Abstract
Cytokinins are important plant hormones, and their biosynthesis most begins with the transfer of isopentenyl group from dimethylallyl diphosphate (DMAPP) to the N6-amino group of adenine by either adenylate isopentenyltransferase (AIPT) or tRNA–IPT. Plant AIPTs use ATP/ADP as an isopentenyl acceptor and bacterial AIPTs prefer AMP, whereas tRNA–IPTs act on specific sites of tRNA. Here, we present the crystal structure of an AIPT–ATP complex from Humulus lupulus (HlAIPT), which is similar to the previous structures of Agrobacterium AIPT and yeast tRNA–IPT. The enzyme is structurally homologous to the NTP-binding kinase family of proteins but forms a solvent-accessible channel that binds to the donor substrate DMAPP, which is directed toward the acceptor substrate ATP/ADP. When measured with isothermal titration calorimetry, some nucleotides displayed different binding affinities to HlAIPT with an order of ATP > dATP ∼ ADP > GTP > CTP > UTP. Two basic residues Lys275 and Lys220 in HlAIPT interact with the β and γ-phosphate of ATP. By contrast, the interactions are absent in Agrobacterium AIPT because they are replaced by the acidic residues Asp221 and Asp171. Despite its structural similarity to the yeast tRNA–IPT, HlAIPT has evolved with a different binding strategy for adenylate.
INTRODUCTION
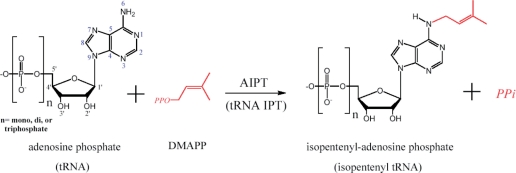
Cytokinins (CKs) are a class of plant hormones that promote cell division. They are primarily involved in seed development, root proliferation, leaf senescence and nutritional balance (1–3). Natural CKs are adenine derivatives with a side chain attached to the N6 atom. They are mainly isoprenoid CKs in higher plants but a few are aromatic CKs (4,5). According to the different types of the substrate, two CK biosynthetic pathways have been described (6). In the first pathway, adenylate isopentenyltransferase (AIPT) transfers the isoprenyl group from dimethylallyldiphosphate (DMAPP) to the N6-amino group of AMP, ADP or ATP (Figure 1). In the second pathway, tRNA–IPT catalyzes the CK formation by isoprenylation of certain tRNA at an adenine adjacent to the 3′-end of the anticodon, and subsequent degradation of the prenylated tRNA generates the CK nucleotide.
Figure 1.
Schematic diagram of the AIPT reaction. The acceptor substrate can also be an adenosine residue of tRNA in tRNA–IPT. Some atoms are numbered for their positions in the adenosine structure.
Each isoprenoid CK is distinguished by the uniqueness of the side chain, which is either an N6 isopentenyl side chain (iP-type) or an N6 hydroxylated isopentenyl side chain (zeatine-type). The side chain of a zeatine-type occurs in either the cis or trans configuration depending on stereoisomeric position of the hydroxyl group at the end of the isopentenyl chain. The activity of CKs may be related to their binding affinity to CK receptors. Studies by Spíchal et al. (7) demonstrated that iP and trans-zeatine have much higher affinities than cis-zeatine with Arabidopsis CK receptors (AHK3 and AHK4/WOL/CRE1).
AIPTs have been found in higher plants and some phytopathogenic microorganisms such as Rhodococcus fascians and Agrobacterium tumefaciens (8,9). The structure of Agrobacterium AIPT has been determined using X-ray crystallography (10). tRNA–IPT is found in a wider range of organisms. Structures of tRNA–IPT from Sacchromyces cerevisiae (yeast) and Escherichia coli, both in the apo-form and in complex with substrates, are also known (11–13). From these structures, a shared mechanism involving a P-loop in binding the donor substrate (DMAPP) and a conserved aspartic acid in abstracting a proton from the acceptor (N6 of adenine) is well established (10,12). Recently, several plant AIPTs from Arabidopsis thaliana, rice, mulberry (Morus alba) and hop (Humulus lupulus L) have been cloned and characterized (14–17). Previous studies have put forth several unsolved problems because plant AIPTs show some special characteristics that are unlike those of microorganisms (18). Regarding the substrate specificity of the enzymes, plant AIPTs belong to ATP/ADP IPTs, prefer ATP or ADP over AMP as the isopentenyl acceptor, and use DMAPP as the isopentenyl donor, whereas bacterial AIPTs use only AMP as the prenyl acceptor but use both DMAPP and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBDP) as donors (19). Due to the preferential usage of HMBDP, bacterial AIPTs can more efficiently synthesize highly active trans-zeatine in the A. tumefaciens-infected plant cells, resulting in tumorigenesis.
Interestingly, plant AIPT from M. alba, which showed 73% amino acid sequence identity to H. lupulus AIPT (abbreviated HlAIPT), is able to use CDP and GDP as the prenyl acceptors to produce non-adenine type CK (16). Since isopentenyltransferase reaction is the key rate-limiting enzyme step in CK biosynthesis, it is important to understand the binding affinity of plant AIPT with different nucleotides. Currently, little is known about the regulation mechanisms of AIPT apart from upregulation by nitrate or phosphate at the expression level (3,20). Finally, with regard to identifying an application in growth control in plants, it is useful to combine all functional and structural characteristics of AIPT in order to design regulators that may modulate CK biosynthetic activity (18). Here we solve the hop AIPT structure and focus on analyzing its interactions with various nucleotides in order to better understand the structural basis of different substrate specificity.
MATERIALS AND METHODS
Sample preparation and crystallization
All chemicals were purchased from Sigma-Aldrich except mentioned otherwise. The pQE80L plasmid containing the HlAIPT gene (329 amino acids) was a kind donation by Professor I. Abe of the University of Shizuoka, Japan. We overexpressed the protein containing the N-terminal hexahistidine tag (17) in Escherichia coli BL21 (DE3) (Novagen) and cultured the cells at 37°C until A600 reached 0.6. They were subsequently induced with 0.3 mM IPTG at 18°C overnight. The cell pellet was suspended in 50 mM potassium phosphate buffer (KPB), pH 8.0, containing 0.15 M NaCl, and disrupted on ice by French press equipment. The cell lysate was centrifuged at 43 667 g for 50 min. The supernatant was loaded onto a column of Ni-NTA resin (Qiagen) which contained Ni2+ as an affinity ligand. After washing with 50 mM KPB, pH 8.0, containing 0.15 M NaCl, the recombinant AIPT was eluted with a gradient of 0–0.25 M imidazole in the wash buffer. The protein-containing fractions were dialyzed at 4°C against a 30 mM KPB (pH 8.0) that contained 1 mM dithiothreitol (DTT). The sample was then applied to a mono-Q anion exchange column (GE Healthcare) and eluted with 1 M sodium chloride. The fractions were collected and dialyzed again with a buffer containing 20 mM HEPES (pH 7.0) and 1 mM DTT, plus 2 mM ATP. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis was performed to ensure purity, which was estimated to be more than 99% by staining, and the protein concentrated to 14 mg/ml by centrifugation using Amicon (10-kDa cutoff, Millipore) and stored at −80°C. We also constructed a vector to express a truncated version of the HlAIPT (aa 27–329), which lacks the N-terminal signal peptide. Site-directed mutagenesis and affinity measurement were based on this truncated HlAIPT.
For crystallization experiment of the full-length HlAIPT in complex with ATP (2 mM) and dimethylallyl S-thiodiphosphate (DMASPP, an analog of DMAPP; 5 mM), the sitting drop-vapor diffusion method was employed, with the protein-to-reservoir ratio being 0.5 µl:0.5 µl. Using crystallization kits for the initial screening, we obtained crystals of the enzyme-substrate complex with 0.2 M ammonium tartrate and 20% PEG 3350. The crystals were grown at 25°C and the reservoir volume was 100 µl. Before the use in X-ray data collection, crystals were soaked for 10 s in the reservoir solution containing 25% (v/v) glycerol as a cryoprotectant.
Data collection, structure determination and refinement
The 25–2.37-Å data set of the P212121 crystal was collected at beamline BL13B1 in NSRRC in HsinChu, Taiwan, using the ADSC Quantum 315 CCD detector. The data were processed and integrated with HKL2000 (21). The detailed statistics are listed in Table 1. Calculation of Matthews coefficient suggested that the asymmetric unit contains one protein molecule. The structure of HlAIPT was determined by molecular replacement method using the CNS program (22) and the yeast tRNA–IPT structure (12; PDB accession number: 2QGN) as a search model. Model building and refinement were carried out by using the programs XtalView (23) and CNS. Improvement of the model was guided by the sigma A-weighted 2Fo–Fc electron density maps.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P212121 |
| Unit cell (Å) | a = 45.42 |
| b = 75.89 | |
| c = 81.13 | |
| Beam line | NSRRC 13B1 |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 25 – 2.37 |
| Number of reflections | |
| Observed/unique | 50 322/12 174 |
| Redundancy | 4.1 (4.3)a |
| Rsym | 0.099 (0.442) |
| I/σ | 22.06 (4.24) |
| Completeness (%) | 99.7 (99.9) |
| Refinement statistics | |
| R/Rfree (%)b | 18.1/23.5 |
| r.m.s.d. | |
| Bond lengths (Å)/bond angles (deg) | 0.018/2.0 |
| Ramachandran plot | |
| Most favored/allowed residues (%) | 92.0/8.0 |
| Average B-factor (Å2)/number of atoms | |
| Protein | 31.1/2277 |
| ATP | 29.0/31 |
| Phosphate | 29.6/5 |
| Water | 38.3/198 |
aValues in parentheses indicate the specific values in the highest resolution shell.
bRfree = R factor calculated using 5% of the resolution data chosen randomly and omitted from the start of refinement.
Isothermal titration calorimetry
ATP and all nucleotides were dissolved in the dialysis buffer. Except for GTP, experiments were conducted at 25°C and consisted of 19 2-µl injections of 0.2 mM nucleotide ligand into a 0.2 ml sample cell containing 16 µM of the truncated HlAIPT (aa 27–329). Injections were made over a period of 4 s with 150-s intervals between injections. For the GTP experiment, 16 0.5-µl injections of 0.3 mM GTP, subsequent 16 1-µl injections and seven 2.5-µl injections were made. The sample cell was stirred at 1000 r.p.m. Data were acquired on an iTC200 microcalorimeter (Microcal, Northhampton, MA, USA) and analyzed with the ORIGIN v.7.0 software provided with the microcalorimeter.
RESULTS AND DISCUSSION
The overall structure of H. lupus AIPT
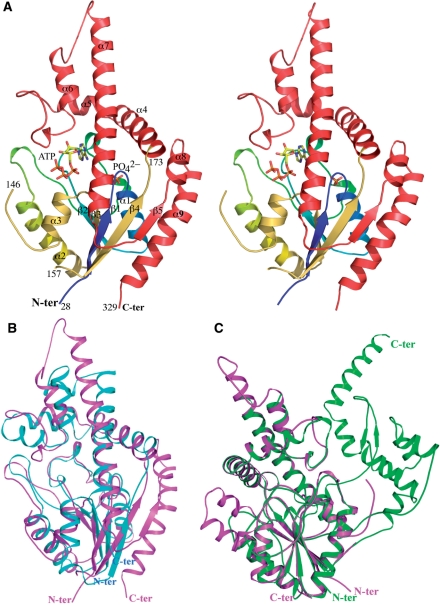
The refined structure at 2.37-Å resolution of the HlAIPT contains one protein molecule composed of amino acid residues 28–146 and 157–329, as shown in Figure 2A. It shares a similar core protein fold with the previously determined structures of Agrobacterium AIPT (10) and yeast and E. coli tRNA–IPT (11–13). In the HlAIPT crystal, the electron densities for residues 1–27 and residues 147–156 were not visible, while the region of residues 1–36 has been considered as the chloroplast targeting sequence (17). Although the crystal was obtained in the presence of both ATP and DMASPP, only ATP but not DMASPP was seen in the active site. However, a phosphate ion in the active site was observed, which presumably corresponds to the α-phosphate location of the donor substrate.
Figure 2.
Overall structure of HlAIPT. (A) The HlAIPT–ATP complex. The N-terminal core domain (aa 1–173) and C-terminal variable domain (aa 174–329) are shown in rainbow and red, respectively. In this structure, residues 1–27 and 146–154 are disordered. The HlAIPT is compared with the structures of Agrobacterium AIPT (B) and S. cerevisiae tRNA–IPT (C). The extra domain of the tRNA–IPT, which is involved in interactions with nucleotides in the anticodon stem of tRNA, does not exist in HlAIPT. The superimposition of structures employed the program Swiss-PdbViewer (27). All structural figures were generated by using PyMol (28).
Attempts to produce co-crystals with different substrates (ATP/Mg2+ and ATP/Mg2+/DMASPP), were not successful. We also used the truncated HlAIPT (aa 27–329), but so far we have not been able to obtain a suitable crystal from this smaller version of protein for X-ray diffraction. Interestingly, we had to include the 2 mM ATP in the dialysis buffer to stabilize the full-length HlAIPT, but the truncated protein appeared to be more stable, for which the addition of ATP was not necessary. The tightly bound ATP molecule was clearly seen in the first Fourier maps, as well as in the omit maps calculated using the refined model (Supplementary Figure S1).
According to the results of a Dali search, the core region (aa 28–173) of HlAIPT adopts a structural fold homologous to that of the NTP-binding kinase family, containing a four-stranded parallel β-sheet (β1–β4) flanked by α (α1–α3) and 310 helices on both sides (Figure 2A). The C-terminal region (aa 174–329) is mainly α-helical (α4–α9), with a β-strand (β5) parallel to the central β-sheet (β1–β4). The C-terminal region is more variable among different AIPT proteins when the structures of Agrobacterium AIPT (10) and yeast tRNA–IPT (12) are compared (Figure 2B and C). Superimpositions of the three structures also reveal that the core region of HlAIPT bears greater similarity to that of the tRNA–IPT than Agrobacterium AIPT. Moreover, the charge distribution on the surface of the three complex structures also shows a similar result (Supplementary Figure S2).
Active site
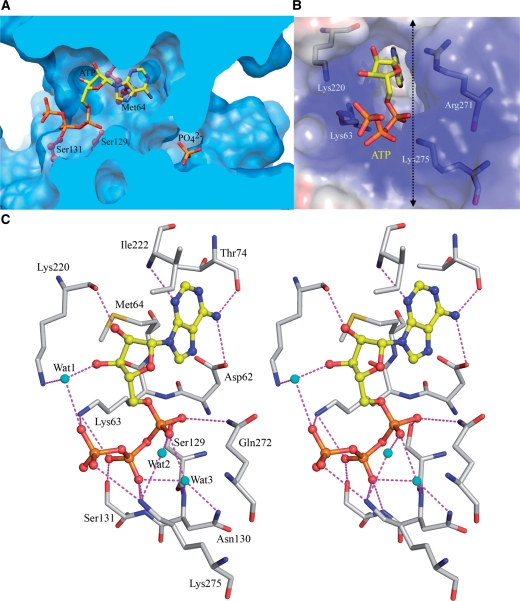
In the complex structure of AIPT, the ATP molecule is located on one side of the channel with the adenine ring buried in a deep pocket (Figure 3A), and the phosphate ion bound to another side of the channel, which involves a so-called P-loop found in all NTP binding kinases (37-GATGTGKS-44 in HlAIPT). The sugar-phosphate moiety of ATP is not deeply buried when bound to the enzyme, but particularly about the phosphate groups the opening of the channel is surrounded by a number of positively charged residues (Figure 3B). There is an extensive hydrogen-bond network binding the substrate with HlAIPT (Figure 3C). The highly conserved Asp62 is directly hydrogen-bonded to the N6 position of adenosine. The backbone N and O of Ile222 and Thr74 form hydrogen bonds with the N1 and N6 positions of adenosine, respectively. The backbone and side chain of Lys220 make direct hydrogen bond and indirect water-mediated interaction with O2' and O3' hydroxyl groups in the ribose of ATP. In addition, the side chains of Met64, Leu268, Ala221 and Ile222 form van der Waals interactions with the adenine ring and the ribose.
Figure 3.
The active site of HlAIPT. (A) Molecular surface of HlAIPT. The enzyme forms a central reaction channel. The cross-sectional surface, which is cut at the dashed line in (B), is colored light blue. The ball-stick model shows the side chains of Met64, Ser129 and Ser131. The ATP molecule and phosphate ion are shown as rods. (B) A cluster of positively charged residues surround the open end of the ATP-binding channel. (C) Stereoscopic view of ATP binding to the HlAIPT. Hydrogen bonds and charge–charge interactions are shown as dashed lines.
The α-phosphate group of ATP forms hydrogen bonds with the side chains of Ser129 and Gln272 as well as two water-mediated interactions. At the N-terminal end of helix α3, the β-phosphate makes two direct hydrogen bonds with the backbone and side chain of Ser131, whereas interactions with Asn130 are mediated by a water molecule (Figure 3C). Furthermore, a cluster of positively charged residues Lys63, Lys220 and Lys275 surrounds the end of ATP binding to the β and γ-phosphate groups (Figure 3B). In general, enzymes need a cofactor like Mg2+ ion to stabilize the β and γ-phosphate groups of bound ATP. However, no metal ion coordinating to ATP in the plant AIPT structure was observed here. Instead, the side chains of Lys63 and Lys275 are employed to simultaneously stabilize the β- and γ-hosphate groups of ATP, replacing the role of Mg2+ ion (Figure 3C). According to the multiple sequence alignment, this cluster of residues is conserved in many plant AIPTs (Supplementary Figure S3). Hence, taken together, most of the interactions between ATP and AIPT are focused on the β- and γ-phosphates.
A previous enzymatic assay showed that the triphosphate or diphosphate moiety is important for the substrate recognition of HlAIPT as shown by the lower Km values of ADP and ATP (19.3 and 16.2 µM, respectively) compared to that of AMP (759 µM) (17). Our ATP-bound complex structure offers a good explanation. The absence of β-phosphate and γ-phosphate moiety of ATP would eliminate their interactions with HlAIPT as described earlier (Figure 3B and C). Hence, the diphosphate or triphosphate moiety is necessary for tighter binding with HlAIPT.
Nucleotide binding assay
It has been reported that other ribonucleotides, such as guanosine monophosphate (GMP), cytosine monophosphate (CMP), or uridine monophosphate (UMP), are not accepted by M. alba and H. lupulus AIPT as substrates (17). However, M. alba AIPT is able to use CDP and GDP as the isopentenyl acceptors, producing non-adenine type CK (16). Hence, it is crucial to investigate the enzyme’s capacity to bind nucleotides other than adenosine phosphates. In order to assess the binding affinities of different nucleotides, we used isothermal titration calorimetry (ITC) to obtain information that could help identify the preferred nucleotides, determine the order of the nucleotides’ binding affinities and quantify the thermodynamic parameters of interactions between AIPT and the nucleotides.
We chose ATP, ADP, dATP (deoxyadenosine triphosphate), CTP, GTP and UTP to measure their binding affinity to the HlAIPT. As shown in Table 2, HlAIPT discriminates among different nucleotides largely in the Ka term. A few typical ITC tracings, showing raw and integrated data, can be found in Supplementary Figure S4. Results of the ITC experiment revealed that HlAIPT favors ATP as its ligand while it also shows some preference for dATP over ADP. We then investigated whether GTP, CTP and UTP are able to bind to HlAIPT. Surprisingly, the data show that these nucleotides display an order of difference in their binding affinity to AIPT. The GTP nucleotide, which is also a purine nucleotide, shows a superior binding affinity to that of CTP or UTP pyrimidine nucleotides. This is the first direct evidence that non-adenosine nucleoside triphosphates are able to bind to HlAIPT. By contrast, GMP, CMP and UMP are not accepted as a substrate (17).
Table 2.
Thermodynamic parameters of interactions between AIPT and nucleotides
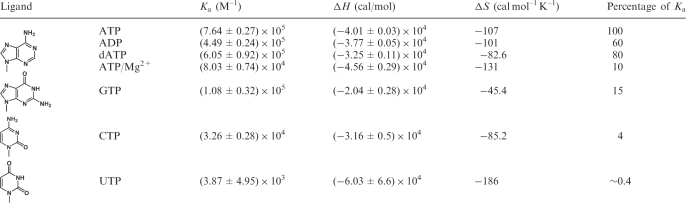 |
Protein and substrate were dissolved in HEPES buffer (20 mM, pH 7.0; 150 mM NaCl, 3 mM DTT).
ITC experiments were performed at 25°C.
Ka: association constant; ΔH: enthalpy changes; ΔS: entropy changes. ATP/Mg2+: ATP binding to HlAIPT under condition containing 2.5 mM magnesium (II).
Plus/minus (±) signed values represent standard error between the observed data and fitted parameters.
These results can be explained based on our crystal structure of the HlAIPT/ATP complex. First, for binding to adenosine derivatives, a decrease in affinity of dATP relative to ATP is caused by the loss of the hydrogen bond between the O2' hydroxyl and the carbonyl group of Lys220 residue (Figure 3C). On the other hand, when the absence of γ-phosphate abolishes the interactions associated with the cluster of positively charged residues, the result is a lower affinity of ADP to HlAIPT than that of dATP. Second, we found that the binding affinity decreases 25-fold when the substrate changes from purine (ATP) to pyrimidine (CTP). Third, if the ligand is replaced by UTP, there is an ∼10-fold decrease in binding affinity. This can be attributed to an unfavorable interaction between the O4 atom of the uridine and the side chain of Asp62.
Finally, for the binding of GTP to HlAIPT, we observed that affinity decreases about 5-fold when compared with ATP. Again, it can be explained by the unfavorable interactions between GTP’s O6 atom and Asp62. Interestingly, a HlAIPT/GTP model based on our crystal structure suggests that the N2-amino group of GTP would fit snugly between the backbone O atoms of Thr73 and Ile222, each making a well-oriented hydrogen bond of 2.9 Å with the GTP’s N2. Hence, the active site of HlAIPT is able to accommodate a larger group than adenine (Figure 3A). This offers us useful information not only concerning the role of GTP, but also about designing modulators analogous to nucleotides. For example, it is expected that the un-natural 2,6-diamino purine riboside triphosphate may be a suitable acceptor for the plant AIPT.
Hence, the above-mentioned results reveal different binding affinities of nucleotides to HlAIPT with an order of ATP > dATP ∼ ADP > GTP > CTP > UTP. As the amino-acid sequences of M. alba and H. lupulus AIPT are 73% identical, and all residues that interact with the nucleotide substrate are conserved (Supplementary Figure S3), these two proteins should also express very similar enzymatic properties. Combining our ITC data with the previous results of activity measurement of M. alba and H. lupulus AIPT (16,17), we can now understand the relationship between binding and catalysis among nucleotides. First, in HlAIPT, the binding affinity of dATP is 80% of the affinity of ATP, generally consistent with the observation that the activity of dATP for AIPT is 44% that of ATP in M. alba (16). This may be caused by a difference in the preferred conformation between ATP and dATP. Here, the ribose of the bound ATP exists in the C3′-endo conformation. The deoxyribose of dATP may have the chance to form a C2′-endo conformation resulting in a change of the base orientation, consequently decreasing the activity. Second, although the binding affinity of GTP is higher than that of CTP, the O6 atom of the guanine base, unlike the N4 atom of a cytosine base, is not a good nucleophile capable of effectively attacking DMAPP. Hence, the relative activity for CTP (30%) is higher than for GTP (6.4%) (16). Finally, UDP displays the weakest binding affinity among all nucleotides tested and does not have a suitable position for the attachment of the isopentenyl group, resulting in a failure to proceed with the reaction.
Structural comparison of the H. lupus AIPT and Agrobacterium AIPT
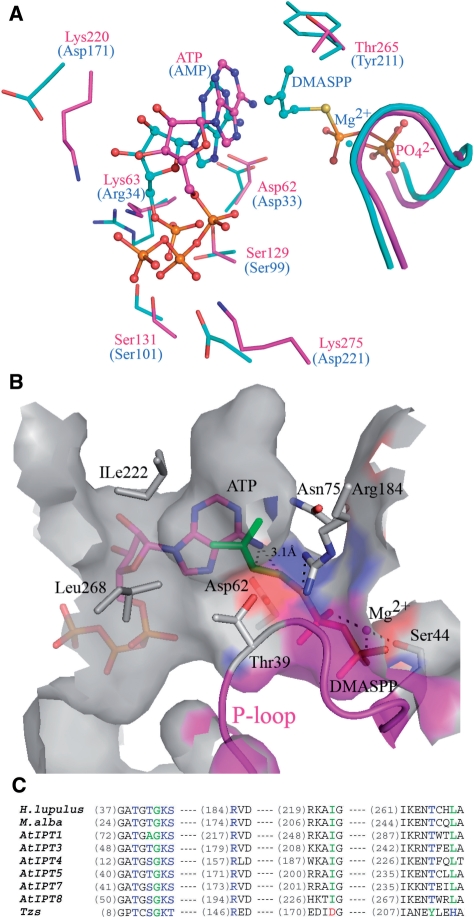
The plant AIPT uses either ADP or ATP as the acceptor; bacterial AIPT uses only AMP (19). In order to identify the controlling residue for different prenyl-acceptor substrates, we superimposed the structure of H. lupus and Agrobacterium AIPT to compare the residues in the active site (Figure 4A). We observed that several residues (Asp62, Ser129 and Ser131), essential for catalysis and binding the adenosine, are conserved between H. lupus and Agrobacterium AIPT. However, interestingly, we found differences in three other residues between the two enzymes. First, two basic residues, Lys220 and Lys275, in HlAIPT, which interact with the β- and γ-phosphate groups of ATP, are substituted by two acidic residues Asp171 and Asp221 in the Agrobacterium AIPT. Apparently, this finding suggests that Agrobacterium AIPT cannot use ADP or ATP because of the repulsive interactions between β- or γ-phosphate and the two acidic residues. Another subtle difference is found in the residue Lys63 of HlAIPT, which interacts with the β- and γ-phosphate groups, whereas its corresponding Arg34 in the Agrobacterium AIPT makes a hydrogen bond with the a α-phosphate group (Figure 4A). This implies that the positively charged residue in this position plays a flexible role for the nucleotide carrying different numbers of phosphate groups.
Figure 4.
Comparison of HlAIPT with Agrobacterium AIPT. (A) Superposition of adenine nucleotide and dimethylallyl group binding sites of HlAIPT (magenta) and Agrobacterium AIPT (cyan). (B) The modeling shows that the pyrophosphate group of DMAPP binds to the P-loop and is coordinated by an Mg2+ ion. Ile222 and Leu268 surrounding the hydrophobic end of the dimethylallyl group may contribute to the substrate specificity. (C) Sequence alignment of AIPTs. The amino acid residues involving prenyl-donor substrate specificity are shown in blue, green and red according to their types. AtIPTs are isozymes from A. thaliana (17). Asp173, Tyr211 and His214 of Agrobacterium AIPT (Tzs) are substituted by Ile222, Thr265 and Leu268 in HlAIPT, respectively.
In order to elucidate the interactions between DMAPP and the AIPT, we made a complex model by computer docking. Superimposition of the two structures shows that the bound phosphate ion in the active site of HlAIPT occupies the binding site for the β-phosphate of a DMASPP molecule in Agrobacterium AIPT. Hence, a DMAPP molecule with a coordinating Mg2+ ion was manually docked into the active site of the AIPT based on the position of the bound phosphate ion. Then, the model was subjected to several cycles of energy minimization, together with the bound ATP (Figure 4B). The resulting model reflects the findings of previous studies in that it shows the pyrophosphate group of DMAPP binds to the P-loop by hydrogen bonds and is coordinated by Mg2+ ion as seen in previous studies (10,12). We compared the residues of the active site surrounding the end of the dimethylallyl group in HlAIPT and Agrobacterium AIPT, and noted two characteristics. First, the substitute of Asp173 of Agrobacterium AIPT, which was proven as a key residue in making contact with the trans-side end of dimethylallyl group (10), should correspond to the residue Ile222 of HlAIPT (Figure 4B and C). Asp173 of Agrobacterium AIPT could interact with the hydroxyl group of the trans-side end of HMBDP resulting in the production of trans-zeatine (10). This information suggests that the substrate specificity may be changed using mutagenesis. Second, surrounding the cis-side end of the dimethylallyl group, Tyr211 of Agrobacterium AIPT is substituted by Thr265 in HlAIPT (Figure 4C). This replacement results in a deeper cavity formation in HlAIPT than Agrobacterium AIPT and may allow it to accommodate a longer prenyl side chain such as C10-geranyl group (Supplementary Figure S5).
Structural comparison of H. lupus AIPT with tRNA–IPT
Based on the analysis of the phylogenic tree of isopentenyltransferases, plant AIPTs are considered to be evolutionally divergent from tRNA–IPTs, which are found in a wide variety of organisms (18). In addition, according to previous studies of bacterial AIPT and tRNA–IPT (10–13), plant AIPT may share a conserved reaction mechanism and critical catalytic amino acid residues. Furthermore, our HlAIPT structure was determined by using the yeast tRNA–IPT as a search model rather than the Agrobacterium AIPT, suggesting a closer kinship between the hop and yeast enzymes. Hence, it is interesting to gain an insight into how the plant AIPT has evolved to use free nucleotides as substrates, in contrast to the macromolecular tRNA substrate of the tRNA–IPTs.
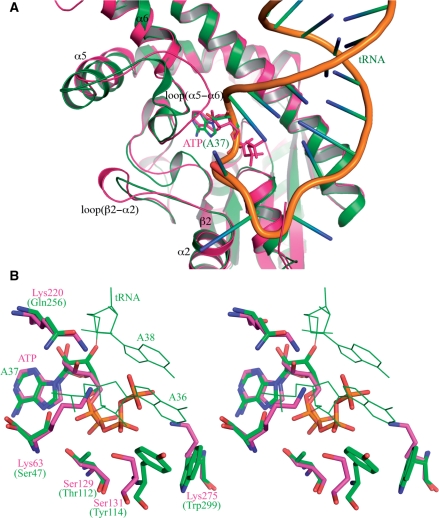
Structural comparison of the H. lupus AIPT and S. cerevisiae tRNA–IPT is shown in Figure 4. Apart from the obvious structural differences found in several loop regions contacting the substrate such as loops β2–α2 and α5–α6 (Figure 5A), they share a similar core structure. The ATP in the HlAIPT complex structure superimposes with the A37 of tRNA, which is flipped out from the anticodon loop of tRNA in the tRNA–IPT complex (Figure 5B; 12). We found that the base, ribose and α-phosphate of ATP are all close to those of A37. For the neighboring A36, it is near the positions of β- and γ-phosphate of ATP located on the open side of the reaction channel. Hence, for different substrate binding, notable changes are found in the residues that interact with the ribose and base of A36 or β- and γ-phosphate of ATP. In the tRNA–IPT complex, Tyr114 and Trp299 cooperatively stabilize the base of A36. With the substrate changing to ATP, Tyr114 and Trp299 are substituted by Ser131 and Lys275 which interact with β- and γ-phosphate of ATP, respectively. Moreover, neighboring residues such as Ser47 and Gln256 in the tRNA–IPT are also substituted by positively charged residues Lys63 and Lys220 of HlAIPT to increase its binding affinity to the phosphate groups of ATP (Figure 5B).
Figure 5.
Comparison of HlAIPT with tRNA–IPT. (A) Structural superposition of the HlAIPT and tRNA–IPT shows prominent conformational differences in the loop regions β2–α2 and α5–α6 adjacent to the substrate. (B) The adenine base of ATP in the HlAIPT complex superimposes with that of the flipped-out A37 in the tRNA–IPT complex. With different substrate binding modes, notable changes are found in the residues that interact with the ribose and base of A36 or β and γ-phosphate of ATP.
However, the binding of tRNA to tRNA–IPT involves an additional helical domain that wraps around the anticodon arm of the tRNA (12,13). The extensive RNA–protein interactions induce significant conformational changes in both macromolecules when they bind to each other. Consequently, although the specific interactions of AIPT and tRNA–IPT with ATP and A37 can be correlated, mutating Ser131 and Lys275 of HlAIPT to Tyr and Trp would not allow it to act on tRNA. Such mutations are not sufficient to provide the required mechanism to flip A37 out of the tRNA molecule. Besides, loop α5–α6 in HlAIPT, on which Lys220 and Ile222 are located, is three-residue longer than its counterpart in yeast tRNA–IPT and is closer to the active site (Figure 5A). It may cause steric hindrance to clash with tRNA, but allows a more secured binding to ATP.
Structural characteristics of nucleotide binding
The Dali search and previous studies show that AIPT and tRNA–IPT are structurally homologous to P-loop-containing nucleotide kinases (10,11). Most nucleotide-binding proteins use P-loop to interact with the phosphate moiety of the ligand, in which the participation of metal ion (e.g., Mg2+) is a common feature (Figure 6A and B; 24). In IPTs, the equivalent P-loop is used to bind to the donor substrate DMAPP and also involves a Mg2+ ion (Figure 6C; 10,12). For its acceptor substrate, tRNA–IPT has an additional domain that embraces the tRNA (12,13). By contrast, bacterial and plant AIPTs lack such extensive interactions as they bind to the rather small molecules of AMP, ADP and ATP. The Agrobacterium AIPT complex structure contains two Zn2+ ions (10). The first Zn2+ is bound to DMASPP and the P-loop and it is supposed to be replaced by an Mg2+ upon catalysis. The second Zn2+ is bound to a site near the phosphate of AMP but its function is unclear (10). Our structure of HlAIPT does not contain any bound metal ion and it suggests that, unlike other homologs, the plant AIPT uses positively charged side chains to bind to the β- and γ-phosphates of ATP, rather than using negatively charged residues and metal ions.
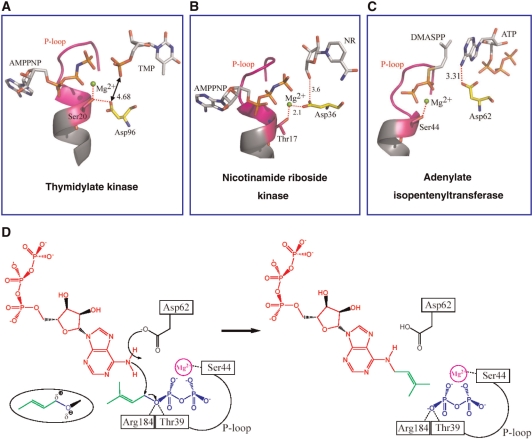
Figure 6.
The role of a conserved aspartic acid. (A) In TK, the Asp96 residue plays a noncatalytic role. (B) In NrK, the Asp36 plays a dual role in stabilizing the bound Mg2+ and activating the nicotinamide riboside oxygen. (C) By contrast, in HlAIPT, the Asp62 plays an essential role as a general base. (D) The proposed catalytic mechanism of HlAIPT involves Asp62, but also Ser44 for binding to DMAPP via Mg2+ ion and Thr39 and Arg184 for stabilizing the negative charge developed in the leaving pyrophosphate group.
In light of this, we performed an ITC analysis of ATP binding to HlAIPT in the presence of 2.5 mM MgCl2, which is approximately the concentration observed under physiological conditions. The results show that the presence of 2.5 mM MgCl2 decreases the binding constant (Ka) of the ATP-HlAIPT complex by about 10-fold (Table 2; see also Figure S4). The slightly larger value of ΔH component suggests that more interactions are established upon binding. The significant decrease in entropy (ΔS) implies that the ensuing structural rearrangements are more extensive in the presence of Mg2+ ion. These results are consistent with the facts that free ATP tends to associate with Mg2+ in solution and that formation of the ATP–HlAIPT complex does not involve the metal ion. Therefore, upon binding to HlAIPT the ATP molecule must dissociate from the Mg2+ ion. Presumably, the released metal ion will interact with the bulk solvent and cause the decrease in entropy.
The ribose moiety of ATP has comparatively weak interactions with the protein. Removal of the 2′-OH group nevertheless decreases its Ka to HlAIPT by about 20% (Table 2). The adenine base makes three direct hydrogen bonds, and these bonds serve to discriminate GTP, CTP and UTP from ATP. GTP may also make three hydrogen bonds with HlAIPT (via its N2 and N3) but its O6 atom cannot make favorable interaction as does the N6 of ATP. Neither is the O6 a good nucleophile. We suspect that the previous difference in the reactivity of inosine diphosphate (IDP) and GDP due to the absence of N2 in IDP (16) was a result of failure in binding to the enzyme, rather than lack of an acceptor for the isoprene group. On the other hand, regardless of the difference between pyrimidine and purine that makes CTP a weaker ligand, its N4 group and the N6 of ATP have similar chemical property. In fact, using HlAIPT as a catalyst, preliminary results of product analysis by mass spectroscopy (data not shown) suggest that ATP and CTP, but not GTP, can react with DMAPP.
The role of a conserved aspartic acid
Asp62 of HlAIPT, which corresponds to Asp33 of Agrobacterium AIPT and Asp46 of yeast tRNA–IPT, is essential for the enzymatic reaction (10,12). A previous site-directed mutagenesis experiment showed that the replacement of Asp62 with Ala resulted in a complete loss of enzyme activity (17). The structural comparisons further indicate that the catalysis-involved residues Thr39, Asp62 and Arg184 are fully conserved among all AIPTs and tRNA–IPTs. These results suggest that the enzymatic reaction of HlAIPT proceeds by the Sn2-reaction mechanism similar to the catalytic reaction of Agrobacterium AIPT and tRNA–IPT (10,12). Taken together, the ATP/DMASPP/Mg2+ model agrees with the proposed catalytic reaction (Figure 6D). N6 of ATP as a nucleophile attacks the C1 carbon atom of DMAPP resulting in the transfer of isopentenyl group from DMAPP to N6 of ATP. The distance between the C1 carbon atom of DMAPP and the N6 nitrogen atom of ATP is 3.1 Å, close to the 3.3 Å observed in the Agrobacterium AIPT complex (10). Asp62 acts as a general base to accept a proton from N6 of ATP. The roles of Thr39 and Arg184 are probably to stabilize the developing negative charge in transition state through interactions with the oxygen atom that connects the hydrocarbon group to the pyrophosphate (10,12). The side chain of Ser44 is supposed to interact with the Mg2+ ion coordinating to the pyrophosphate of DMAPP.
Interestingly, the role of an Asp evolutionally changes from a noncatalytic to a catalytic residue (Figure 6). Superposition of the three enzyme structures including thymidylate kinase (TK; 25), nicotinamide riboside kinase (NrK) (26) and HlAIPT, reveals that Asp96 of TK is structurally equivalent to Asp36 of NrK and the Asp62 of HlAIPT. However, in the protein structures, which have a kinase-like fold, the participating functional role of Asp is diverse. In TK, the Asp96 residue interacts with the Ser20 residue of the P-loop, while remaining uninvolved in catalysis or interaction with any substrate or metal ion (Figure 6A; 25). In NrK, Asp36 plays a dual role in stabilizing the ATP-bound Mg2+ ion and activating the nicotinamide riboside oxygen (Figure 6B; 26). Finally, the Asp62 residue in HlAIPT acts as a general base to accept a proton from the N6-amino group of ATP (Figure 6C). In short, the role of Asp changes from structural stabilizer to catalysis participant as the location of the Asp becomes closer to the acceptor binding site.
ACCESSION NUMBER
3A8T.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charges: Academia Sinica and Core Facility for Protein Production and X-ray Structural Analysis (grant NSC97-3112-B-001-035-B4 to A.H.-J.W.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. I. Abe (University of Shizuoka) for kindly providing us with the pQE80L plasmid encoding the AIPT gene. We also thank Mr. T.-H. Chang for experimental discussion and Dr. S.-C. Jao (Biophysics Core Facility, Scientific Instrument Center, Academia Sinica) for technical assistance with the ITC experiment. The synchrotron data collections were conducted with the Biological Crystallography Facilities (Beamline BL13B1 at NSRRC in HsinChu).
REFERENCES
- 1.Werner T, Motyka V, Strnad M, Schmulling T. Regulation of plant growth by cytokinin. Proc. Natl Acad. Sci. USA. 2001;98:10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- 3.Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008;59:75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 4.Kakimoto T. Biosynthesis of cytokinins. J. Plant Res. 2003;116:233–239. doi: 10.1007/s10265-003-0095-5. [DOI] [PubMed] [Google Scholar]
- 5.Strnad M. The aromatic cytokinins. Physiol. Plant. 1997;101:674–688. [Google Scholar]
- 6.Yevdakova NA, von Schwartzenberg K. Characterization of a prokaryote-type tRNA-isopentenyltransferase gene from the moss Physcomitrella patens. Planta. 2007;226:683–695. doi: 10.1007/s00425-007-0516-0. [DOI] [PubMed] [Google Scholar]
- 7.Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov G, Strnad M, Schmulling T. Two cytokine receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 2004;45:1299–1305. doi: 10.1093/pcp/pch132. [DOI] [PubMed] [Google Scholar]
- 8.Crespi M, Messens E, Caplan AB, van Montagu M, Desomer J. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992;11:795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyoshi DE, Klee H, Amasino RM, Nester EW, Gordon MP. T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 1984;81:5994–5998. doi: 10.1073/pnas.81.19.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugawara H, Ueda N, Kojima M, Makita N, Yamaya T, Sakakibara H. Structural insight into the reaction mechanism and evolution of cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 2008;105:2734–2739. doi: 10.1073/pnas.0707374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie W, Zou C, Huang RH. Structure of tRNA dimethylallyltransferase: RNA modification through a channel. J. Mol. Biol. 2007;367:872–881. doi: 10.1016/j.jmb.2007.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc. Natl Acad. Sci. USA. 2008;105:16142–16147. doi: 10.1073/pnas.0805680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seif E, Hallberg BM. RNA–protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe) J. Biol. Chem. 2009;284:6600–6604. doi: 10.1074/jbc.C800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransfearse. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- 15.Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransfearse, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- 16.Abe I, Tanaka H, Abe T, Noguchi H. Enzymatic formation of unnatural cytokinin analogs by adenylate isopentenyltransferase from mulberry. Biochem. Biophys. Res. Commun. 2007;355:795–800. doi: 10.1016/j.bbrc.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Sakano Y, Okada Y, Matsunaga A, Suwama T, Kaneko T, Ito K, Noguchi H, Abe I. Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop (Humulus lupulus L.) Phytochemistry. 2004;65:2439–2446. doi: 10.1016/j.phytochem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Kamada-Nobusada T, Sakakibara H. Molecular basis for cytokinin biosynthesis. Phytochemistry. 2009;70:444–449. doi: 10.1016/j.phytochem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl Acad. Sci. USA. 2005;102:9972–9977. doi: 10.1073/pnas.0500793102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004;45:1053–1062. doi: 10.1093/pcp/pch119. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.McRee DE. XtalView/Xfit – a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 24.Leipe DD, Koonin EV, Aravind L. Evolution and classification of p-loop kinase and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 25.Ostermann N, Schlichting I, Brundiers R, Konrad M, Reinstein J, Veit T, Goody RS, Lavie A. Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structure of enzyme complexes along the reaction coordinate. Structure. 2000;8:629–642. doi: 10.1016/s0969-2126(00)00149-0. [DOI] [PubMed] [Google Scholar]
- 26.Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedylkova L, Yang T, Sauve AA, Park HW, et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS Biol. 2007;5:2220–2230. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA: DeLano Scientific, LLC; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.