Abstract
Misfolded proteins are caused by genomic mutations, aberrant splicing events, translation errors or environmental factors. The accumulation of misfolded proteins is a phenomenon connected to several human disorders, and is managed by stress responses specific to the cellular compartments being affected. In wild-type cells these mechanisms of stress response can be experimentally induced by expressing recombinant misfolded proteins or by incubating cells with large concentrations of amino acid analogues. Here, we report a novel approach for the induction of stress responses to protein aggregation. Our method is based on engineered transfer RNAs that can be expressed in cells or tissues, where they actively integrate in the translation machinery causing general proteome substitutions. This strategy allows for the introduction of mutations of increasing severity randomly in the proteome, without exposing cells to unnatural compounds. Here, we show that this approach can be used for the differential activation of the stress response in the Endoplasmic Reticulum (ER). As an example of the applications of this method, we have applied it to the identification of human microRNAs activated or repressed during unfolded protein stress.
INTRODUCTION
Quality control of the proteome begins during protein synthesis (1–3) and ends with stress responses that manage misfolded proteins throughout the cell (4,5). Accumulation of misfolded proteins in the lumen of the endoplasmic reticulum is managed by the unfolded protein response (UPR) (6) and unfolded polypeptides in the cytosol induce the heat-shock response (7,8). Unrecoverable misfolded proteins in different cellular compartments are tagged for degradation by the ubiquitin–proteasome system (UPS) (9,10). In addition to the UPR and the UPS, a third unfolded protein response has been described in the mitochondria (11–13). A detailed understanding of the coordination among proteome quality control mechanisms, and of relative hierarchies among existing stress control pathways, is lacking.
Typically, stress responses to the accumulation of unfolded proteins have been studied via two different experimental strategies: the utilization of single mutant proteins known to aggregate, or the incubation of cells with analogues of proteinogenic amino acids. Historically, the use of amino acid analogues like azetidine carboxylic acid or canavanine has been extremely useful for the identification of components of the heat-shock response and the UPS (14,15). Canavanine and ACA cause cell toxicity in a large number of experimental systems. These toxic effects are generally attributed to their ability to infiltrate the proteome, although the incubation of cells with amino acid analogues probably can affect other cellular pathways.
Generally speaking unfolded protein accumulation in the Endoplasmic Reticulum (ER) causes the release of the ER-resident chaperone Grp78 (BiP) from its membrane localization (16). Release of BiP induces three adaptive responses that require activation of Xbp-1, phosphorylation of eIF2α and proteolytic cleavage of ATF6 (16,17). If adaptive responses fail to reduce ER stress pro-apoptotic pathways are induced through the activation of the transcription factor CHOP. The mechanisms that control the necessary balance between adaptive and pro-apoptotic responses are still poorly understood (18–20).
Studies of the UPR, or of the ER-associated protein degradation pathway (ERAD), typically rely on protein mutants that are aberrantly glycosylated in the ER and are thus targeted for degradation by the proteasome. An example of such a protein is human α1-antitrypsin variant null Hong Kong (21). Studies of the UPR-ERAD pathways have revealed the extraordinary complexity of the glycosylation machinery of the ER, and the co-existence of parallel stress mechanisms that respond to different glycosylation signals (16,22–25). Thus, the use of single protein markers, albeit extremely informative, may not be enough to activate the whole set of UPR-ERAD components and interactions.
In this work, we present a new approach to generating proteome errors and unfolded protein stress in cells. We have engineered a battery of mutagenic tRNAs that introduce a range of 10 different mutations in a human cell type and in a vertebrate embryonic model. In order to quickly follow the effect of each tRNA, we have constructed a GFP protein that is not affected by the mutagenic tRNAs and can be used as a marker of the overall physiological state of the cells. This method allows for the controlled induction of generalized proteome defects in a direct manner, without the potential for other secondary effects. This strategy also permits the uniform introduction of different types of mutations throughout the proteome, which can be applied to the analysis of the timing and grade of different stress responses, as well as to the identification of new links between those responses.
Here, we show that these mutagenic tRNAs efficiently and specifically cause randomized proteome mutations and increasing levels of stress response activation depending on the mutation being introduced. As an example, we describe the utilization of this method for the identification of human microRNAs (miRNAs) that may be involved in the induction of apoptosis during late stages of the unfolded protein response.
MATERIALS AND METHODS
Construction of tRNA chimeras
A DNA fragment of 0.7 kb corresponding to the gene encoding human wild-type tRNASer and its flanking regions was amplified by PCR from genomic DNA and cloned into vector pCR2.1. Chimeras of tRNASer were built by QuikChange site-directed mutagenesis from Stratagene by substituting the anticodon wild type by 18 other nucleotide triplets (to amber stop codon UAG or 17 different anticodons, Supplementary Table S1). Similarly, QuikChange site-directed mutagenesis was used to replace codon 65 in GFP by 18 other nucleotide triplets. To restrict the decoding specificity of the chimeras to codon 65, we took advantage of the degeneracy of the genetic code and chose anticodon/codon combinations that were unique and not represented elsewhere within the sequence of the GFP. Since methionine and tryptophan are encoded by a single codon, these two chimeras were purposely removed from our study.
Amino acids quantitative analysis
Samples were hydrolysed in 6 M HCl for 16 h at 110°C and filtered. Excess HCl was evaporated to complete dryness and the pellet was resuspended in 20 mM HCl. Filtered samples were derivatized using AccQ-Tag (Waters®) (26,27), and derivatized amino acids were quantified by HPLC (WATERS600, Waters®) [UV detection λ = 254 nm, detector W-2487 (Waters)].
Cell culture
Hek293 cells were grown in DMEM with 10% fetal bovine serum (FBS), 100 U/ml penicillin, an 100 µg/ml streptomycin at 37°C, 5% CO2. For transient transfections cells (60–70% confluence) were transfected with 10 µg DNA/ml using lipofectamine (Invitrogen), following the manufacturer’s instructions.
Chick in ovo electroporation
Eggs from White-Leghorn chickens were incubated at 38.5°C in an atmosphere of 70% humidity. Embryos were staged according to Hamburger and Hamilton (28). Chick embryos were co-electroporated with plasmids encoding chimeric tRNAs together with GFP. Briefly, plasmid DNA was injected into the lumen of HH Stage 11–12 neural tubes, electrodes were placed either side of the neural tube and electroporation carried out using an Intracel Dual Pulse (TSS10) electroporator delivering five 50 ms square pulses of 30–40 V. Transfected embryos were allowed to develop to the specific stages, then dissected, fixed and processed for immunohistochemistry or in situ hybridization. One hour prior fixation BrdU was injected into the lumen of neural tubes.
Metabolic labelling
Four hours after transfection with the plasmids encoding the chimeras, Hek293 cells were pulsed with l-azidohomoalanine 200 μM (methionine’s analogue) for 1 h in DMEM without methionine (Invitrogen). Cells were lysed and labelled with the Click-iT Protein Analysis Detection kit (Invitrogen). Labelled proteins were loaded on a 10% denaturing polyachrylamide gel and visualized by UV.
Cell proliferation and cell death
Cells were incubated in 96-well plate with 10 μl of WST-1 (Roche). The formazan dye produced by metabolically active cells was quantified by a scanning multiwell spectrophotometer at 450 nm after 1 h WST-1 addition. For quantification of cell death, cells were grown in 96-well plate and incubated with 5 μg/ml of propidium iodide (Sigma Aldrich), and measured by flow cytometry.
Statistical analysis
The list of ER proteins was obtained from the HERA database (http://www.mcb.mcgill.ca/∼hera/). Lists of proteins from the cytoplasm, mitochondria and nucleus families were obtained from the Gene Ontology project Consortium (2000) (2 July 2008 release), using the following GO identifiers: GO:0044428, GO:0005739, GO:0044444, respectively. To count codon frequencies, we used the CCDS database (2 July 2008 release, http://www.ncbi.nlm.nih.gov/ccds/), which provides the nucleotide sequences for human proteins. Finally, the protein family lists were mapped to the CCDS database using NCBI RefSeq identifiers. To assess the effect of several factors on fluorescence, we used general additive models, as implemented in the gam function in the R package mgcv S.N. (2004)17. The smoothing parameters were set via cross-validation. The threshold for statistical significance was 0.05.
RT–PCR analysis of XBP-1 splicing
Total RNA from Hek293 cells was extracted using Trizol reagent (Invitrogen) and treated with DNase 30 min at room temperature to remove traces of genomic DNA. RNAs were purified on RNAeasy columns (Qiagen) before being reverse transcribed using the Reverse Transcription System (Promega). cDNA was used as a template for PCR amplification across the fragment of the Xbp-1 cDNA bearing the intron target of IRE1α ribonuclease activity. Primer and PCR conditions used were those described by Lin et al. (19). A 289 base pair amplicon was generated form unspliced Xbp-1; a 263 base pair amplicon was generated from spliced Xbp-1. PCR products were resolved on a 2% agarose/0.5X TBE gel.
Semi-quantitative RT–PCR analysis
Total RNA was collected and cDNA retro-transcribed as described above. Primers used to amplify BiP, Chop and Rpl19 cDNA are those described by Lin and collaborators (19). Rpl19 mRNA encodes a ribosomal protein, whose transcription is not regulated by ER stress. It served as a marker of equal loading. PCR conditions for these three species were: 1 × 95°C for 1 min; 23 × 95°C for 1 min, 55°C for 1 min, 72°C for 1 min 30 s. Primers to amplify GFP cDNA are: 5′-CCACCTACGGCAAGCTGACC-3′ and 5′-TGCTGGTAGTGGTCGGCGAG-3′. PCR conditions were: 1 × 95°C for 1 min; 23 × 95°C for 30 s, 60°C for 1 min, 72°C for 1 min 30 s.
Western blot analysis
Hek293 cells were placed in cold lysis buffer (300 mM NaCl, 20 mM Tris, 10 mM EDTA, 2% Nonidet NP-40, plus protease inhibitors), scraped and centrifuged. Fifty to sixty μg of total cell lysate were separated by 10% SDS–PAGE and transferred to Immobilon-P (Millipore), followed by incubation with primary antibodies and the appropriate peroxidase-conjugated secondary antibodies and ECL detection (Pierce). Anti-phospho-eIF2α was from Invitrogen; anti-ATF-4 was from Affinity BioReagents; anti-GFP was from Immunokontact (IK); anti-β-tubulin was from Chemicon; monoclonal anti-Ubiquitin was from Biomol; anti-ATF-6 and polyclonal anti-Hsp72 were from Stressgen.
Immunohistochemistry and in situ hybridization
For immunostainings, chicken embryos were fixed 2–4 h at 4°C in 4% paraformaldehyde in PB, rinsed, and sectioned in a Leica vibratome (VT 1000S). Immunostainings were performed following standard procedures. For bromo-desoxy-uridine (BrdU) detection, sections were incubated in 2N HCl for 30 min followed by 0.1 M Na2B4O7 pH 8.5 rinses further PBT rinses and anti-BrdU incubation. The following antibodies were used: anti-green fluorescence protein (GFP) (Molecular Probes), anti-caspase3 (BD) and anti-BrdU (G3G4 from the Developmental Studies Hybridoma Bank). Alexa488- and Alexa555-conjugated anti-mouse or anti-rabbit antibodies (Molecular Probes) were used. After single or double staining, sections were mounted, and imaged using a Leica confocal microscope. Cell counting were done on 10–40 different sections of at least four different embryos after each experimental condition (n > 4). For in situ hybridization, embryos were fixed overnight at 4°C in 4% paraformaldehyde in PB, rinsed and processed for whole mount RNA in situ hybridization following standard procedures using probes for chick BIP (form the chicken EST project, UK-HGMP RC). Hybridization was revealed by alkaline phosphatase-coupled anti-digoxigenin Fab fragments (Boehringer Mannheim). Hybridized embryos were post-fixed in 4% paraformaldehyde, rinsed in PBT and vibratome sectioned.
Analysis of variations in microRNA populations caused by mutagenic tRNAs
Total RNA was isolated from Hek293 cells 48 h upon transfection with chimeric tRNAs. Five micrograms of labelled RNA from each treatment was hybridized to a microRNA microarray with 866 human miRNAs (Agilent Technologies, ref. 12.0 GA4471A) and imaged using GenePixPro (Axon Instruments). To remove differences in probe intensity between chips we used quantile normalization (29), and the bias was removed effectively. Median pixel intensity values were background subtracted and log-transformed. To compare the miRNA expression levels on each tRNA substitution versus wild-type tRNASer, we performed the moderated t-test (30) implemented in the Bioconductor LIMMA Library software (www.bioconductor.org/limma), and the P-values were adjusted following the Benjamini–Yekutieli procedure (31). Statistically significant changes in miRNA expression in cells transfected with tRNASer(Lys), tRNASer(His) or tRNASer(Ile) compared to wild-type tRNASer-transfected cells were determined (P-value threshold < 0.05). There is no significant variation in miRNA expression between non-transfected Hek293 cells and the same cells trasnfected with wild-type RNASer.
RESULTS
Design of a battery of mutagenic tRNAs
To construct the mutagenic tRNAs used in this study, we took advantage of the recognition mechanism between seryl-tRNA synthetase (SRS) and its cognate substrate tRNASer. Unlike most other cases, the specific recognition of tRNASer by SRS does not depend on the sequence of the anticodon of the tRNA (32,33). Thus, it is possible to modify the decoding sequence (the anticodon) of tRNASer without interfering with its capacity to be serylated (33) (Figure 1a). The result of such a modification is a chimeric tRNA that will be readily aminoacylated with serine by SRS, but will be utilized by the ribosome to translate codons complementary to the engineered anticodon, thus generating ‘X’ to S mutations randomly in the proteome.
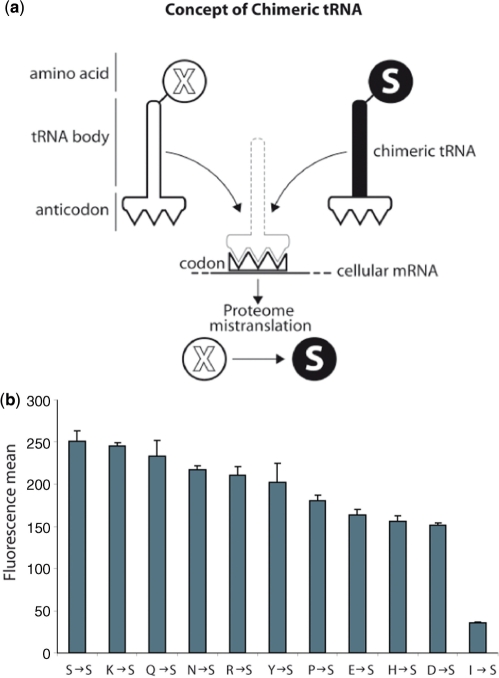
Figure 1.
Chimeric tRNA as adaptors to mistranslate specific codons. (a) Schematic figure of chimeric tRNA concept: chimeric tRNAs are engineered by substituting the wild-type anticodon in tRNASer to specific triplets coding for other amino acids. The chimeric tRNASer will only be charged with serine by seryl-tRNA-synthetase, no matter what the anticodon sequence is. Therefore, expression of chimeric tRNASer will produce mistranslated proteins. (b) GFP fluorescence was used as a marker to measure the impact of chimeric tRNA expression on the cell. Fluorescence was analysed by flow cytometry. A histogram depicting the different effect of each chimeric tRNA is shown. Mean values + standard deviations from three independent transfections are given.
We constructed a battery of engineered tRNASer that could be expressed and were active in human cells, and in a chick embryo neural system, after transfection with a plasmid carrying a gene coding for the modified tRNASer (Supplementary Figure S1). We initially constructed 17 tRNA variants designed to cause all possible X-S mutations with exception of W-S and M-S (Supplementary Table S1). We tested all the tRNAs for their ability to restore fluorescence of a reporter GFP coded by a gene where the essential residue serine-65 (S65) had been substituted for each of the 18 codons recognized by our engineered tRNAs. Ten out of the eighteen tRNAs could be shown to cause an increase in GFP fluorescence in cells expressing the corresponding GFP variant (Supplementary Figure S2).
In order to evaluate the relative impact of the mutagenic tRNAs in the cell, we constructed a new GFP variant that did not contain any of the codons being mutated by our tRNAs. In this way, cell fluorescence could be used as an indirect measure of the effect of each tRNA variant. Since tryptophan and methionine only use one codon, we could not use this strategy to evaluate tRNAs causing W-S, or M-S mutations. In cells containing this reporter GFP protein, and transfected with our battery of tRNAs, reductions of fluorescence ranging from 4 to 85% depending on the tRNA chimera used were seen (Figure 1b). Based on these initial results chimeric tRNAs harbouring anticodons decoding isoleucine [tRNASer(Ile)], histidine [tRNASer(His)] and lysine [tRNASer(Lys)], were chosen as representatives of, respectively, high, medium and low effect constructs and used for all further studies.
We performed amino acid quantitative analysis to confirm the introduction of amino acid substitutions in the proteome. We used two different proteins as markers to quantify the misincorporation efficiency of the chimeric tRNAs. One of the proteins was cytosolic GFP, and the second was human epidermal growth factor receptor (EGFR), which undergoes obligatory ER transit. Strikingly, amino acid analyses of purified GFP and EGFR from cells transfected with tRNASer(Lys), tRNASer(His) and tRNASer(Ile) showed that all the mutable residues had been replaced by serine (Table 1). Thus, our method is extraordinarily efficient at the introduction of mutations in the proteome. This is most likely due to the high cytosolic concentrations reached by the engineered tRNAs. These results, and the statistical analysis performed later (see below), show that all our tRNAs are equally active and efficiently used by the ribosome.
Table 1.
Amino acid quantitative analyses of reporter proteins EGFR (synthesized in ER-bound ribosomes) and GFP (synthesized in cytosolic ribosomes)
| Number of codons theoretically targeted | Decrease of targeted amino acid | Number of original serine residues | Increment in serine residues | |
|---|---|---|---|---|
| EGFR (ER transit) | ||||
| tRNASer Lys | 35 | 34.9 | 84 | 34.9 |
| tRNASer His | 11 | 11 | 84 | 11 |
| tRNASer Ile | 17 | 16.73 | 84 | 17.9 |
| GFP (no ER transit) | ||||
| tRNASer Lys | 19 | 19 | 16 | 18.7 |
| tRNASer His | 2 | 2 | 16 | 2 |
| tRNASer Ile | 5 | 4.96 | 16 | 5.81 |
‘Codons theoretically targeted’ refers to the total number of codons complementary to the anticodon of each chimeric tRNA. ‘Decrease of targeted amino acid’ values are obtained by subtracting the experimentally determined number of each of the amino acids whose codons are targeted by the three chimeric tRNAs (Lys, His and Ile) from the total number of the same residue expected from the gene sequence.
Similarly, the ‘Number of original serine residues’ refers to the number of serines expected in the protein from the gene sequence, while ‘increment in serine residues’ refers to the increment of serine residues experimentally determined in the reporter proteins after transfection with each chimeric tRNA. Close coincidence between the values in columns ‘Decrease of targeted amino acid’ and ‘Increment in serine residues’ indicates that chimeric tRNAs cause a complete substitution in their targeted codons after transfection.
Effects upon cell physiology and growth of mutagenic tRNAs
We evaluated the effects of the chimeric tRNAs upon general cell parameters such as transcription, protein synthesis levels, growth rate, and cell viability. GFP mRNA levels were found to be equal in all transfected cells, but GFP levels varied in correlation with the measured fluorescence (Figure 2a). Thus, as expected, the effect of chimeric tRNAs occurs at the level of gene translation. Incorporation of l-azidohomoalanine (a labelled derivate of methionine) confirmed that the observed down-regulation of translation affected the entire proteome in the same linear manner (Figure 2b).
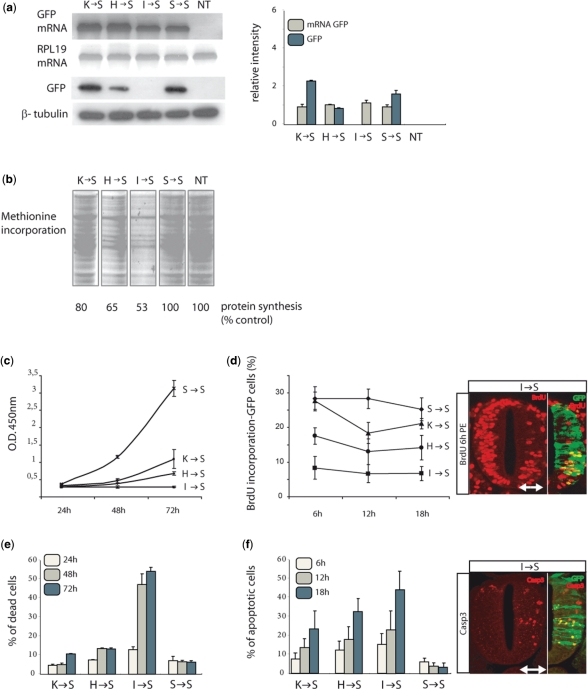
Figure 2.
General physiological effects of chimeric tRNAs in vitro and in vivo. (a) Semi-quantitative RT–PCR of wt GFP mRNA shows that transcription is not affected by the expression of chimeric tRNAs. RPL19 mRNA was used as a control. Immunoblotting with an α-GFP antibody shows that GFP synthesis is reduced proportionally to the reduction observed in cell fluorescence (Figure 1b). β-Tubulin was used as a control. Error bars represent standard deviations from five independent experiments. (b) Total protein synthesis analysed by pulse-label experiments shows a reduction in total protein production after expression of chimeric tRNAs proportionally to the reduction in cell fluorescence (Figure 1b). Controls from cells not transfected and cells transfected with wild-type tRNASer are shown. (c) Effect upon cell division of the chimeric tRNAs. Cells were counted at different times using flow cytometry. The data shows a reduction in growth rate for each tRNA that is to the reduction in cell fluorescence caused by the same molecule. (d) Effect in vivo of tRNASer(Ile) upon cell division. BrdU incorporation was measured in vivo in chicken embryos electroporated with chimeric tRNASer(Ile). A significant decrease in BrdU incorporation was observed in transfected embryos, indicating that the expression of chimeric tRNAs blocks cell proliferation during neural tube development. (e) Cell death and apoptosis measured in cultures transfected with chimeric tRNAs. Apoptosis was monitored by staining with propidium iodide, and the proportion of dead cells in each culture was measured by flow cytometry. The induction of cell death and apoptosis by each of the chimeric tRNAs tested was proportional to the reduction observed in cell fluorescence for the same tRNAs. (f) Induction of apoptosis in a chick neural cells expressing tRNASer(Ile) was monitored by caspase-3 immunostaining. Sections from electroporated embryos were stained with an α-caspase-3 antibody (6, 12 and 18 h after electroporation) and the number of labelled cells was quantified. A significant increase of caspase-3-positive cells is detected. The white arrow indicates the electroporated side of the embryo. GFP shows transfected cells. Data are means + SD (graph).
We then studied the effect of the mutagenic tRNAs upon cell viability and growth, both in mammalian cells and in chick embryos (28). The effect of the three chimeric tRNAs in the human cell line was again linear, and correlated with their relative effect upon protein synthesis. In agreement with our previous observations, chimeric tRNASer(Ile) caused the strongest inhibition of cell division in vitro (Figure 2c) and was active in vivo (Figure 2d). Similarly, the three tRNAs caused increasing levels of cell death in human cells (Figure 2e), and tRNASer(Ile) caused the strongest apoptosis induction in chick embryos 6 h after electroporation (Figure 2f).
Statistical analysis of the physiological effects caused by the mutagenic tRNAs
To further characterize our approach, we wanted to compare the observed effects of our tRNAs with the expected impact of each engineered mutation. We reasoned that the impact of each mutation would depend on the frequency of the mutated codon and on the nature of the amino acid substitution. If all the tRNAs used are functionally equal, values in sequence substitution matrices (PAM or BLOSUM, for instance) should correlate with the intensity of the effect of each mutagenic tRNA. Equally, the impact of the tRNAs should correlate with the frequency of the mutated codons in the genome.
To verify these hypotheses, we analysed the correlation of these two parameters (substitution likelihood and codon frequency) with the decrease in fluorescence caused by the ten tRNA chimeras. The substitution likelihood values were obtained from the substitution matrix Blosum62 (34). The GenBank database was used to calculate the frequency of the mutated codons in the human genome and in genes coding for proteins associated with the ER, nucleus, mitochondria and cytosol (35–37).
As expected, a significant correlation was found between the effects of the chimeric tRNAs and the substitution values for their respective mutations determined by the Blosum62 matrix (Figure 3a). This is evidence that all the tRNA variants are equally active in the cell, and that the relative effects that they cause are determined predominantly by the nature of the mutation that they generate.
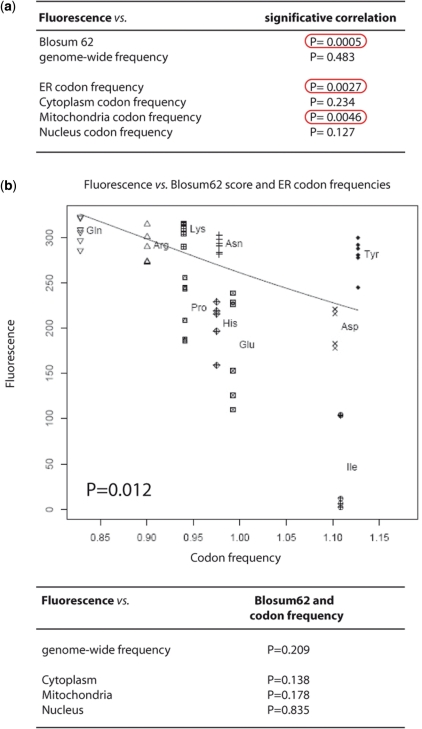
Figure 3.
Statistical analysis. (a) Univariate analysis between the effect of each of the 10 tRNAs tested (in fluorescence values) and BLOSUM62 matrix substitution values, genome-wide codon frequency and, ER-, mitochondria-, nucleus- and cytoplasm-codon frequencies in related genes. The P-values for statistical significance are shown. BLOSUM62 matrix values were significantly associated with fluorescence. A significant association was also found for the ER- and mitochondria-related genes. No significant association was found for the cytoplasm and nucleus protein families. (b) Multivariate analysis graph. Fluorescence versus BLOSUM62 score and ER codon frequencies (P = 0.012) are represented. Fluorescence values determined for each of the ten mutagenic tRNAs are plotted against the frequency of the codons recognized by the same tRNAs in ER-related genes. For each tRNA the results of five independent transfections are shown. In this analysis, only the ER codon population statistically correlates with the profile of differential effects. The P-values calculated for the comparison with genome-wide, cytoplasmic, mitochondrial and nuclear codon frequencies are shown.
We then compared the effect of the tRNAs tested with frequencies of their respective codons in the whole human genome. Surprisingly, fluorescence variation was not associated with genome-wide codon frequencies. We then identified genes coding for ER-, mitochondria-, nucleus- and cytoplasm-associated proteins. No significant association was found with cytoplasm- and nucleus-related genes, but a significant association was found with genes coding for ER- and mitochondria-targeted proteins. Importantly, these correlations are not caused by an over-representation in the ER of codons recognized by high-effect chimeric tRNAs, or by differences in the overall number of substitutions expected for each section of the proteome (Supplementary Table S2).
In a multivariate analysis of our data that compared codon frequencies and Blosum62 substitution values with fluorescence values a significant correlation was found only between the decrease in fluorescence caused by our tRNAs and the codon composition of ER-associated family of proteins (Figure 3b). Thus, the statistical analysis performed show that our tRNAs cause effects that correlate with the nature of the substitution that they generate. Moreover, in human Hek293 cells the effect of the tRNAs correlates better with the codon composition of ER-associated genes, indicating that the ER may be the principal agent of the cellular reaction to the mutagenic stress caused by the mutations in this cell type.
Induction of the unfolded protein response
We tested the activation of UPR caused by the chimeric tRNAs in our two experimental models. In cells expressing the chimeric tRNAs, a linear increment in activated forms of UPR components was observed. Figure 4a shows the activation of BiP and transcription factor CHOP in human cells after transfection with tRNASer(Lys), tRNASer(His) and tRNASer(Ile). Figure 4b shows the activation of Xbp-1 and ATF6, and phosphorylation of eIF2-α. Thus, chimeric tRNAs activate all the major components of the UPR. Moreover, in all cases the relative increases caused by the different mutating tRNAs are linear, and the activation of the response takes place between 10 and 24 h after transfection. In all cases, the relative increases caused by the different mutating tRNAs are equivalent, and the activation of the response takes place between 10 and 24 h after transfection.
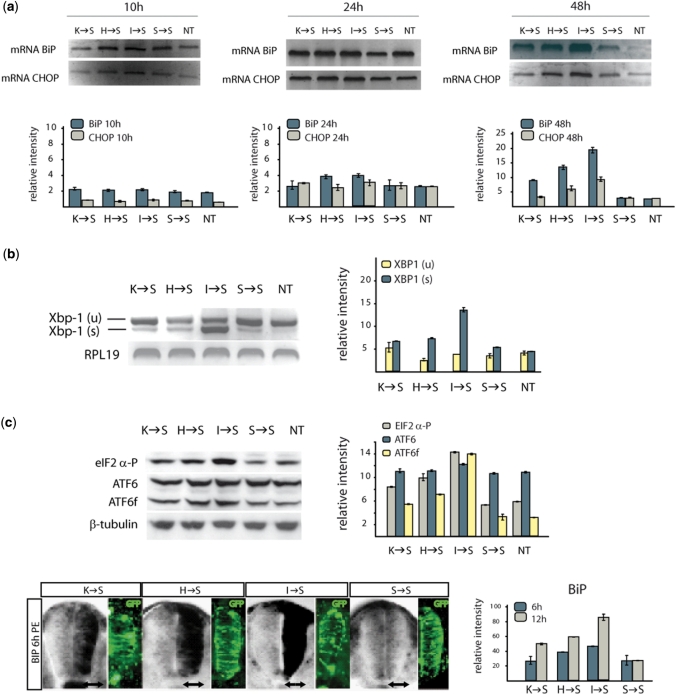
Figure 4.
Activation of UPR by chimeric tRNAs. (a) Kinetics of BIP and CHOP with persistent ER stress in vitro. Semi-quantitative RT–PCR (SQ-RT–PCR) was performed to check BIP and CHOP mRNA levels at the indicated different time points. (b) Activation of Xbp-1 and ATF6, and phosphorylation of eIF2α in cells expressing chimeric tRNAs. The presence of full-length (u) and spliced (s) Xbp-1 forms was checked by PCR. (c) BiP transcription induction in vivo. The expression levels of BiP (black staining) induced by each chimeric tRNA in a chicken embryo are proportional to the decrease in wt GFP fluorescence caused by the same tRNAs. Images are representative of three independent experiments 6 h post-electroporation (PE). GFP signal indicates the co-electroporated cells.
To obtain a second experimental reference, we monitored the activation of the UPR in a chicken embryo model. Figure 4c shows the time-dependent activation of BiP in the neural system of chick embryos electroporated with the three chimeric tRNAs. BiP was activated differentially by the three different tRNAs 6 h after electroporation. The activation pattern found is equivalent to the response detected in human cells.
Together, our results show that the UPR is reactive to the accumulation of translation errors in the proteome caused by the mutagenic tRNAs. The mutations caused by the tRNA chimeras promptly induce an activation that is linear and depending on the nature of the mutations being introduced. This responsiveness of the ER is coherent with the statistical correlation found between the effects of the tRNAs and the codon composition of ER-associated genes.
We also tested whether components of the heat-shock response and UPS were activated by monitoring levels of HSP72 and ubiquitin in the same cell type and in chicken embryos. As expected, ubiquitin and HSP72 levels increased significantly in both systems (Supplementary Figure S3). The activation of ubiquitin and HSP72 was intense, but equal for all tRNAs. Both components were activated later and uniformly by the three chimeric tRNAs (Supplementary Figure S3).
Thus, our method readily activates the two main stress responses known in eukaryotic cells. We could detect a linear activation of the UPR depending on the mutation being introduced, which may indicate a role of the UPR in the early response to proteome damage. We reasoned that we could take advantage of that response profile of the UPR to identify new UPR-related factors that may be induced with a similar pattern.
Identification of microRNA families responsive to proteome stress
Having observed that, in Hek293 cells, the components of the UPR reacted proportionally to the extent of proteome stress induced by our tRNAs we decided to use this response pattern to identify miRNAs that might be repressed or expressed as a consequence of the induced proteome stress. microRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression (38,39). They have been implicated in a number of biological processes (40,41) and have been recently emerged as important regulators of translation and cell stress responses (42,43).
Total RNA was isolated from Hek293 cells transfected with the chimeric tRNAs (tRNASer(Lys), tRNASer(His), tRNASer(Ile) and the control wild-type tRNASer). Five micrograms of labelled RNA purified from each transfected culture were hybridized to a microRNA array with 866 human miRNAs. We profiled global miRNA expression in Hek293 cells transfected with the chimeric tRNAs and we compared the miRNA differential expression with cells transfected with wild-type tRNASer. Importantly, no variation in miRNA expression signature was found between non-transfected Hek293 cells and the same cells after transfection with wild-type tRNASer.
The effect of the three chimeric tRNAs upon the microRNA population of transfected cells was identical to that observed for UPR components. A graded change in the concentration of specific miRNAs was seen, with the strongest effect being caused by tRNASer(Ile). Thus, expression of tRNASer(Lys), tRNASer(His) and tRNASer(Ile), respectively, induced 3, 131 and 205, and repressed 2, 22 and 43 miRNAs (Figure 5a). Although the number of miRNAs being affected by tRNASer(Lys) was modest, those identified were also affected by tRNASer(His) and tRNASer(Ile), and their relative change in concentration was significant and proportional to the physiological effects of the same tRNAs (Figure 5b).
Figure 5.
Microarray screen identifies miRNAs linked to UPR induction. Up- and down-regulated miRNAs populations in cells transfected with the mutagenic tRNAs. Number of miRNAs activated or repressed in cells transfected with tRNASer(Lys), tRNASer(His), tRNASer(Ile), compared with cells transfected with wild-type tRNASer. Graded activation and repression of miRNAs can be observed.
DISCUSSION
The identification of all the components of cellular stress responses and the elucidation of the functional and spatial connections between these pathways will require the application of different experimental approaches to induce proteome stress. We have implemented a controlled proteome-wide mutagenesis system through the use of engineered tRNAs that induces unfolded protein stress responses. This system is based in the recognition characteristics between tRNASer and SRS, and allows for the introduction of at least ten different ‘X to S’ substitutions throughout the proteome. Strikingly, amino acid analysis of proteins isolated from cells transfected with chimeric tRNASSer(Ile) indicate a complete substitution of the isoleucine residues coded by the targeted codon by serine in two different reporter proteins.
This substitution efficiency is likely due to the high expression of the tRNAs transfected, which can be detected in the cell up to five days after transfection (Supplementary Figure S1d). It is likely that chimeric tRNA levels could be experimentally regulated through modifications of their gene context or transfection conditions. It should also be noticed that the recognition of tRNAAla by alanyl-tRNA synthetase, or of tRNALeu by leucyl-tRNA synthetase, are also insensitive to the anticodon sequences of the tRNAs and could be used in a similar way to increase the range of mutations that can be introduced in the proteome (44–47).
Both in human cells and in chicken, the mutagenic effect of chimeric tRNAs elicits a response 6 h after transfection, which increases progressively up to 72 h. Interestingly, not all the stress pathways respond identically. Our results indicate that the proteasome-ubiquitin system responds to our set of mutagenic tRNAs uniformly, whereas the UPR reacts proportionally to the severity of the errors being accumulated in the whole proteome. Similarly, the UPS reaction is slower than the activation of the unfolded protein response. This late activation of the UPS may reflect a late onset of ERAD in our experimental models. Although the reasons for this differential activation of UPR and UPS are not clear they suggest that modifications in the method (for instance down regulating the levels of tRNAs) may allow for the differential activation of different stress responses.
As we have shown, the response profile that our approach generates can be useful in the identification of potential interactions or components of the induced stress pathways. Our approach can be applied to any cell type or organism susceptible of transfection. The extreme conservation of components of the translation apparatus facilitates the utilization of human tRNAs in other vertebrate systems, as seen in this work. Variations in the response to the stress caused by mutagenic tRNAs among different cell types could help identify the reasons behind the idiosyncratic sensitivities to proteome damage of different tissues, and may help explain why certain organs appear to be particularly susceptible to damage caused by protein aggregation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
BIO2006-01558 (to L.R.de.P.); BFU2007-60487 (to E.M.). Funding for open access charge: Spanish Ministry of Science and Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Leslie Nangle (Atyr Pharma, San Diego), Drs Manuel Palacín and Antonio Zorzano (IRB Barcelona) and Dr Alfred Goldberg (Harvard Medical School, Cambridge) for helpful discussions and suggestions.
REFERENCES
- 1.Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- 2.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 3.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 6.Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 7.Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Kostova Z, Wolf DH. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 10.Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 13.Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg AL, Howell EM, Li JB, Martel SB, Prouty WF. Physiological significance of protein degradation in animal and bacterial cells. Fed. Proc. 1974;33:1112–1120. [PubMed] [Google Scholar]
- 15.Knowles SE, Gunn JM, Hanson RW, Ballard FJ. Increased degradation rates of protein synthesized in hepatoma cells in the presence of amino acid analogues. Biochem. J. 1975;146:595–600. doi: 10.1042/bj1460595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 17.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J. Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 19.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Swulius MT, Moremen KW, Sifers RN. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc. Natl Acad. Sci. USA. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Despa F, Orgill DP, Lee RC. Molecular crowding effects on protein stability. Ann. NY Acad. Sci. 2005;1066:54–66. doi: 10.1196/annals.1363.005. [DOI] [PubMed] [Google Scholar]
- 24.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Cabral CM, Liu Y, Moremen KW, Sifers RN. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol. Biol. Cell. 2002;13:2639–2650. doi: 10.1091/mbc.E02-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen SA, Michaud DP. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 27.Bosch L, Alegria A, Farre R. Application of the 6-aminoquinolyl-N-hydroxysccinimidyl carbamate (AQC) reagent to the RP-HPLC determination of amino acids in infant foods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006;831:176–183. doi: 10.1016/j.jchromb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 31.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 32.Lenhard B, Orellana O, Ibba M, Weygand-Durasevic I. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 1999;27:721–729. doi: 10.1093/nar/27.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cusack S, Yaremchuk A, Tukalo M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S, Henikoff JG. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott M, Lu G, Hallett M, Thomas DY. The Hera database and its use in the characterization of endoplasmic reticulum proteins. Bioinformatics. 2004;20:937–944. doi: 10.1093/bioinformatics/bth010. [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 40.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 41.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 42.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 43.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 44.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 45.McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 46.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 47.Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M. Recognition nucleotides of Escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr) J. Mol. Biol. 1993;231:219–229. doi: 10.1006/jmbi.1993.1277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.