Abstract
Retroviruses rely on host RNA-binding proteins to modulate various steps in their replication. Previously several animal retroviruses were determined to mediate Dhx9/RNA helicase A (RHA) interaction with a 5′ terminal post-transcriptional control element (PCE) for efficient translation. Herein PCE reporter assays determined HTLV-1 and HIV-1 RU5 confer orientation-dependent PCE activity. The effect of Dhx9/RHA down-regulation and rescue with siRNA-resistant RHA on expression of HIV-1NL4–3 provirus determined that RHA is necessary for efficient HIV-1 RNA translation and requires ATPase-dependent helicase function. Quantitative analysis determined HIV-1 RNA steady-state and cytoplasmic accumulation were not reduced; rather the translational activity of viral RNA was reduced. Western blotting determined that RHA-deficient virions assemble with Lys-tRNA synthetase, exhibit processed reverse transcriptase and contain similar level of viral RNA, but they are poorly infectious on primary lymphocytes and HeLa cells. The results demonstrate RHA is an important host factor within the virus-producer cell and within the viral particle. The identification of RHA-dependent PCE activity in cellular junD RNA and in six of seven genera of Retroviridae suggests conservation of this translational control mechanism among vertebrates, and convergent evolution of Retroviridae to utilize this host mechanism.
INTRODUCTION
Retroviruses rely on host RNA-binding proteins to modulate post-transcriptional control of viral gene expression. Several animal retroviruses contain a RNA structural element within their 5′ untranslated region (UTR) that is necessary for efficient viral protein synthesis (1). Viral proteins are not required. Instead this retroviral post-transcriptional control element (PCE) requires interaction with host Dhx9/RNA helicase A (RHA) (2). PCE was originally identified in the 5′ long terminal repeat (LTR) of avian spleen necrosis virus (3) and subsequently in retroviruses that infect feline, bovine, catarrhine, and human hosts (4–6). These viruses represent five genera of the Retroviridae (alpharetrovirus, betaretrovirus, deltaretrovirus, gammaretrovirus and spumavirus). The possibility of PCE activity in the lentivirus genus was addressed in this study.
RHA recognizes functionally redundant structural features encoded by the RU5 regions of the SNV LTR (2,7). Experiments with human T-cell leukemia type 1 (HTLV-1) provirus determined that RHA down-regulation reduces the polysome association of HTLV-1 gag RNA and severely attenuates virion production, indicating that RHA is an important host factor for HTLV-1 replication (5). Reporter assays determined the HTLV-1 LTR is sufficient for PCE activity although a sufficient role for the RU5 regions remains to be evaluated. Subsequent to identification in Retroviridae, PCE activity was identified in the complex 5′ UTR of rat and human junD (2). RHA is necessary for efficient translation of endogenous junD and results in the reporter assay demonstrated that RHA down-regulation eliminates junD PCE activity (2).
Recently RHA/Dhx9 was identified as an important host factor in HIV-1 replication in meta-analysis of genome-wide studies (8). Previous RHA overexpression studies detected increased HIV-1 gene expression as measured by HIV-1 LTR-luciferase reporter gene activity (9); an effect on balanced expression of the unspliced HIV-1 gag RNA; and increased virion protein production (10). The results suggested RHA affects HIV-1 transcription and/or post-transcriptional expression. Biochemical analysis has revealed that RHA can act as a scaffold to bridge the association of CREB-binding protein and RNA polymerase II (11). And a role for interaction with HIV-1 RNA was invoked from Northwestern analysis detecting RHA interaction with the HIV-1 trans-activation response element (TAR) within the R region of the HIV-1 5′ LTR (9); TAR is the target for the essential HIV-1 Tat transcriptional trans-activator. Recently, Liang and colleagues determined that RHA interacts with HIV-1 Gag and is packaged into HIV-1 particles in an RNA-dependent manner. RHA down-regulation by siRNA decreased HIV-1 infectivity that was attributed in part to lower reverse transcriptase (RT) activity (12). In summary, RHA appears to influence HIV-1 gene expression and possibly the process of virus assembly. A possible role for RHA in translation of the virus and possible involvement of the ATP-dependent helicase activity remains an open issue.
Given the identification of RHA-dependent PCE activity in the RU5 of several animal retroviruses, we evaluated the RU5 regions of human retroviruses, HTLV-1 and HIV-1, for PCE reporter activity. We also examined the effect of RHA down-regulation and rescue with siRNA-resistant RHA on expression of HIV-1 provirus, virion production, content and infectivity in PBMC. The results demonstrated that RU5 of HIV-1 and HTLV-1 confer orientation-dependent PCE reporter activity, and that RHA affects two steps in replication of HIV-1: (i) translation of the viral RNA and (ii) infectivity of progeny virions.
MATERIALS AND METHODS
Cell culture, transfection, infection
HEK 293 and HeLa TZM-bl (13–16) were cultured in Dulbecco’s modified eagle medium (Invitrogen), 10% FBS (Invitrogen) and 1% antibiotic (Gibco) at 37°C/5% CO2. Peripheral blood mononuclear cells (PBMCs) were isolated as described previously (17) and were stimulated by 5 μg/ml PHA and 100 U/ml recombinant human Interleukin-2 (Roche) for 2 days in RPMI 1640 medium (Invitrogen), 10% FBS, 1% antibiotic and then with supplementation of 50 U/ml Interleukin-2 at 37°C/5% CO2.
Cultures of 2.5 × 105 HEK 293 cells per 35 mm dish were incubated overnight in 2 ml medium and then transfected with 2 µg indicated reporter plasmid, 150 ng pGL3 firefly luciferase plasmid in 2 × HBS as described (3) for 2 days. Cells were harvested in PBS, resuspended in 50 µl NP40 lysis buffer and 5 µl lysate was tested for Luciferase activity (Promega) as described (18) or diluted for HIV-1 Gag p24 ELISA (Zeptometrix). Construction of PCE reporter plasmids is provided in Supplementary Data. HTLV-1 reporters contain the Tax-dependent U3 reporter and were co-transfected with 500 ng HTLV-1 tax-1 expression plasmid. Methods for RNA isolation and analysis are provided in Supplementary Data.
For siRNA transfection, 1 × 106 HEK 293 cells per 10 cm dish were cultured overnight and treated with 50 µl Oligofectamine (Invitrogen) and 20 µM indicated siRNA (Dharmacon) in 2 ml OptiMEM media (Gibco) and 5 ml DMEM without serum. Previously described (2) RHA siRNA: RHA1 UAGAAUGGGUGGAGAAGAAUU and RHA2 GGCUAUAUCCAUCGAAAUUUU and non-silencing scrambled (Sc) siRNA UAGACUAGCUGACGAGAAAUU were transfected for 4 h, and the cultures were supplemented with 3.5 ml DMEM containing 30% FBS, 1% antibiotic and incubated for 48 h. These cells were split, cultured overnight. After second siRNA treatment, cells were cotransfected with 10 µg pNL101 HIV-1NL4–3 and 5 µg siRNA-resistant FLAG-tagged RHA (FL-RHA) or 5 µg siRNA-resistant FL-RHA in 1:3 DNA to FuGene6 (Roche) ratio by manufacturer protocol. RHA down-regulation was verified by immunoblot as described in Supplementary Data and ref. (2).
For infection experiments, HIV-1NL4–3 was propagated by transfection of cultures of 1 × 106 HEK 293 cells per 10 cm dish with 10 µg pNL101 in FuGene6 for 48 h. Virus-containing cell-free supernatant was used for Gag p24 ELISA, and 2 ng HIV-1 p24 was used to infect 1 × 106 PBMC or 2 × 105 HeLa TZM-bl cells (13–16). As described previously (19), virus growth on PBMC target cells was monitored by Gag ELISA on cell-free supernatant. HeLa TZM-bl cultures were harvested 48 h post-infection, extracted in the NP40 lysis buffer, 10 µg aliquots determined by Bradford assay, and used for Luciferase assay.
Statistical analyses
Four independent PCE reporter transfections were performed and data were log-transformed for the statistical analyses. ANOVA model was used to study the differences in Gag protein level and gag RNA level. Dunnett’s method was used to adjust for multiple comparisons. Data are presented as mean fold change with corresponding P-values (Figures 1 and 2). A one sample t-test compared the fold change differences in HIV-1 Gag production between RHAi and Sci groups (Figure 4). Three replicates were obtained for the HIV-1 infection assays. Data were analyzed using an ANOVA model that included group, time and their interaction term. Holm’s method was used to adjust for multiplicity (Figures 3 and 8).
Figure 1.
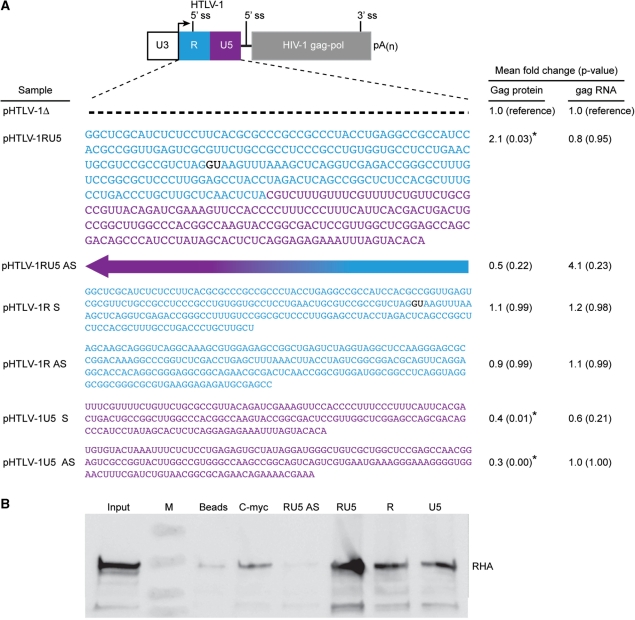
HTLV-1 RU5 is necessary for PCE activity and interaction with RHA. (A) Drawing of HTLV-1 PCE-gag reporter plasmid with HTLV-1 R depicted in blue and U5 in purple with corresponding sequence shown below. Arrow indicates transcription start site; 5′ ss, splice site; pA(n), polyadenylation signal. Dashed line denotes deletion of RU5 and reverse arrow indicates antisense orientation (AS) of RU5. HEK 293 cells were transfected with indicated reporter plasmid and HTLV-1 tax-1 expression plasmid to trans-activate the HTLV-1 U3 promoter for 48 h. Gag protein production was measured by Gag p24 ELISA and gag mRNA was quantified by RT-real-time PCR in four independent experiments. ANOVA model was used to study the differences in Gag protein level and gag RNA level and data are presented as mean fold change with corresponding P-values relative to pHTLV-1Δ with P-value indicated in parentheses. Asterisks indicate statistically significant difference (P ≤ 0.05). (B) RNA affinity chromatography was performed with HeLa nuclear extract and equimolar HTLV-1 RU5, RU5 AS, R, U5 or c-myc RNA bait. After sequential washes with KCl, high affinity proteins were eluted in 2 M KCl and immunoblotted with RHA antiserum lanes: input lysate; M, marker; Beads, reaction with no RNA control and indicated RNA bait.
Figure 2.
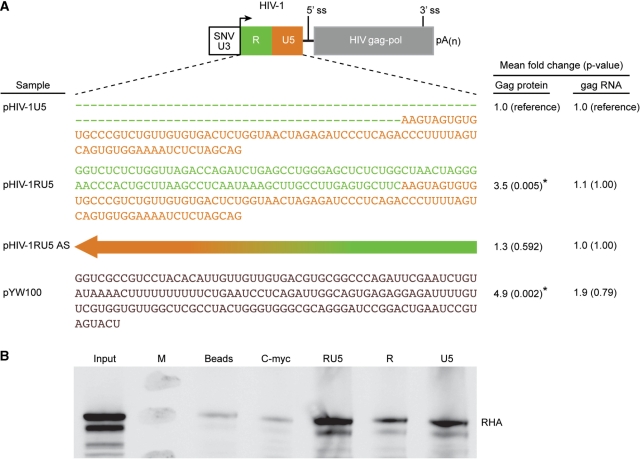
HIV-1 RU5 is necessary for PCE activity and interaction with RHA. (A) Drawing of HIV-1 PCE-gag reporter plasmid with HIV-1 R depicted in green and U5 in orange with corresponding sequence shown below. Arrow indicates transcription start site; 5′ ss, splice site; pA(n), polyadenylation signal. Dashed line denotes deletion of R and reverse arrow indicates antisense orientation (AS) of RU5. HEK 293 cells were transfected with the indicated plasmid, Gag production was measured by Gag p24 ELISA and gag mRNA was quantified by RT-real-time PCR. ANOVA model was used to study the differences in Gag protein level and gag RNA level. Data are presented as mean fold change with corresponding P-values. Results are presented as mean fold change relative to pHIV-1U5 with P-value indicated in parentheses. Asterisks indicate statistically significant difference (P ≤ 0.05). (B) RNA affinity chromatography was performed with HeLa nuclear extract and equal moles of indicated HIV-1 RNA or c-myc RNA bait. High affinity proteins were eluted in 2 M KCl and immunoblotted with RHA antiserum. Lanes: input lysate; M, marker; Beads, reaction with no RNA control and indicated RNA bait.
Figure 4.
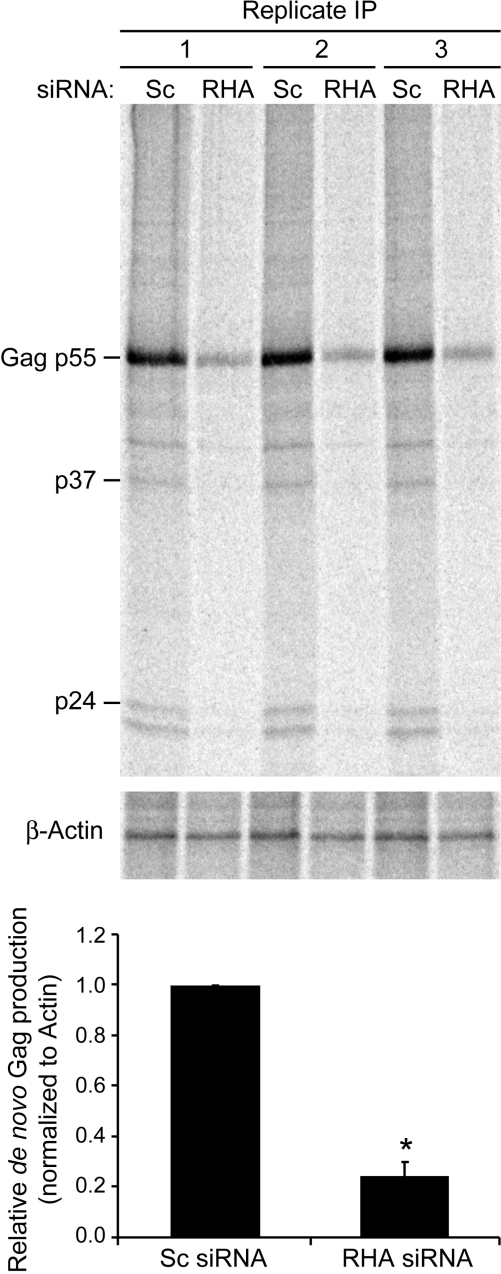
RHA is necessary for efficient translation of HIV-1 gag RNA. HEK 293 cells were transfected consecutively with non-silencing (Sc) or RHA siRNAs (RHA) and HIV-1NL4–3 for 48 h, labeled for 1 h with [35S]-cysteine/methionine and immunoprecipitation was performed in triplicate. Graph summarizes densitometry of [35S]-labeled Gag and β-Actin protein. Asterisks indicate statistically significant difference from Sc siRNA control (P = 0.002).
Figure 3.
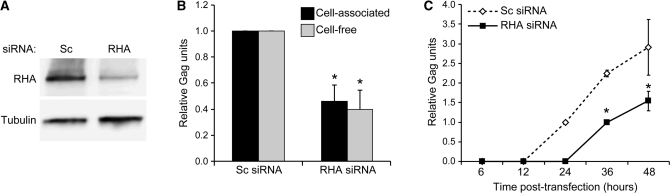
RHA is necessary for efficient production of HIV-1 Gag. HEK 293 cells were transfected consecutively with non-silencing (Sc) or RHA siRNAs (RHA) and HIV-1NL4–3. (A) Immunoblot of total cell protein with antiserum to RHA or α-Tubulin verified RHA down-regulation. (B) Cell-associated (n = 5) and cell-free (n = 16) Gag levels measured by Gag immunoblot or Gag ELISA, respectively. Asterisks indicate statistically significant difference from Sc siRNA (P = 0.0006 for cell-associated and P < 0.0001 for cell-free Gag, respectively). (C) Cell-free Gag production measured by Gag ELISA (n = 3). Asterisks indicate statistically significant difference from Sc siRNA at indicated time point (P ≤ 0.0001).
Figure 8.
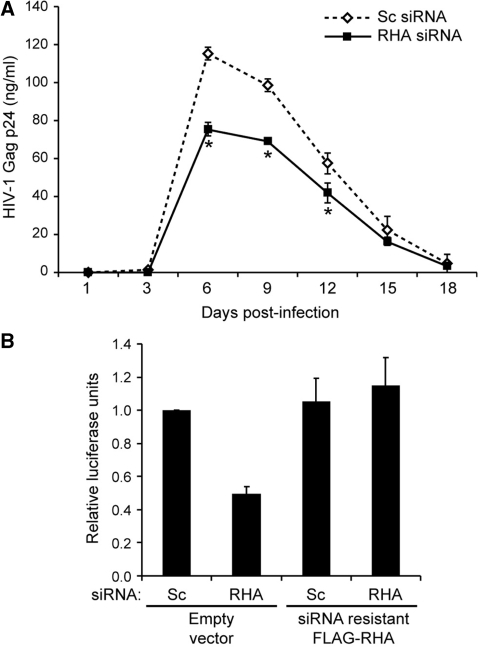
RHA down-regulation reduces infectivity of progeny HIV-1. (A) HEK 293 cells were transfected with scrambled (Sc) or RHA (RHA) siRNAs, and then second dose of siRNA and HIV-1NL4–3 for 48 h. Cell-free virus equivalent to 2 ng Gag was used to infect activated PBMCs. Virus growth on PBMC was measured by Gag ELISA (n = 3). Asterisks indicate statistically significant difference from Sc siRNA control was observed at day 6, 9 and 12 (P ≤ 0.0005, 0.0008, 0.0174). (B) HEK 293 cells were transfected with Sc or RHA siRNAs, and then second dose of siRNA with either empty vector or siRNA-resistant FLAG-RHA and HIV-1NL4–3. Cell-free virus equivalent to 2 ng Gag was used to infect TZM-bl cells and luciferase activity determined at 48 h (n = 3).
RESULTS
HTLV-1 RU5 provide orientation-dependent PCE activity
Previous results demonstrated that the 5′ LTR of HTLV-1ACH confers PCE activity in reporter assays (5). PCE reporter activity is defined by Rev/Rev responsive element (RRE)-independent expression of intron-containing HIV-1 gag RNA (3–5). Site-directed mutagenesis of SNV RU5 have determined that two functionally redundant stem-loop structures encoded by RU5 provide orientation-dependent PCE activity (5,6). To determine whether the R and U5 regions of HTLV-1 are necessary or sufficient for PCE activity, PCE reporter plasmids were generated (Supplementary Data) that contain the HTLV-1 U3 promoter and lack or contain R (nucleotides 1–220, relative to the transcription start site) and U5 (nucleotides 237–402) in the sense and antisense orientations (Figure 1A). The plasmids were cotransfected with tax-1 transcriptional trans-activator of the HTLV-1 U3 promoter. HIV-1 Gag levels were measured by ELISA. Results of four independent transfection experiments were evaluated for statistical trends (Figure 1A). Compared to pHTLV-1Δ that lacks RU5, pHTLV-1RU5 exhibited low but detectable Rev/RRE-independent expression of HIV-1 gag RNA. Gag levels increased 2-fold in response to RU5 in the sense but not the antisense orientation (pHTLV-1RU5, P = 0.03 and pHTLV-1AS, P = 0.22). HTLV-1 R in either the sense or antisense orientation did not increase Gag production. HTLV-1 U5 was inhibitory to Gag protein production in either sense (P = 0.01) or antisense orientation (P ≤ 0.01) orientation, which may be attributable to RNA structural features that impede ribosome scanning (20,21). To examine the abundance of steady-state gag RNA, polyA+ RNA was prepared from these samples; reverse transcribed and subjected to real-time PCR with gag and β-actin primers. Significant differences in gag mRNA levels were not observed among the samples, indicating the increase in Gag production is not attributable to increased mRNA accumulation (Figure 1A). The results indicated that HTLV-1 R or U5 are not sufficient for PCE activity and the combination of R and U5 function in an orientation-dependent manner to provide HTLV-1 PCE reporter activity.
Association with RHA was evaluated by RNA affinity chromatography and immunoblot by the protocol used to identify specific interaction of RHA with SNV PCE (2). Biotinylated HTLV-1 RU5, R and U5 transcripts or c-myc control RNA were synthesized with T7 RNA polymerase and purified on G-25 columns. Equimolar amounts (15 µM) of each transcript were bound to streptavidin agarose beads, incubated with HeLa nuclear lysate, and washed with binding buffer. Low affinity RNA-binding proteins were eluted by progressive treatment with elution buffer with 40, 100 and 200 mM KCl. High affinity RNA-binding proteins were eluted in 2 M KCl and immunoblotted with antisera to RHA (Figure 1B). RHA co-precipitated with HTLV-1 RU5. RHA was less abundant in reactions with equimolar amounts of R or U5, which correlated with the dearth of PCE activity. RHA was not enriched on beads alone, antisense RU5 (AS-RU5) or c-myc 5′ UTR RNA, indicating that RHA interaction with RU5 was not attributable to non-specific interaction.
HIV-1 RU5 confer PCE activity
A similar approach was used to examine HIV-1 RU5 for PCE activity. As summarized in Figure 2A, PCE reporter plasmids that encode the SNV U3 promoter and HIV-1 leader and gag open reading frame were evaluated in four independent transfections. Baseline Gag production was observed from pHIV-1U5 that lacks R and pHIV-1RU5AS that contains RU5 in the antisense orientation did not change Gag production (P = 1). The pHIV-1RU5 that contains RU5 in the sense orientation exhibited Rev/RRE-independent Gag production at a level 3.5-fold greater than baseline (pHIV-1RU5, P = 0.005). A similar level of PCE activity is observed from SNV PCE reporter pYW100 that contains SNV RU5 (4.9-fold). RT-real-time PCR determined steady-state gag RNA levels were similar among the samples (Figure 2A), indicating the increase in Gag production is not attributable to increased accumulation of the mRNA.
RNA affinity chromatography and RHA immunoblot surveyed association of HIV-1 RU5 with RHA. RHA co-precipitated with HIV-1 RU5 but was less abundant in equimolar HIV-1 R or U5 (Figure 2B). RHA was not enriched in reactions with beads alone or c-myc RNA, indicating that RHA interaction with RU5 was not attributable to non-specific interaction. To examine physical interaction in cells, in addition, RNA co-IP assays were performed on cells co-transfected with pHIV-1 RU5 and pCMV-FL-RHA or empty pFL plasmid. Cytoplasm was harvested in aliquots for IP with FLAG antibody followed by RNA isolation from the immunoprecipates, or isolation of cytoplasmic RNA. RNA from the immunoprecipates was treated with random hexamers in reactions with RT and without RT, and reactions with primers specific for gag or junD control were subjected to real-time PCR (2). The copy number of gag in the cytoplasmic RNA was similar between samples with FL-RHA or empty FL (2.7 × 105 and 3.9 × 105, respectively). The gag copy number in FL-RHA co-precipitate (3.5 × 104) was enriched >3-fold over the empty FL control, which provided the detection limit of the assay (1.0 × 104). In sum, the RNA co-precipitation results agree with the RNA affinity chromatography and further demonstrate that gag RNA interacts with RHA in vivo. We conclude that HIV-1 U5 is not sufficient for PCE activity and RU5 function in an orientation-dependent manner to provide PCE reporter activity.
RHA modulates the cytoplasmic utilization of HIV-1 gag RNA
To evaluate whether RHA is necessary for efficient translation of HIV-1, RHA was down-regulated with two previously characterized siRNAs or non-silencing scrambled control siRNA (Sc) for 48 h (2) (data not shown). These cells were transfected with HIV-1NL4–3 and the same siRNA for 48 h and immunoblot verified RHA down-regulation (Figure 3A). Total cellular protein was subjected to Gag immunoblot and cell-free supernatant medium was subjected to Gag ELISA. The results indicate significant reductions in cell-associated (P = 0.0006) and cell-free Gag (P < 0.0001; Figure 3B). The equivalent reductions in cell-associated and cell-free Gag indicated that RHA down-regulation affects the rate of Gag expression, but does not impair particle assembly or release. ELISAs were also performed on cell-free supernatant collected sequentially over 48 h. These results also indicated that RHA down-regulation delayed the onset of virion production by twelve hours and reduced absolute Gag production (Figure 3C).
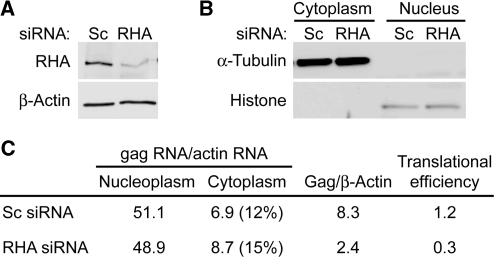
Metabolic labeling measured the effect of RHA down-regulation on Gag protein synthesis. After 1 h incubation with [35S]-cysteine/methionine, IP was performed with Gag or β-Actin antibody. As shown in Figure 4, RHA down-regulation significantly reduced de novo Gag protein synthesis, but did not reduce β-Actin protein synthesis (P = 0.002). To quantify the effect of RHA down-regulation on steady-state gag RNA abundance and cytoplasmic accumulation, nuclear and cytoplasmic RNA were isolated and subjected to RT-real-time PCR. Immunoblotting verified RHA down-regulation (Figure 5A), and antisera to α-Tubulin and Histone H1 verified effective nucleus and cytoplasm fractionation (Figure 5B). The gag and β-actin RNA levels were neither reduced by RHA down-regulation nor by the cytoplasmic accumulation of gag RNA (Figure 5C). Similar to the results of Figure 4, Gag IP on these samples determined that RHA down-regulation reduced de novo Gag protein synthesis but did not affect Actin synthesis (data not shown). Given the similar levels of HIV-1 gag mRNA, the results indicate that RHA down-regulation reduces the translational efficiency of HIV-1 gag RNA. As summarized in Figure 5C, gag translation efficiency was reduced by a factor of 4.
Figure 5.
RHA down-regulation does not affect cytoplasmic accumulation of HIV-1 gag RNA. HEK 293 cells were transfected consecutively with non-silencing (Sc) or RHA siRNAs (RHA) and HIV-1NL4–3 for 48 h and aliquots harvested for fractionation or 1 h labeling with [35S]-cysteine/methionine. (A) Immunoblot of total cell protein with antiserum to RHA or β-Actin. (B) Immunoblot of nuclear or cytoplasmic extracts with Histone H1 or Tubulin antibody. (C) RT real-time PCR measured nuclear or cytoplasmic gag RNA and β-actin RNA. Cytoplasmic accumulation of gag RNA relative to β-actin is expressed as percentage. Translational efficiency is expressed as ratio of [35S]-labeled Gag protein to cytoplasmic gag RNA.
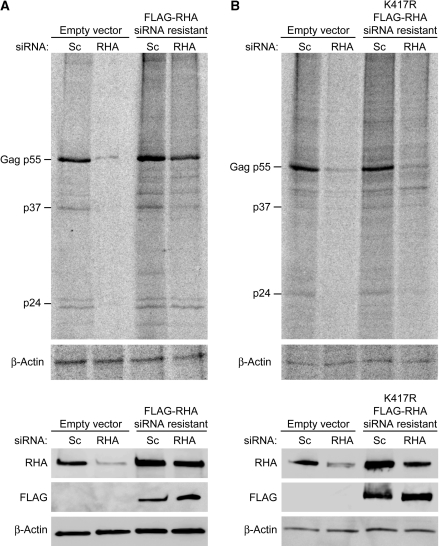
Next, we evaluated whether exogenous expression of siRNA-resistant FLAG-RHA is sufficient to rescue Gag protein synthesis. HEK 293 cells were treated with siRNAs and immunoblot determined that RHA was down-regulated (data not shown). These cells were transfected for 48h with siRNA-resistant FL-RHA and HIV-1NL4–3 and second treatment of siRNA, and then subjected to IP. RHA immunoblot verified RHA down-regulation and also detected the FL-RHA (Figure 6A, bottom). FLAG immunoblot verified expression of siRNA-resistant FL-RHA and Actin immunoblot controlled for protein loading (Figure 6A, bottom). Similar to results of Figure 4, Gag IP indicated that RHA down-regulation severely impaired de novo Gag protein synthesis (Figure 6A, top). Furthermore, expression of the siRNA-resistant FL-RHA rescued Gag protein synthesis (Figure 6A, top). Actin protein synthesis was not affected by RHA down-regulation or FL-RHA expression, consistent with previous results (2). To examine the role for the ATP-dependent helicase (22), we evaluated rescue by siRNA-resistant FL-RHA bearing K417R point mutation previously shown to eliminate ATP hydrolysis (11,22). FLAG immunoblot verified expression of K417R (Figure 6B, bottom). Gag IP determined that the K417R mutant did not rescue Gag protein synthesis, suggesting that the ATP-dependent helicase activity is necessary for efficient translation of HIV-1 gag (Figure 6B, top). As expected, Actin synthesis was not affected by expression of K417R. The results demonstrated that RHA is necessary for efficient translation of gag RNA and the ATP-dependent helicase domain is involved.
Figure 6.
ATPase domain of RHA is necessary for efficient translation of HIV-1 gag RNA. HEK 293 cells were transfected consecutively with non-silencing (Sc) or RHA siRNAs (RHA) and HIV-1NL4–3 for 48 h. Cells were labeled with [35S]-cysteine/methionine for 1 h for IP with Gag and β-Actin antibody. (A) Evaluation of rescue by siRNA-resistant FLAG-RHA by IP (top panel). Immunoblot of total cell protein with indicated antiserum verified RHA down-regulation, expression of siRNA-resistant FLAG-RHA and equal protein loading, respectively (bottom panel). (B) Evaluation of rescue by siRNA-resistant FLAG-RHA K417R by IP (top panel). Immunoblot of total cell protein with indicated antiserum verified RHA down-regulation, expression of siRNA-resistant FLAG-RHA K417R and equal protein loading, respectively (bottom panel).
RHA is not necessary for packaging of virion RNA or LysRS
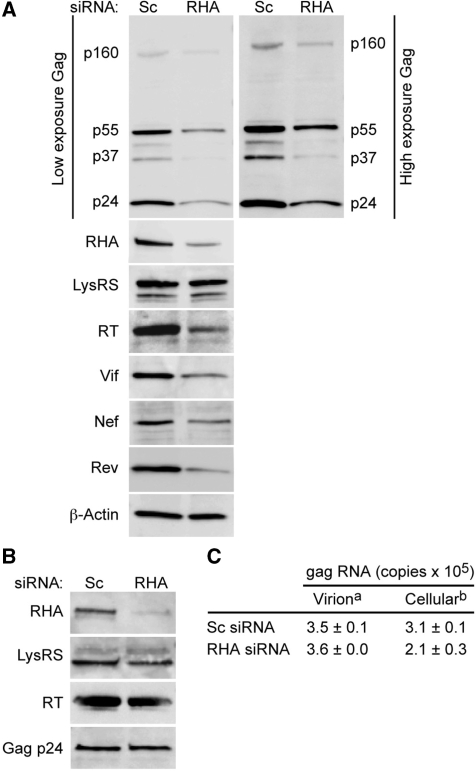
Next, we examined virions produced from cells treated with RHA siRNA or non-silencing control siRNA. As described in the last section, RHA was down-regulated, verified by immunoblot, HIV-1NL4–3 was transfected with a second treatment of siRNA and after 48 h, progeny virions were harvested by centrifugation and virion-associated RNA was quantified by RT-real-time PCR. Immunoblotting verified down-regulation of RHA but no change in Actin or cell-associated Lys-tRNA synthetase (LysRS) (Figure 7A). However, HIV-1 Gag p55 and the processed forms of the Gag precursor protein p37 and p24 were reduced by RHA down-regulation. High exposure of the Gag immunoblot revealed that Gag-Pol polyprotein p160 is also reduced (right panel). As expected RT immunoblot detected similar reduction in RT p66, which is proteolytic processed from Gag-Pol p160. Vif, Rev and Nef immunoblot revealed concordant reduction in these virus proteins, suggesting that RHA is important for translation of genome-length, single-spliced and double-spliced mRNA.
Figure 7.
RHA down-regulation reduces virus protein but not HIV-1 RNA packaging. HEK 293 cells were transfected consecutively with scrambled (Sc) or RHA (RHA) siRNAs and HIV-1NL4–3 provirus and cellular protein and RNA were isolated. Gag p24 ELISA on cell-free medium was performed. (A) Immunoblot of total cell protein (10 µg) with antiserum to Gag, RHA, LysRS, Vif, Nef, Rev or β-Actin. (B) Immunoblot of virion preparation (equivalent to 5 ng of p24) with antiserum to RHA, LysRS, RT, Gag. (C) RT-real-time PCR determined copy number. aGag copy number in virion preparation equivalent to 25 ng of p24. bCellular gag RNA copy number standardized to β-actin RNA.
Next, equivalent quantities of virions were subjected to immunoblot (Figure 7B) and extraction of virion-associated RNA (5 ng Gag for immunoblot and 25 ng Gag for RNA extraction as measured by Gag ELISA). Gag immunoblot verified equivalent virion loading (Figure 7B, bottom panel). As expected, virions produced from cells treated with RHA siRNA are deficient in RHA (Figure 7B, top panel). However they are not deficient in cellular cofactor LysRS (23) nor processed RT (Figure 7B, middle panels). RT-real-time PCR with gag primers detected similar amounts of virion-associated RNA (Figure 7C). In this particular experiment, RT-real-time PCR on total cellular RNA detected a minor reduction in cell-associated gag RNA, Figure 7C. The results indicate that viral RNA packaging efficiency was not reduced by RHA down-regulation. In sum, analysis of HIV-1NL4–3 demonstrated that RHA down-regulation reduces the efficiency of HIV-1 RNA translation. However, virion associated LysRS and viral RNA are not reduced, suggesting that RHA is not necessary for their packaging into RHA-deficient particles.
RHA is important for HIV-1 infectivity on primary cells
To examine the effect of RHA down-regulation on HIV-1 infectivity, equivalent cell-free virion preparations were used for infection of human PBMCs or a HeLa-based Luciferase reporter cell line, TZM-bl integrated with HIV-1 LTRluc reporter gene (13–16). Gag ELISA was performed on the supernatant medium from producer cells treated with RHA siRNA or Sc siRNA, and infections were performed with 2 ng Gag aliquots of virions deficient in RHA or containing RHA. Virus growth on PBMC in triplicate experiments determined that RHA-deficient virions exhibit reduced Gag production in PBMC (Figure 8). Reduced virus growth was observed at days 6, 9 and 12, indicating virus replication is impaired (P < 0.0005, 0.0008, 0.0174, respectively, Figure 8A). Similarly, virus infectivity on TZM-bl cells was reduced and expression of siRNA-resistant RHA was sufficient to rescue HIV-1 infectivity (Figure 8B). The results demonstrate that RHA down-regulation impaired the infectivity of progeny virions. In conclusion, RHA plays two critical roles in the virus producer cell: (i) RHA is necessary for efficient HIV-1 translation; (ii) RHA assembled into HIV-1 virions promotes infectivity on primary human cells.
DISCUSSION
RHA is an important host factor
This report determined that RHA is necessary for two aspects of HIV-1 biology: RHA present in the infected cell facilitates efficient translation of viral mRNA and RHA assembled into virus particles promotes infectivity of progeny virions. The virions are not deficient in viral RNA, processed RT, nor the cellular cofactor LysRS. The results open the new issue of the possible interrelationship of RHA promoting viral mRNA translation and production of infectious virions.
First, the results demonstrated that translation of HIV-1 gag mRNA is reliant on the ATP-dependent helicase activity of RHA. A straightforward model is that RHA interaction promotes RNP rearrangement to open the complex 5′ UTR to efficient ribosome scanning (Figure 9). We invoke ribosome scanning model instead of an internal ribosome entry site (IRES)-related mechanism because RHA has been shown to promote cap-dependent, but not cap-independent translation (5) and because the retroviral PCEs do not substitute for IRES in a bicistronic reporter RNA (5). We speculate that all HIV-1 transcripts require RHA for efficient translation for two reasons: (i) all HIV-1 transcripts contain the similar highly complex 5′ UTR (1), which includes the RU5 region that conveys PCE reporter activity and interacts with RHA (Figure 2) and was previously shown to be sufficient for RHA-dependent PCE activity (5); (ii) Vif, Rev and Nef protein levels are reduced by RHA down-regulation, consistent with translation down-regulation (Figure 7). These observations open new questions. First, what features of HIV-1 RU5 and possibly distal sequences are sufficient for the specific interaction with RHA. Second, is the ATP-dependent helicase activity of RHA necessary to promote virus infectivity. Attractive speculation is that helicase-dependent rearrangement of the 5′ UTR involving RHA changes the activity of other cis-acting replication sequences in the 5′ UTR, including the packaging signal.
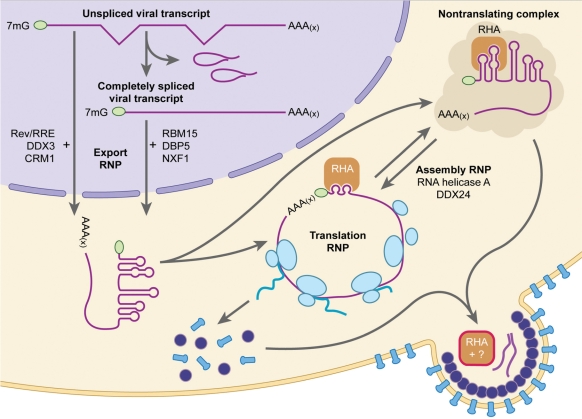
Figure 9.
Model summarizing that DExD/H proteins contribute to many steps in retroviral posttranscriptional gene expression and coordination of virus assembly. Nuclear export of HIV-1 Rev/Rev responsive element (RRE)-dependent HIV-1 RNA requires DDX3 with CRM1 nuclear export receptor (27). Alternatively, the nuclear export ribonucleoprotein (RNP) of completely spliced viral RNA involves superfamily members RBM15 and DBP5 and NXF1 nuclear export receptor (28,29). Efficient viral translation requires DHX9/RNA helicase A [herein and (5)]. RNA helicase A recognizes structural features of virus 5′ RNA terminus and possibly distal structures, and facilitates efficient protein synthesis in an ATP-dependent manner (2). RNA helicase A also assembles into virions and supports virus infectivity, perhaps by promoting rearrangement of the viral RNP for reverse transcription or by carrying in a host cofactor (12). DDX24 promotes packaging of HIV-1 RNA (30).
A simple model invokes an inverse relationship between translation and packaging efficiency. Herein RHA neutralizes structural features of the 5′ UTR to promote efficient ribosome scanning. However, the structural rearrangement could compromise the integrity of the RNA packaging signal. The predicted outcome is reduced virion RNA content that engenders lower virion infectivity. Unexpectedly, the prediction of less viral RNA content was not validated by RHA down-regulation assays with highly sensitive quantitative RT-real-time PCR (Figure 7) or RNase protection assay (12); viral RNA packaging was not impaired. A caveat to the experimental approaches is the down-regulation of Gag protein synthesis changes the intracellular ratio of assembling virions to viral RNA. The reduction in Gag is not in proportion to viral RNA level; the increased steady-state virion precursor RNA and may eliminate the need for efficiency in the RNA assembly process, and erase detection of a packaging phenotype. In sum, alternative experiments are needed to measure whether or not the ATP-dependent helicase activity affects the activity of the packaging signal, and possibly other cis-acting replication sequences in the 5′ UTR.
Particle-associated RHA may be important for infectivity by affecting the efficiency of reverse transcription, as first suggested by Liang (12). In our report, immunoblots verified that RHA down-regulation did not reduce processed RT in the RHA-deficient particles (Figure 7). Possibly, RHA helicase activity may be necessary for proper conformation of the primer binding site, accessibility to tRNA, or primer annealing. Our immunoblots demonstrated that RHA down-regulation does not eliminate packaging of the LysRS, which is involved in tRNA packaging (23). Another option is RHA supports virus infectivity independently of helicase activity; in this model RHA acts as a scaffold for accessory host protein or RNA cofactor (Figure 9).
RHA may also operate a mechanistic link between HIV-1 RNA trafficking and infectious virion assembly. Results with Ty3 show that trafficking the viral RNP through processing bodies promotes assembly of virus-like particles (24) and yeast strains deficient in DhhP1, a DEAD box protein that is a central protein for translation repression and targeting mRNAs to these RNA storage granules, are deficient in retrotransposition (24,25). Likewise, RHA trafficking between polysomes (2,5) and stress granules (Bolinger and Boris-Lawrie, unpublished results) may coordinate viral RNA fates as mRNA template and translationally inactive virion precursor RNA that is assembled into nascent particles (Figure 9). This notion is supported by observations that translation of cytoplasmic HIV-1 RNA is not necessary for its assembly into progeny virions and may be recruited in a translationally repressed form (26).
The role of yeast DhhP1 and metazoan RHA supports the growing realization that DExD/H proteins contribute to many, if not all steps in retroviral posttranscriptional gene expression and coordination of virus assembly. As summarized in Figure 9, DExD/H superfamily members DDX3, RBM15 and DBP5 are components of ribonucleoprotein (RNP) complexes that promote nuclear export of viral RNA (27–29). Superfamily members DhhP1, RHA and DDX24 facilitate viral RNA trafficking and coordination of virus assembly (12,24,30). In conclusion, further study of RHA in retroviruses is expected to illuminate the poorly understood issue of how the translation and assembly processes are coordinated. Historically the virus-host interactions that contribute to coordination of the translation and assembly processes have been of great interest (31).
RHA response element is conserved across Retroviridae
PCE reporter assays determined that the RU5 region of the HIV-1 5′ LTR and the HTLV-1 LTR facilitate HIV-1-based reporter mRNA translation. These new findings suggest that previous studies of reporter constructs containing RU5 sequences may be explained by RHA translational stimulation. In particular, reports of increased HIV-1 LTR-luciferase activity (9,32) and virion protein production by RHA overexpression (10) may be attributed to increased translation. Likewise, HTLV-1 RU5 has been used in retrovirus vectors to increase translation efficiency (33,34) and we speculate that this activity is attributable to increased RHA-dependent PCE activity. SNV PCE has been employed in HIV-1 based lentivirus vector to increase transgene translation (35).
The identification of PCE activity in the RU5 region of HTLV-1 and HIV-1 identifies conservation across six of the seven genera of the Retroviridae (3–5,36). Future experiments with 5′ LTR sequences from the Epsilon genus are necessary to address possible conservation of PCE activity across all genera of this virus family. In conclusion, the identification of RHA-dependent PCE activity in cellular junD RNA and in six of seven genera of Retroviridae suggests conservation of this translational control mechanism among vertebrates, and convergent evolution of Retroviridae to utilize this host mechanism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health National Cancer Institute RO1CA108882 and P30CA100730 to K.B.L., P01CA16058 to Ohio State University Comprehensive Cancer Center, and American Heart Association predoctoral fellowship to A.S. Funding for open access charge: OHIOlink.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Tim Vojt for illustrations and J. Marcela Hernandez for RNA IP experiments, Karin Musier-Forsyth for LysRS antiserum, NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for reagents summarized in Supplementary Data.
REFERENCES
- 1.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 3.Butsch M, Hull S, Wang Y, Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hull S, Boris-Lawrie K. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 2002;76:10211–10218. doi: 10.1128/JVI.76.20.10211-10218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, Forget M, Green PL, Boris-Lawrie K. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007;35:2629–2642. doi: 10.1093/nar/gkm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell RA, Zeng Y, Erlwein O, Cullen BR, McClure MO. The R region found in the human foamy virus long terminal repeat is critical for both Gag and Pol protein expression. J. Virol. 2001;75:6817–6824. doi: 10.1128/JVI.75.15.6817-6824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts TM, Boris-Lawrie K. Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J. Virol. 2003;77:11973–11984. doi: 10.1128/JVI.77.22.11973-11984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman FD, Malani N, Fernandes J, D'O;rso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T. A role of RNA Helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, Wong-Staal F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl Acad. Sci. USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 12.Roy BB, Hu J, Guo X, Russell RS, Guo F, Kleiman L, Liang C. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J. Biol. Chem. 2006;281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 13.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'B;rien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol. 2008;82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie L, Yamamoto B, Haoudi A, Semmes OJ, Green PL. PDZ binding motif of HTLV-1 Tax promotes virus-mediated T-cell proliferation in vitro and persistence in vivo. Blood. 2006;107:1980–1988. doi: 10.1182/blood-2005-03-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hull S, Boris-Lawrie K. Analysis of synergy between divergent simple retrovirus posttranscriptional control elements. Virology. 2003;317:146–154. doi: 10.1016/j.virol.2003.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 20.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 21.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierro-Monti I, Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 23.Cen S, Khorchid A, Javanbakht H, Gabor J, Stello T, Shiba K, Musier-Forsyth K, Kleiman L. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 2001;75:5043–5048. doi: 10.1128/JVI.75.11.5043-5048.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beliakova-Bethell N, Beckham C, Giddings TH, Jr, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 2005;15:641–654. doi: 10.1101/gr.3739005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butsch M, Boris-Lawrie K. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 2000;74:11531–11537. doi: 10.1128/jvi.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Lindtner S, Zolotukhin AS, Uranishi H, Bear J, Kulkarni V, Smulevitch S, Samiotaki M, Panayotou G, Felber BK, Pavlakis GN. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006;281:36915–36928. doi: 10.1074/jbc.M608745200. [DOI] [PubMed] [Google Scholar]
- 29.Zolotukhin AS, Uranishi H, Lindtner S, Bear J, Pavlakis GN, Felber BK. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp782. [Epub 28 September, 2009] PMID: 19786495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J, Rong L, Zhou Y, Roy BB, Lu J, Abrahamyan L, Mouland AJ, Pan Q, Liang C. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology. 2008;375:253–264. doi: 10.1016/j.virol.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 31.Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadler AJ, Latchoumanin O, Hawkes D, Mak J, Williams BR. An antiviral response directed by PKR phosphorylation of the RNA helicase A. PLoS Pathog. 2009;5:e1000311. doi: 10.1371/journal.ppat.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attal J, Theron MC, Kann G, Bolifraud P, Puissant C, Houdebine LM. The stimulation of gene expression by the R region from HTLV-1 and BLV. J. Biotechnol. 2000;77:179–189. doi: 10.1016/s0168-1656(99)00221-7. [DOI] [PubMed] [Google Scholar]
- 34.Attal J, Theron MC, Taboit F, Cajero-Juarez M, Kann G, Bolifraud P, Houdebine LM. The RU5 (‘R’) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 1996;392:220–224. doi: 10.1016/0014-5793(96)00815-0. [DOI] [PubMed] [Google Scholar]
- 35.Yilmaz A, Fernandez S, Lairmore MD, Boris-Lawrie K. Coordinate enhancement of transgene transcription and translation in a lentiviral vector. Retrovirology. 2006;3:13. doi: 10.1186/1742-4690-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 2000;74:8111–8118. doi: 10.1128/jvi.74.17.8111-8118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.