Sci. 2007;32:415–424. doi: 10.1016/j.tibs.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wuchty S, Fontana W, Hofacker I, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–164. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Flamm C, Hofacker IL, Maurer-Stroh S, Stadler PF, Zehl M. Design of multistable RNA molecules. RNA. 2001;7:254–265. doi: 10.1017/s1355838201000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss B, Meyer C, Giegerich R. Evaluating the predictability of conformational switching in RNA. Bioinformatics. 2004;20:1573–1582. doi: 10.1093/bioinformatics/bth129. [DOI] [PubMed] [Google Scholar]
- 20.Glover F, Laguna M. Tabu Search. Massachusetts: Kluwer Academic Publishers, Norwell; 1997. p. 376. [Google Scholar]
- 21.Van Batenburg FH, Gultyaev AP, Pleij CW. Pseudobase: structural information on RNA pseudoknots. Nucleic Acids Res. 2001;29:194–195. doi: 10.1093/nar/29.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sneath P, Sokal R. Numerical Taxonomy: the Principles and Practice of Numerical Classification. San Francisco: Freeman; 1973. [Google Scholar]
- 23.Cormen T, Leiserson C, Rivest R. Algorithms. MIT Press, Cambridge, Massachusetts; 1990. p. 1028. [Google Scholar]
- 24.Mandal M, Boese B, Barrick J, Winkler W, Breaker R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 25.Freyhult E, Moulton V, Clote P. Boltzmann probability of RNA structural neighbors and riboswitch detection. Bioinformatics. 2007;23:2054–2062. doi: 10.1093/bioinformatics/btm314. [DOI] [PubMed] [Google Scholar]
- 26.Göbel U, Forst C. RNA Pathfinder - global properties of neutral networks. Z. Phys. Chem. 2002;216:1–18. [Google Scholar]
- 27.Hofacker I. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro BA, Bengali D, Kasprzak W, Wu JC. RNA folding pathway functional intermediates: their prediction and analysis. J. Mol. Biol. 2001;312:27–44. doi: 10.1006/jmbi.2001.4931. [DOI] [PubMed] [Google Scholar]
- 29.Franch T, Gultyaev AP, Gerdes K. Programmed cell death by hok/sok of plasmid r1: processing at the hok mRNA 3H-end triggers structural rearrangements that allow translation and antisense RNA binding. J. Mol. Biol. 1997;273:38–51. doi: 10.1006/jmbi.1997.1294. [DOI] [PubMed] [Google Scholar]
- 30.Moulton V, Zuker M, Steel M, Pointon R, Penny D. Metrics on RNA secondary structures. J. Comput. Biol. 2000;7:277–292. doi: 10.1089/10665270050081522. [DOI] [PubMed] [Google Scholar]