Abstract
Chz1p is a histone chaperone that interacts physically and functionally with the histone variant Htz1p, which has been implicated in establishing and maintaining boundaries between transcriptionally inactive heterochromatin and active euchromatin. To investigate the role of Chz1p in chromatin organization, we performed genome-wide expression arrays and chromatin immunoprecipitations of SIR complex components and modified histones in a CHZ1 deletion strain. Deletion of CHZ1 led to reduced ubiquitination of subtelomere-associated H2B, reduced subtelomeric H3K79 di-methylation, and increased binding of Sir3p, and Sir4p at telomere-distal euchromatin regions, correlating with decreased gene expression in subtelomeric regions. This anti-silencing defect appears to be mediated by enhanced association of de-ubiquitinase Ubp10p with subtelomeric DNA, as detected by chromatin immunoprecipitation analysis. In support of this, we show that deletion of UBP10 can antagonize the subtelomeric silencing phenotype of Δchz1. Taken together, the results demonstrate a novel role for Chz1p in epigenetic regulation, through H2B de-ubiquitination by Ubp10p.
INTRODUCTION
Eukaryotic genomes are structured into two distinct, interchangeable states. Heterochromatin is distinguished from euchromatin by its condensed nature and its relative inaccessibility to the transcriptional apparatus. Thus, euchromatin is readily transcribed; whereas, heterochromatin forms transcriptionally repressed regions of the genome. Understanding the mechanisms underlying these different epigenetic states, and the transitions between them, is fundamental to understanding expression of the genome and is known to involve at least three unique, but not mutually exclusive, activities: DNA methylation, posttranslational modification of histones and nucleosome remodeling. While there are some differences between silencing and activating mechanisms in different organisms, the major epigenetic mechanisms controlling histone modifications and nucleosome remodeling are extremely well conserved between yeast and higher organisms (1–3).
Although most of the genome of Saccharomyces cerevisiae can be characterized as transcriptionally accessible, S. cerevisiae also contains heterochromatin-like regions that include silent mating type loci (HML and HMR), the rDNA (encoding ribosomal RNA) and subtelomeric regions. These loci have been studied comprehensively to gain insight into gene silencing mechanisms (4) and have revealed a process whereby Rap1p recruits Sir4p, which in turn forms a complex with Sir2p and Sir3p. NAD+-dependent histone de-acetylase activity of Sir2p, leads to histone H4 tail de-acetylation, transcriptional silencing and the recruitment of additional SIR complex proteins. However, of the Sir2-4 proteins, only Sir2p is required for rDNA silencing (5–7). At this locus, Sir2p-mediated silencing involves the RENT complex (8).
However, the molecular mechanisms of chromatin silencing extend beyond this simple model. For example, Dot1p is a histone lysine methyltransferase that methylates histone H3K79, and absence of Dot1p leads to a complete loss of H3K79 methylation and defects in heterochromatin-mediated silencing (9). Loss of Hst3p and Hst4p results in hyperacetylation of H3K56 and subsequent telomeric silencing (10). Sas2p is the catalytic subunit of the yeast histone acetyltransferase SAS complex, which is essential for maintaining the accurate silencing at subtelomeric regions (11–13). The ubiquitin protease Ubp10p regulates the global balance of H2B ubiquitination and loss of Ubp10p causes decreased silencing in subtelomeric regions (14). Ubp10p association with chromatin reduces both H2B Lys123 ubiquitination at subtelomeric regions and is also known to reduce H3 Lys4 and Lys79 methylation. Maintenance of low histone ubiquitination by Ubp10p correlates with SIR association in subtelomeric regions (15). However, disruption of any one of the above mechanisms has only incomplete consequences on gene expression, signifying that these anti-silencing factors likely have redundant roles.
If not for the presence of anti-silencing factors to antagonize the ‘local spread’ of Sir-mediated silencing, genes at adjacent loci would be inappropriately silenced. The histone H2A variant Htz1p plays a central role in preventing ectopic heterochromatin spread. The loss of HTZ1 results in the spreading of SIR proteins at the mating locus HMR, and at a subset of chromosome ends (16–18). Histone chaperones are also now thought to play key roles in the maintenance of chromatin dynamics (19); several studies have shown that misregulation of telomere heterochromatin results from inactivation of histone chaperones, such as CAF-1 and RTT106 (20). Chz1p is a histone chaperone that shows a preference for H2AZ-H2B over H2A-H2B and cooperates with the SWR1 complex in the exchange of H2A for Htz1p (21,22). NMR structure analysis has revealed that Chz1p might also have an important role in H2A-H2B eviction from nucleosomes during the SWR1-catalyzed replacement reaction (23).
We previously demonstrated a role for Htz1p in chromatin dynamics and the regulation of oleate-responsive promoters (24); the association of Htz1p with these promoters was Chz1p dependant. As Htz1p is involved in the regulation of heterochromatin spreading, physical and functional interaction between Htz1p and Chz1p led us to test whether Chz1p is also involved in subtelomeric gene expression. In this study, we used genome-wide expression arrays to uncover an uncharacterized function of the histone variant chaperone Chz1p in the maintenance of subtelomeric anti-silencing. Defects in anti-silencing seen in mutant Δchz1 yeast strains are not due to altered acetylation of histone H4K16 (like Δhtz1), but instead appear to be linked to reduced levels of histone H3K79 di-methylation at subtelomeric regions, resulting in the redistribution of SIR proteins and heterochromatin formation within normally euchromatic regions. Moreover, we present evidence showing that Chz1p mediates the reduction of Ubp10p association with subtelomeric regions. Absence of CHZ1 results in increased association of ubiquitinated H2B with chromatin, which leads to the observed decrease in H3K79 di-methylation in subtelomeric regions. Thus, this study reveals a novel role for Chz1p in heterochromatin formation.
MATERIALS AND METHODS
Yeast stains
Genotypes of yeast strains used in this study are described in Table 1. Yeast genetic manipulations were performed following standard methods. Myc-epitope tags (9 repeats) were added at the C-termini of Sir2p, Sir3p and Sir4p as described previously (24). The tagged Sir2-4 strains had similar mating efficiencies as the corresponding wild-type strain. Δsir3 cells failed to mate in a quantitative mating assay (data not shown), suggesting that the Sir2-4 fusion proteins were functional.
Table 1.
Relevant yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YWY003 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Open |
| YWY004 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, htz1::kanMX4 | Open |
| YWY161 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, chz1::kanMX4 | Open |
| YWY679 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, dot1::hphMX | This study |
| YWY682 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, dot1::hphMX chz1::kanMX4 | This study |
| YWY731 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 | This study |
| YWY812 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 chz1::natMX | This study |
| YWY813 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rad6::kanMX4 | Open |
| YWY814 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rad6::kanMX4 chz1::natMX | This study |
| YWY815 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubp10::hphMX | This study |
| YWY816 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubp10::hphMX chz1::natMX | This study |
| YWY723 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubp8::kanMX4 | Open |
| YWY796 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, ubp8::kanMX4 chz1::natMX | This study |
| YWY822 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 ubp10::hphMX | This study |
| YWY823 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 ubp10::hphMX chz1::natMX | This study |
| YWY824 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rad6::kanMX4 ubp10::hphMX | This study |
| YWY825 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rad6::kanMX4 ubp10::hphMX chz1::natMX | This study |
| YWY826 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 rad6::hphMX | This study |
| YWY827 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, bre1::kanMX4 rad6::hphMX chz1::natMX | This study |
| YWY013 | MATa, leu2Δ0, ura3Δ0, HA-HTZ1 | This study |
| YWY0165 | MATa, leu2Δ0, ura3Δ0, HA-HTZ, chz1::kanMX4 | This study |
| YWY423 | MATa, leu2Δ0, ura3Δ0, HA-HTZ, nap::hphMX | This study |
| YWY284 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR2-9MYC::natMX | This study |
| YWY286 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR3-9MYC::natMX | This study |
| YWY296 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR4-9MYC::natMX | This study |
| YWY292 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR2-9MYC::natMX chz1::kanMX4 | This study |
| YWY294 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR3-9MYC::natMX chz1::kanMX4 | This study |
| YWY412 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, SIR4-9MYC::natMX chz1::kanMX4 | This study |
| YWY671 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 UBP10-GFP:his5 | Open |
| YWY696 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 UBP10-GFP:his5 chz1::kanMX4 | This study |
| YWY750 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 | (39) |
| pRS314 [Flag-HTB1-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-URA3 Integrative] | ||
| YWY759 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 | This study |
| pRS314 chz1::kanMX [Flag-HTB1-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-URA3 Integrative] | ||
| YWY751 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 | (39) |
| pRS314 [Flag-htb1-KR-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-UBA3 Integrative] | ||
| YWY760 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 | This study |
| pRS314 ubp10::hphMX [Flag-HTB1-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-URA3 Integrative] |
In vivo histone H3K79 di-methylation assay
For the analysis of in vivo bulk histone H3K79 di-methylation modifications in different mutant strains, whole-cell extracts were prepared as previously described with minor modifications (14). Briefly, 50 ml cultures were grown overnight at 30°C to mid-log phase. Cells were harvested by centrifugation and then lysed in 200 μl of lysis buffer [50 mM HEPES pH 7.9, 2 mM EDTA, 0.25 M NaCl, 0.1% SDS, 0.1% sodium deoxycholate (NaDOC), 1% Triton X-100 and 1 × Protease Inhibitor Cocktail (PIC)]. For immunobloting, aliquots of cell lysates containing equal protein amounts were resolved by SDS–PAGE on 12% polyacrylamide gels, the proteins were transferred to nitrocellulose and immunoblotted with an H3K79 di-methyl antibody.
Isolation of ubiquitinated H2B
The relative levels of ubiquitinated histone H2B (ubH2B) in different strains (wild-type, Δchz1, Δubp10 and htb1-KR) were determined by purifying histone H2B using an N-terminally Flag-tagged histone H2B and subsequently detecting the different forms of H2B using rabbit anti-Flag-HRP antibody as described previously (25).
Chromatin immunoprecipitation
ChIP analyses were performed as described previously (24). For successive chromatin immunoprecipitation from the same sample (ChDIP). Chromatin was isolated and incubated with an anti-Flag antibody. Precipitated protein complexes were subjected to a second immunoprecipitation with an anti-HA antibody, and eluted under standard ChIP conditions. The purified ChIP samples were used for qPCR analysis.
Real-time qPCR was performed by using an iCycler instrument (ABI 7900) and a DyNAmo Flash SYBR green qPCR kit. The average of the results of three independent replicates is reported as the relative amplification of each target of interest compared to a normalization control amplicon within the non-promoter IGRi YMR325W. The occupancy level was determined by dividing the relative abundance of an experimental target by the relative abundance of a control target. This ratio represents the enrichment of ChIP DNA over the input DNA for a specific target versus the control. All oligonucleotides sequences are listed in Supplementary Table S1.
Transcript DNA microarray analysis
Total RNA was prepared from exponentially growing yeast cells grown in YPD using hot acid phenol extraction. Expression microarray analyses were performed as described previously (24). All experiments were performed with duplicate experimental and duplicate technical replicates of each condition, and the log10 of the average mRNA abundance ratios are reported. Differentially expressed genes were identified by maximum-likehood analysis (λ > 100) and significantly affected genes in the mutants were identified by a change in expression of 2-fold or more compared to the expression in the relevant wild-type strains. Significantly differentially expressed genes were analyzed as previously described (26) by using Excel (Microsoft) for chromosome position analysis.
RESULTS AND DISCUSSION
Chz1p regulates transcription of telomere-proximal genes
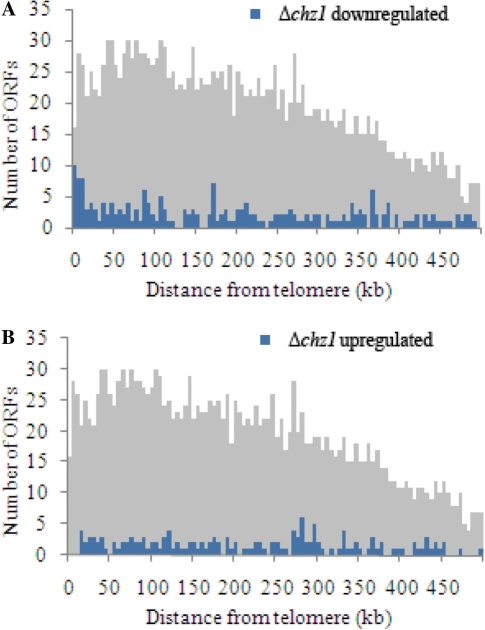
The previously reported physical interaction between Htz1p and Chz1p suggests that Chz1p may have a role in Htz1p-mediated boundary activity that antagonizes the spread of subtelomeric silencing. We predicted, therefore, that boundaries in Δchz1 cells would not be stably maintained and these cells would display different transcriptional profiles relative to their wild-type counterparts. To test this hypothesis, we compared the global steady-state mRNA levels between logarithmically growing Δhtz1 and Δchz1 null cells using DNA microarrays (representing 6271 yeast ORFs) (24) and analyzed the chromosomal locations of the significantly affected genes. This analysis yielded 162 and 217 genes that significantly increased and decreased in expression, respectively, in the absence of CHZ1. Of the genes that decreased in expression in this experiment, 75 also decreased in expression in the absence of HTZ1 (Supplementary Figure S1A).
Visual examination of the list of Δchz1 downregulated genes revealed that many are located in subtelomeric regions (Figure 1A). Similar results have been reported for an HTZ1 deletion strain (12), but the magnitude of the effect of Δchz1 was less than that observed for Δhtz1. As in the case of Htz1p, genes within ∼20 kb of telomeres appear to be enriched for those upregulated by Chz1p; 10% of genes between 0 and 20 kb of telomeres are activated by Chz1p, whereas genes >60 kb from telomeres are depleted for Chz1p-activated genes (Table 2). We also note that genes upregulated by deletion of CHZ1 appear to be randomly distributed throughout the genome (Figure 1B). These results are consistent with our predicted role for Chz1p in Htz1p-mediated anti-silencing at subtelomeric regions; however, upon closer examination of the data, we found that the expression profiles of specific genes within ∼50 kb of the telomeres were different for the two deletion strains (Δchz1 and Δhtz1). The majority of genes appeared to be specifically down-regulated by either Δhtz1 or Δchz1―not both (Supplementary Figure S1B). Overall, these data suggest a steady-state alleviation of telomeric repression in cells lacking CHZ1, supporting a biological role for Chz1p in maintaining chromosomal boundaries in these regions; however, the distinct subsets of genes regulated by Htz1p and Chz1p suggest functional and mechanistic differences in their anti-silencing roles.
Figure 1.
Genes exhibiting aberrant expression in cell lacking CHZ1 map to distinct chromosome regions. Chromosome position analysis was done on genes with significant differential expression in Δchz1 that were repressed (A) or induced (B) more than 2-fold. For each gene, the distance to the nearest telomere was determined and these distances were grouped into 5-kb bins and plotted as a function of telomeric distance. The shaded histograms indicate the distribution of telomeric distances for all ORFs plotted at one-third scale on the y-axis.
Table 2.
Genes requiring Chz1 for normal expression are enriched near telomeres
| Interval (kb) | Δhtz1 |
Δchz1 |
||
|---|---|---|---|---|
| Fraction | P | Fraction | P | |
| 0–10 | 0.368 | 8.5E-57 | 0.135 | 1.91E-09 |
| 10–20 | 0.190 | 5.82E-12 | 0.077 | 2.09E-02 |
| 20–30 | 0.086 | 0.248 | 0.050 | 0.615 |
| 30–40 | 0.114 | 0.007 | 0.036 | 0.998 |
| 40–50 | 0.073 | 0.539 | 0.034 | 0.996 |
| 50–60 | 0.040 | 0.0763 | 0.034 | 0.997 |
| > 60 | 0.040 | 2.86E-05 | 0.031 | 0.280 |
Htz1p association with subtelomeric DNA is independent of Chz1p
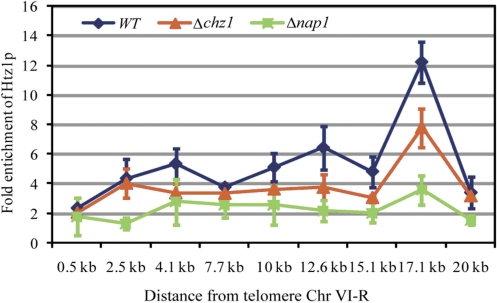
Htz1p has been shown to bind preferentially near telomeres and at regions flanking the silent mating type loci, where it antagonizes the effects of gene silencing by heterochromatin (18). The physical and functional connections between Htz1p and Chz1p suggest that Chz1p might facilitate the assembly or remodeling of Htz1p-containing nucleosomes in subtelomeric regions, leading to changes in the transcription of targeted genes. Accordingly, we compared the subtelomeric binding of Htz1p in wild-type and Δchz1 mutant cells using formaldehyde cross-linking and chromatin immunoprecipitation (ChIP). ChIP assays were used to monitor Htz1p occupancy on the right arm of chromosome VI in vivo. In wild-type cells, levels of Htz1p increased from telomere proximal probes to telomere distal probes, consistent with previous studies (Figure 2) (12). Surprisingly, following deletion of CHZ1, the association of Htz1p with telomere-proximal regions was not dramatically reduced. Therefore, we concluded that Chz1p is not required for Htz1p incorporation to subtelomeric regions. These results were similar at all other telomere-proximal loci examined (n = 6; data not shown).
Figure 2.
Chz1p is not involved in Htz1p incorporation near telomeres. Anti-HA antibodies were used to pull down HA-tagged Htz1p to determine its enrichment by ChIP assays. Each primer set is ∼2.5 kb apart, interspersed along the 20-kb region from the right telomere of chromosome VI. Relative enrichment values (y axes) are the averages of the results from three independent ChIPs with qPCR performed twice per biological replicate. Non-promoter IGRi YMR325W was used as an internal control to normalize signals of enrichment.
It has been previously shown that a fraction of unincorporated Htz1p–H2B dimers in cell extracts is associated with Nap1p, and that Nap1p–Htz1p–H2B complexes are capable of providing Htz1p for SWR1-mediated histone replacement in vitro (21,22). Interestingly, unlike the CHZ1 deletion, disruption of NAP1 can dramatically affect the deposition of Htz1p on subtelomeric regions (Figure 2). Therefore, in agreement with the microarray data, Htz1p deposition on chromatin (and consequent gene expression) does not depend on Chz1p, further supporting distinct roles for Chz1p and Htz1p in chromatin function in subtelomeric regions. Further genome-wide localization analysis of Htz1p in the absence of Chz1p may provide additional insights into their functional relationships.
An absence of Chz1p leads to aberrant binding of Sir3p and Sir4p at subtelomeric regions
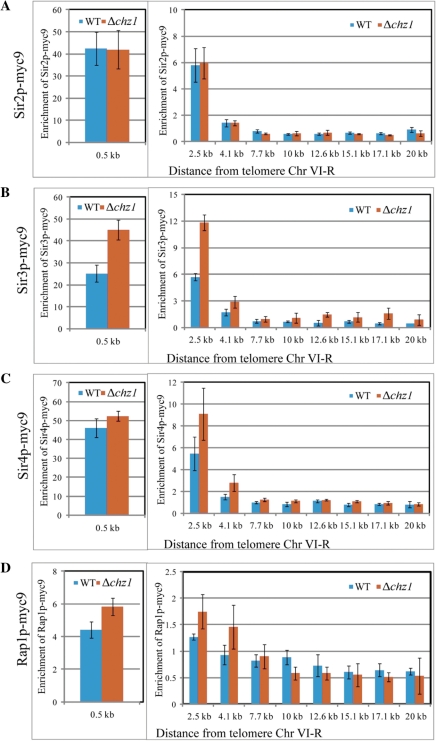
To investigate whether Chz1p regulates subtelomeric silencing through interaction with chromatin, we immunopurified Chz1p and examined its associated chromatin. However, these ChIP-chip experiments failed to detect (stable) association of Chz1p with chromatin (data not shown). This suggested that Chz1p indirectly influences the formation of silent chromatin at subtelomeric regions, perhaps by mediating the interaction between silencing factors and subtelomeric chromatin. To investigate this possibility, we investigated SIR proteins and telomere-associated histone modifications in cells lacking Chz1p. SIR proteins are restricted to telomere proximal heterochromatin by modification of histones located in euchromatin, such as acetylation of H4K16 by the histone acetyltransferase Sas2p (27–29). To determine if the subtelomeric anti-silencing defect in Δchz1 mutant is caused by an ectopic spread of components of the SIR complex, ChIP was used to compare the patterns of Sir2p, Sir3p and Sir4p association in wild-type and Δchz1 strains. Primer sets targeting DNA from 0 to 20 kb from telomere ends were used to amplify the immunoprecipitated DNA. Results from ChIP experiments revealed considerable enrichments of Sir2p (42-fold), Sir3p (25-fold) and Sir4p (47-fold) with regions adjacent to telomeres (within 0.5 kb), whereas the association of the SIR complex at several telomere-distal euchromatic genes appeared to be at background levels (Figure 3). As shown in panel A, the association of Sir2p with heterochromatin was not affected by the absence of CHZ1 in either telomere-distal or telomere proximal regions (Figure 3A).
Figure 3.
Deletion of Chz1p leads to increased binding of Sir3p and Sir4p to subtelomeric chromatin. The association of Sir2p (A), Sir3p (B), Sir4p (C) and Rap1p (D) with subtelomeric chromatin regions was determined by ChIP using anti-myc antibodies, followed by qPCR. The relative enrichment ratio for each is plotted. ACT1 was used as an internal control to normalize signals of enrichment. Error bars show the standard deviation from three independent biological replicates with two technical replicates of each.
Binding of Sir3p and Sir4p at telomere-distal locations was observed in the Δchz1 strain (Figure 3B–C). Indeed, the association of Sir3p and Sir4p with subtelomeric chromatin in the Δchz1 mutant was significantly increased compared to wild-type cells within 4 kb of the telomere end. This does not appear to be a result of transcriptional misregulation of SIR genes in the Δchz1 mutant, as we did not observe altered transcript levels of any of the known Sir silencing proteins by microarray analyses (data not shown). These data suggest that in the absence of Chz1p, Sir3p and Sir4p binding may spread from telomeres into subtelomeric regions, increasing silencing of normally expressed genes. It is also possible that Sir3p and Sir4p do not redistribute from the telomeres to other sites of the chromosome, but rather may come from a SIR complex pool within nucleus (30,31). This suggestion is supported by ChIP assays with Rap1p. Rap1p initiates silencing by recruitment of the SIR complex (4) and in the absence of Chz1p, Rap1p binding to telomere proximal regions was increased (Figure 3D). We interpret this to suggest that increased association of Rap1p contributes to the maintenance of Sir3p and Sir4p near telomere regions in the absence of Chz1p and that Chz1p normally functions to inhibit SIR complex protein binding at the euchromatin regions adjacent to silenced regions.
Di-methylation of H3K79 at subtelomeric regions requires Chz1p
It has been demonstrated that histones, and predominantly histone modifications, play an important role in transcriptional silencing, promoted in part by a complex of SIR proteins that bind to the modified nucleosomes. We therefore used ChIP and quantitative PCR to examine four major histone modifications present in subtelomeric chromatin to illuminate the specific defect imparted by the absence of Chz1p: acetylation of H4K16 and K3K56 and methylation of H3K4 and K3K79. H4K16 acetylation, is a target of Sir2p de-acetylation, and as expected, was found enriched in euchromatic compared to heterochromatic regions (Supplementary Figure S2A). However, no dramatic changes in the abundance or distribution of H4K16 acetylation were observed in Δchz1 cells. As a control for antibody specificity, and consistent with previous reports (12), H4K16 acetylation at subtelomeric regions showed only background occupancy in a Δsas2 strain (Supplementary Figure S5A). Similarly, H3K56, another recently identified target of Sir2p (13), was found to be more abundant distal to the telomere end, but this normal distribution and abundance was unaltered in Δchz1 cells (Supplementary Figure. S2B). These data are consistent with our observation that there are no defects in Sir2p distribution across a subtelomeric region of Δchz1 cells (Figure 3), and suggest that the anti-silencing defect observed in Δchz1 cells is not due to alterations in H4K16 or H3K56 acetylation.
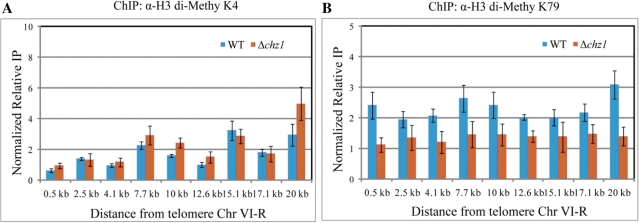
There are also two well-characterized histone methylation modifications important for the organization and maintenance of specialized chromatin regions like subtelomeres in yeast: Set1p-mediated H3K4 methylation and Dot1p-mediated H3K79 methylation (32). Di-methylated K4 and K79 on histone H3 are lost from subtelomeric regions upon establishment of silencing. In Δchz1 cells, the level of di-methylated K4 on histone H3 was not greatly reduced relative to wild-type (Figure 4A); however, di-methylation of K79 on histone H3 was dramatically reduced in Δchz1 cells throughout the subtelomeric region examined (Figure 4B). As a control experiment, the same subtelomeric region was analyzed in a Δdot1 mutant and H3K79 di-methylation was not detected (Supplementary Figure S5B), confirming the antibody’s specificity to its target. Consistent with previous reports (9,33), complete loss of this modification in the Δdot1 mutant resulted in diminished Sir3p association to subtelomeric regions (Supplementary Figure S4).
Figure 4.
Di-methylation of H3K79 near telomeres is dramatically reduced in the absence of Chz1p. ChIP was performed on wild-type and Δchz1 mutant strains. Formaldehyde cross-linked protein-DNA complexes were immunoprecipitated by using antibodies against individual di-methylated H3 lysine residue 4 (A) and 79 (B). Relative enrichment values (y axes) are the averages of the results from three independent ChIPs with qPCR determination performed twice per biological replicate. Non-promoter IGRi YMR325W was used as an internal control to normalize signals of enrichment.
It has been proposed that the region surrounding H3K79 is a direct target for Sir3p and/or Sir4p binding (34). H3K79 methylation may, therefore, prevent Sir3p and/or Sir4p from binding to this region of the nucleosome surrounding H3K79 and the redistribution of Sir3p and Sir4p in the Δchz1 mutant may result from decreased H3K79 methylation. Together, these data suggest that Chz1p function is specifically linked to enhancing H3K79 di-methylation in subtelomeric regions. This interpretation is consistent with the increased binding of Sir3p and Sir4p to telomere proximal regions in the absence of Chz1p, as H3K79 methylation is known to interfere with Sir3p and Sir4p binding (35,36).
To understand how the loss of Chz1p leads to decreased subtelomeric H3K79 di-methylation, we first asked if the defect was local at chromosome ends, or was due to a reduction in cellular levels of di-methyl H3K79. There was no detectable change in the cellular levels of H3K79 di-methylation in Δchz1 cells (Supplementary Figure S3). Controls were consistent with previous observations (14,15); that is, a complete absence of detectable H3K79 di-methylation in Δdot1 and increased levels of di-methyl H3K79 in Δubp10 strains (which indirectly leads to increased H3K79 as discussed below) (Supplementary Figure S3). Therefore, abnormal SIR protein binding in Δchz1 cells is not a result of overall decreased levels of Dot1p-dependent di-methylation of K79 on histone H3.
Chz1p is necessary for ubiquitination of chromatin-associated histone H2B
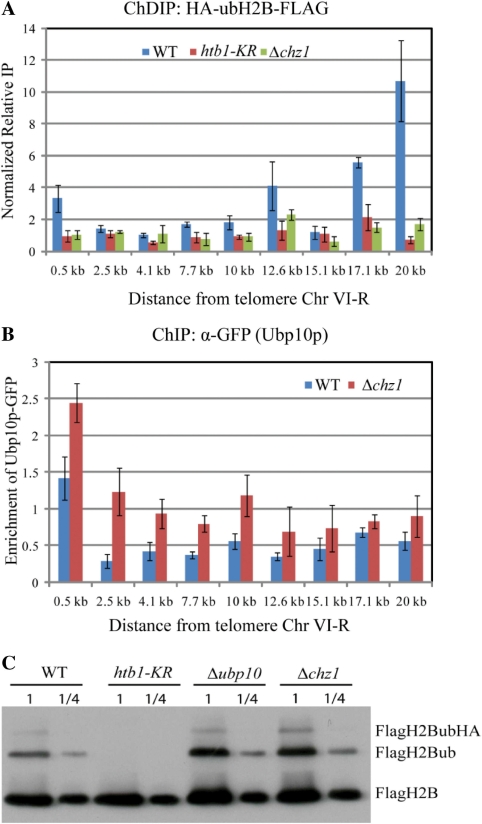
It has been previously shown that H2B ubiqitination leads to increased histone H3 methylation and normal levels of subtelomeric H3 methylation require the ubiquitin conjugase, Rad6p and the ubiqutin ligase Bre1p monoubiquitinate histone H2B at Lys123 (ubH2B) (20,37,38). We therefore asked whether decreased subtelomeric H3K79 methylation in Δchz1 mutant cells was a result of reduced H2B ubiquitination. To do so, we compared levels of ubH2B in subtelomeric regions of wild-type and Δchz1 mutant cells expressing FLAG tagged H2B and HA-tagged ubiquitin using a chromatin double immunoprecipitation (ChDIP) assay. H2B was first enriched via the FLAG tag and the subpopulation of ubH2B was further enriched by subsequent immunoprecipitiation using anti-HA. The levels of HA-ubH2B-FLAG were also determined in htb1-KR, a strain bearing a substitution mutation of the H2B ubiquitination site, which serves as a negative control for the assay. Consistent with previous studies (39), wild-type cells had low ubH2B levels near the chromosome end, and higher levels at regions distal to the telomere (Figure 5A). In contrast, Δchz1 cells had very low levels of HA-ubH2B-FLAG at chromosome regions both proximal and distal to the telomere end. The levels of chromatin associated ubH2B in Δchz1 were statistically indistinguishable from that of the negative control htb1-KR.
Figure 5.
Deletion of CHZ1 leads to enhanced association of Ubp10p near telomeric regions. (A) Double chromatin immunoprecipitation (ChDIP) of ubH2B. Formaldehyde cross-linked chromatin was obtained from wild-type, Δchz1 and htb1-KR mutant cells bearing Flag-tagged H2B and HA-tagged ubiquitin. Relative enrichment values (y axes) are the averages of the results from three independent ChIPs with qPCR determination performed twice per biological replicate. Non-promoter IGRi YMR325W was used as an internal control to normalize signals of enrichment. (B) ChIP assays of Ubp10p were performed using anti-GFP antibody specific for GFP tag fused to the C terminal of Ubp10p. All values are the averages of the at least three independent experiments. Errors bars represent the standard deviation. (C) Chz1p negatively regulates histone H2B ubiquitination. Global steady-state H2B ubiquitination levels from whole-cell lysates were assayed using anti-FLAG antibodies that recognize the FLAG epitope placed on the N terminus of H2B expressed from a plasmid and replacing the native HTB1 gene. Δubp10 and htb1-KR serve as positive and negative control strains for H2B ubiquitination, respectively. The loading equivalents (one and one-fourth) are indicated above each lane.
It is possible that the reduced levels of chromatin associated ubH2B in the Δchz1 mutant are a result of a global reduction in cellular levels of ubH2B. To test this, the cellular levels of ubiquitinated H2B in a Δchz1 strain were compared to those in a wild-type strain and in a strain with a deletion of UBP10 de-ubiquitinase (Figure 5C). Δubp10 was chosen as a control because Ubp10p is known to mediate H2B de-ubiquitination, and has been shown to function directly at silenced chromatin (15). Surprisingly, a noticeable increase of global levels of ubiquitinated H2B was detected in the Δchz1 mutant. The levels were comparable to those observed in the deletion of UBP10. As expected, ubiquitination was not detected in the htb1-KR strain containing a mutated target of H2B ubiquitination. This increased global ubH2B in Δchz1 mutant does not appear to associate with chromatin directly, as we have shown a decreased level of chromatin-associated ubH2B in this strain (Figure 5A). Perhaps there is a pool of ubH2B in the nucleus that is not chromatin associated. It is unclear why global levels of ubH2B increase in CHZ1 mutant, whereas chromatin-associated levels decrease. One possibility is that decreased chromatin-associated ubH2B in the Δchz1 strain is a result of increased chromatin association of the de-ubiquitinase Ubp10p, which sequesters it from ubH2B substrates that are free in the nucleus. Elucidation of this global phenotype requires further analysis.
In conclusion, deletion of CHZ1 results in decreased abundance of chromatin-associated ubH2B, which is not a result of decreased cellular levels of ubH2B. In subtelomeric regions, this defect appears to result in decreased di-methylation of H3K79, leading to dysregulation of subtelomeric silencing.
Chz1p antagonizes the association of Ubp10p with subtelomeric regions
We hypothesized that deletion of CHZ1 causes a defect in the association of regulators of H2B ubiquitination in subtelomeric regions. We specifically focused on whether Chz1p function is associated with Ubp10p binding for three reasons: Ubp10p mediates H2B de-ubiquitination, and has been shown to function directly at silenced chromatin (15), Chz1p does not appear to associate stably with chromatin (data not shown); and Chz1p appears to have a chaperone function (21,23). We used GFP-tagged Ubp10p as a bait and performed ChIP assays in wild-type and Δchz1 cells. As shown in Figure 5B, the association of Ubp10-GFP at subtelomeric regions was dramatically higher in Δchz1 mutant cells than wild-type cells. This does not appear to be due to increased expression of UBP10 because microarrays and western blot analyses failed to detect differences in the UBP10 mRNA and protein levels (Supplementary Figure S7). The increased recruitment of Ubp10p to subtelomeric regions and the concomitant reduction of chromatin-associated ubH2B in Δchz1 mutant cells indicate that an important function of Chz1p is regulating subtelomeric anti-silencing through mediating the disassociation of Ubp10p, and as a result, controlling levels of ubH2B in subtelomeric regions. It will be interesting to determine if the association of Ubp10p or other H2B de-ubiquitinases is altered at other chromosomal locations.
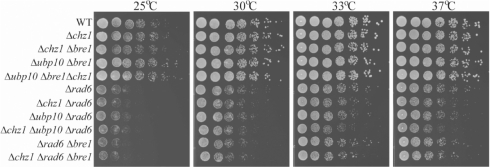
In order to characterize the role of Chz1p in ubiquitination of chromatin-associated H2B, we performed a genetic analysis to understand functional interactions between Chz1p and factors involved in H2B ubiquitination, including Rad6p (an E2 ubiquitin ligase) and Bre1p (required for Rad6p association with H2B) (38), and Ubp10p. While deletion of RAD6 alone led to a growth defect compared to wild-type, double deletion of RAD6 and CHZ1 resulted in a more severe defect at 37°C, which was subtle, but reproducible (Figure 6; Supplementary Figure S6). Moreover, the growth defect is more severe when UBP10 was deleted from rad6/chz1 double mutant (Figure 6). CHZ1 deletion did not show an increased growth defect when combined with double deletion of BRE1 and UBP10. In addition, we also did not observe genetic interaction between CHZ1 and DOT1 (Supplementary Figure S6). These results indicate that Chz1p has genetic interactions with factors involved in H2B ubiquitination. This does not appear to involve a stable physical interaction of Chz1p with Ubp10p, Bre1p or Rad6p, as we did not detect association by immunoprecipitation experiments. In addition, deletion of Chz1p does not appear to affect cellular levels of these proteins by western blotting (Supplementary FiguS7).
Figure 6.
Genetic interaction between CHZ1 and factors involved in H2B ubiquitination. Five-fold serial dilutions of each indicated haploid strain were spotted on complete medium containing 2% glucose for 3 days at 25°C, and for 2 days at 30, 33 and 37°C and photographed.
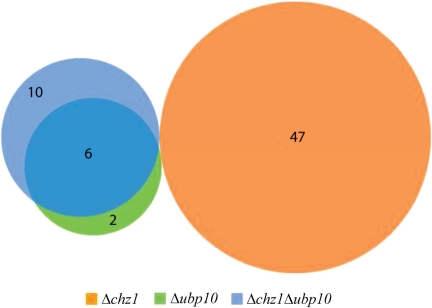
To further investigate the potential relationship between Chz1p and Ubp10p, we compared the gene silencing profiles in subtelomeric regions of Δchz1, Δubp10 and Δchz1 Δubp10 double mutant strains using microarray analysis. The results are summarized in Figure 7. For each strain, the group of subtelomeric genes (within 50 kb of telomeres) that is down-regulated by 2-fold is depicted as a circle. Overlap between datasets is represented by overlap of the circles. Consistent with previous reports, we identified 54 genes in Δubp10 cells that had decreased expression of 2-fold or greater, 8 out of the 54 genes are within 50 kb of telomeres (green circle). In contrast, deletion of CHZ1 resulted in reduced expression of 47 subtelomeric genes within 50 kb of telomeres (orange circle). Interestingly, the downregulated genes in the double mutant (blue circle) shared no overlap with the down-regulated genes in Δchz1; but most of the downregulated genes in the Δchz1/Δubp10 double mutant are the same as those downregulated in the Δubp10 single deletion strain, suggesting the UBP10 mutant is epistatic to the Δchz1 mutant. Thus, Δubp10 can antagonize the subtelomeric expression phenotype in Δchz1, and suggesting that Chz1p functions to reduce association of Upb10p with subtelomeric regions.
Figure 7.
Comparison of genes with altered transcriptional profiles in Δchz1, Δubp10 and Δchz1, Δubp10 mutants. Venn diagram depicting the numbers of genes affected in their transcriptional levels (downregulated ≥ 2-fold) within 50 kb from telomeres as detected by microarray in Δchz1, Δubp10 and Δchz1, Δubp10 mutant strains.
Here, we have characterized a function for Chz1p in the control of subtelomeric anti-silencing in yeast. We identified decreased gene expression in subtelomeric regions in the absence of CHZ1. These regions are normally blocked from the spread of silencing factors that dominate the telomeric regions; but, this anti-silencing activity is compromised by the absence of Chz1p. Cells lacking HTZ1 have a similar phenotype, and Htz1p has a well-characterized boundary function and prevents the ectopic spread of silencing factors to subtelomeric loci (18). In addition, previous reports have suggested that Chz1p acts as a molecular chaperone for Htz1p (H2A.Z) (21). However, while Htz1p and Chz1p can form a complex (21), characterization of Δchz1 cells revealed that they appear to function in distinct physiological pathways to regulate subtelomeric anti-silencing.
Htz1p deletion leads to the spread of Sir2p (and Sir3p) to adjacent euchromatin regions, whereas loss of Chz1p leads to increased binding of Sir3p and Sir4p, but not Sir2p within normally euchromatic regions. This difference may be responsible for the differences in the histone H4 acetylation patterns in these chromatin regions; the pattern appears normal in Δchz1 cells, but in Δhtz1 cells H4 becomes hypoacetylated (18). The reduced expression of subtelomeric loci in Δchz1 cells corresponds to hypomethylation of H3K79. We interpret this to be a result of Chz1p regulating ubiquitination of H2B, which in turn, maintains optimal H3K79 di-methylation in active chromatin. In the absence of Chz1p, there is an increased association of Ubp10p with chromosome ends, which leads to increased de-ubiquitination of H2B, decreased H3K79 di-methylation, and abnormal binding of SIR proteins. Taken together, these data suggest a novel function for Chz1p in anti-silencing near telomeres. Further investigation into the molecular mechanisms by which Chz1p modulates histone H2B ubiquitination will yield greater insight into the function of this histone variant chaperone in chromatin regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (GM067228, RR022220 and GMO76547).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to Shelley Berger for yeast strains and Kenny Lee and Fan Mo for technical assistance and helpful discussions.
REFERENCES
- 1.Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 2.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 3.Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 5.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, Miller A, Kirchmaier AL. HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol. Biol. Cell. 2008;19:4993–5005. doi: 10.1091/mbc.E08-05-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijsing SH, Ehrenhofer-Murray AE. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 17.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 18.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 19.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan Y, Saleem RA, Ratushny AV, Roda O, Smith JJ, Lin CH, Chiang JH, Aitchison JD. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell Biol. 2009;29:2346–2358. doi: 10.1128/MCB.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 28.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 29.Sutton A, Shia WJ, Band D, Kaufman PD, Osada S, Workman JL, Sternglanz R. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 2003;278:16887–16892. doi: 10.1074/jbc.M210709200. [DOI] [PubMed] [Google Scholar]
- 30.Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 32.Qin S, Wang Q, Ray A, Wani G, Zhao Q, Bhaumik SR, Wani AA. Sem1p and Ubp6p orchestrate telomeric silencing by modulating histone H2B ubiquitination and H3 acetylation. Nucleic Acids Res. 2009;37:1843–1853. doi: 10.1093/nar/gkn1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 35.Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat. Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 38.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 39.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.