Abstract
The rate of mutation refers to the probability that a unit length of DNA (generally a base pair) mutates with time. Fluctuation analysis or mutant accumulation assays applied to phenotypic changes measure mutation rates of cells. However, only a few phenotypic changes indicative of mutations are known thus limiting the analysis to those rare genes. Direct sequencing overcomes the limitations imposed by phenotypic analysis but is limited by the extensive number of clones or cells that have to be analyzed in fluctuation or mutant accumulation assays. We propose a strategy to determine the rate of mutation of a gene by limited direct sequencing of a few single cells of a defined lineage. To accomplish this, we determined the average number of mutations per position in each DNA length sequenced from the proportion of the non-mutated positions, according to the Poisson process and/or the Taylor series. Measuring the rate of mutation by direct sequencing of genes does not require ascertaining a phenotype and can be applied to any area of the genome in a cell. The approach avoids fluctuation errors.
INTRODUCTION
Mutations are changes in the nucleotide sequence of DNA. Such changes can be punctual i.e. confined to one base pair or involve multiple base pairs including deletion, addition, amplification and recombination. The rate of mutation is the probability that a given base pair or a larger region of DNA changes with time. For practical reasons mutations are usually detected by changes in phenotype per unit of time indicated as cell generations (1) or days (2). For the purpose of this manuscript, we will consider only punctual changes in the determination of mutation rates.
The mutation rate of stable genomes is estimated to be 10−10/bp per cell generation (3). However, in certain physiologic conditions the rate of mutation increases dramatically. As one example, the immunoglobulin (Ig) genes can undergo mutation at a rate that exceeds the basal rate by more than a million-fold (1,4,5). In another example, a lac I transgene in mice (in the ‘Big Blue’ transgenic mouse) undergoes mutations more frequently than expected assuming a basal mutation rate (6,7). Cancer cells are thought to mutate at a high rate. However, whether mutators drive cancer owing exclusively to the phenotypic manifestations of mutations is controversial (8). Whether the rate of mutation changes under certain physiologic conditions or in response to certain stimuli is not known in part because of the difficulties of examining mutations in cell lineages, throughout time. Hence, determining whether the rate of mutation underlies physiologic or pathophysiologic conditions requires reliable and feasible approaches to measure the rate of mutation.
The rate of mutation in organisms or cells can be determined by fluctuation analysis or by mutation accumulation assays. Fluctuation analysis was invented by Luria and Delbruck (9) to determine whether bacteria acquire resistance to viral infection by spontaneous mutation. The rate of mutation in bacteria could not be determined by simply counting directly resistant organisms because that number could reflect the offspring of one mutant or alternatively the late occurrence of many independent mutations. To overcome this problem, Luria and Delbruck determined the average number of mutations per culture by plotting the frequency of cultures containing no resistant bacteria in the equation describing the zero order of the Poisson process. The probability that zero mutations occurred per culture reduces the Poisson process to P(0)= e−λ, where λ is the average number of mutations per culture (9). The mutation rate was obtained by dividing the average number of mutations per culture by the total number of bacterium per cell generation (9). Lea and Coulson (10), Sandri and Sarkar (11,12) and Zheng (13) added statistical analysis to the application of fluctuation assays in bacteria.
Wabl et al. (1) applied fluctuation analysis to determine the rate of mutation of the immunoglobulin genes in murine B cells. Wabl et al. (1) determined the rate of reversion of the amber STOP codon in the immunoglobulin heavy chain variable exon in B cells. Fluctuation analysis was applied to clones grown from single or very few cells in a compartmentalization assay. The average frequency of mutation (λ) per cell was determined by the zero order of the Poisson process, according to e−λ = N0/N. The rate of mutation was calculated by dividing the average frequency of mutation by the estimated number of generations elapsed during the clone growth and yielded 1.1 − 4.2 × 10−5 per base pair per cell generation (1). The rate of mutation of the immunoglobulin genes in B cells was confirmed independently by Zhu and colleagues (14). This high rate of mutation, about a million times greater than the basal rate of mutation, was termed ‘hypermutation’ (1) and provided a mechanism for the observed somatic diversification of the Ig genes (15,16). The approach used to determine the rate of mutation at immunoglobulin loci is not generally applicable since it is applied to a defined locus with a phenotypic marker for mutations.
The rate of mutation can also be determined by ‘mutant accumulation’ assays. In ‘mutant accumulation’ assays, the rate of mutation is calculated from the rate of change in the fraction of mutants overtime in a culture started from a relatively large population that is mutant free (17,18). ‘Mutant accumulation’ assays are thus limited by the difficulty of establishing large enough mutant-free cultures that generally require the availability of a phenotype associated with a mutational target.
While in principle sequencing might determine the rate of mutation independently of expression of a phenotype, fluctuation dictates the need for analysis of many independent clones. We investigated the feasibility of measuring the rate of mutation by directly sequencing a gene of interest over time. Assuming that every base pair in a given length of DNA has equal probability of mutating, independently of each other and almost no chances of mutating twice, then the average number of mutations can be calculated from the proportion of the non-mutated positions according to the Poisson process and/or the Taylor series. This strategy decreases the number of clones that have to be analyzed to determine the rate of mutation compared to examining changes in phenotype due to a single base pair mutation.
We measured the rate of mutation of the HIV-1 envelope gene integrated into the genome of a line of B cells in which the rate of spontaneous mutation has been determined rigorously at several loci (1,19). Because each envelope gene sequence was obtained from the consensus of all the sequences present in a clone, we could eliminate from the analysis clones with founder mutations present in every clone, thus decreasing fluctuation in the number of mutations considerably. The strategy requires the analysis of relatively few clones if the length of nucleic acid sequenced is at least 1 kb, and is independent of phenotypic analysis; thus it can be extended to any loci in the genome.
METHODS
Cells
To calculate mutation rate in a gene, we exploited the 18.81 murine B cell line because the rate of mutation at the immunoglobulin loci, and other loci, has been rigorously determined (1,19). This cell line was generated by several rounds of subcloning BALB/c bone marrow cells that had been infected with Abelson virus (20,21). We chose a subclone of the 18.81.A3.43 cell line (1). This particular clone of cells expresses activation induced cytidine deaminase (AID) (22) and undergoes somatic hypermutation (1,22).
Gene of interest
We chose to measure the mutation rate of an exogenously introduced foreign gene driven to be expressed by immunoglobulin regulatory elements. To this end, we exploited a gene encoding a modified version of the HIV-1 YU2 strain’s (clade B) envelope protein, provided by Dr João Gonçalves (School of Pharmacy, University of Lisbon, Portugal). This modified HIV-1 envelope gene encodes gp120 and only the extra-cellular portion of gp41 (to avoid membrane insertion and permit secretion), producing a protein of 140 kDa, which we refer to as gp140. The gene was modified as referenced by Bower et al. (23). Briefly, the DNA encoding the gp140 protein is 2004 bp. It was optimized for eukaryotic translation by codon optimizing the sequence encoding the gp140 protein, and by replacing the first 32 amino acids of the gp140 protein with the leader sequence for tissue plasminogen activator. Additionally, a tag of three copies of murine C3d (891 bp each NCBI #BC043338 nucleotides 3058–3948) was added 3′ to gp140 to ensure detection of protein variants that may arise due to mutation of the HIV sequence (24). The entire sequence was ∼4.7 kb in length.
Generation of gp140 DNA vector
To express the gp140 gene in the 18.81 B cell line, the immunoglobulin light chain λ1 promoter (λ1p) was directionally cloned between the HindIII and BamHI sites in the MCS of pcDNA3.1/Hygro + (Invitrogen, Carlsbad, CA, USA), and a Nhe1 restriction cut site was added onto the 5′ end of the µ heavy chain major intronic enhancer (Eµ) by PCR (using primer set Nhe1Add, see below). The enhancer was directionally cloned upstream of the light chain promoter in the MCS of pcDNA 3.1 between the Nhe1 and HindIII sites. The gp140 gene segment was inserted 3′ to the λ1p between the BamHI and Xho sites in the MCS of pcDNA3.1. To do this, the gp140 was digested with EcoRI, filled in with Klenow and digested with Xho1 to generate a 5′-blunt end and a 3′-sticky end. pcDNA 3.1 was BamHI restricted and filled in with Klenow to generate a 5′-blunt end, followed by Xho1 restriction, generating a 3′-sticky end.
The Eµ was a 678 bp sequence (NCBI #M12827 pos 7–683), and the λ1p was a 1625 bp sequence (NCBI #AC140201 pos 32 137–30 517). Both were generous gifts from Dr Young (Rochester, NY, USA) (25).
Transfection of 18.81 B cell line and cell culture
To create a stable B cell line expressing the gp140 protein, we electroporated 2 × 107 cells of clone 1B5 of the 18.81.A3.43 cell line (1) at 315 V with 20 µg of the gp140 vector that had been linearized by digestion with Nru1. Cells were rested for 2 days at 37°C in RPMI with 10% FCS, 50 mM 2-mercapto ethanol, with penicillin and streptomycin (100 U/ml) and 2 mM l-glutamine supplemented in the medium. After 2 days, transfected cells were selected by resistance to 1 mg/ml hygromycin. Thereafter, transfected cells were always maintained in culture with hygromycin.
Western blot
To identify the gp140 protein produced by the transfected B cells, we performed western blots on the cell extracts. This was accomplished by PAGE of 5–20 µg of cell lysate under reducing conditions (5% 2Mε) on 7.5% Tris–HCl READY GELs (Bio-Rad, Hercules, CA, USA) followed by transfer to an Immobilon PVDF membrane (Millipore, Billerica, MA, USA). On the blots, gp140 protein was detected with human HIV Immunoglobulin (HIVIg) (NIH AIDS Research and Reference Reagent Program Division of AIDS, NIAID, NIH: from NABI and National Heart Lung and Blood Institute—from Dr Luiz Barbosa) (1:3000–1:5000) followed by goat α human–HRP (Southern Biotech, Birmingham, AL, USA) (1:2000). C3d was detected using goat α murine C3d antibody (R & D Systems, Minneapolis, MN, USA) (1:1000) followed by rabbit α goat IgG–HRP (Novus Biologicals, Littleton, CO, USA).
Positive controls
Recombinant gp120 produced in a Baclovirus system from the YU-2 strain (Immunodiagnostics, Woburn, MA, USA) was used as a positive control.
Isolation of cellular RNA and RT-PCR
RNA was obtained from cells using the RNeasy kit (Qiagen, Hilden, Germany), and cDNA was created using oligo dT primers and Thermoscript RT-PCR system (Invitrogen, Carlsbad, CA, USA).
Calculation of mutation rate
To detect mutations in the gp140 gene, we examined clones derived from single B cells. These were created by subjecting 18.81 cells to rounds of successive cloning by limiting dilution (1.6 cells/ml, 100 µl per well in 96-well plates). If plates contained less than 10 clones visible after 9 days of growth, the clones were picked and transferred to 4 ml of media and grown for an additional 4 days. After 13 days of growth, the cells in each clone were counted and RNA extracted.
cDNA was produced by RT-PCR and β2 microglobulin, VH81X variable region, gp140, and AID DNA were amplified by PCR (using primer sets β2, VH, gp140-1, gp140-2, AID) followed by gel purification by QIAEXII (Qiagen, Hilden, Germany). The purified DNA except from AID, was directly sequenced at least four times by the Mayo Clinic Core DNA Sequencing and Synthesis Facility. Sequences were aligned by the Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA), and all unique mutations were scored if they did not occur in a primer region and were not present in the lineage founder but present in a daughter clone.
The sequences obtained by sequencing bulk cDNA represented the consensus sequence of all cells present which grew up from one single cell, and thus represented the sequence of the gene in the clone founder (assuming no mutation occurred in the first or second division of those cells).
The number of base pairs sequenced depended on the gene analyzed. For gp140, 2000 bp were sequenced as the gene is 2004 bp long. For VH81X 300 bp were sequenced as that is the approximate length of the variable region gene. Finally for β2 microglobulin, 800 bp were sequenced.
On average, the number of cell generations (i.e. divisions) for any given cell in culture which occurred in the 13 days between the time of the original cloning and the subsequent re-cloning of the cells was calculated by counting the cells in a given clone after 13 days of growth and assuming that they all started from a single cell. So, therefore, if X cells were present after 13 days and the number of generations which occurred was n, that number could be found by solving: 2n = X.
Primer sets
Nhe1 Add: 5′-GATC GCTA GCGA GGTC TGGT GGAG CC-3′ and 5′-GGTA TCGA TAAG CTTG ATAT CGAA TTC-3′. β2 5′-TGGC TCGC TCGG TGAC CCTG-3′ and 5′-GCAG AAGT AGCC ACAG GGTT G-3′. VH 5′-GTGC AGCT GGTG GAGT CTGG-3′ and 5′-CCAG AAGT TACC ATAC TAGT C-3′. Gp140 1 5′-CAAT TCGA TATC AAGC TTG-3′ and 5′-TCTC CCAC TGGG TCTT GCTC A-3′. Gp140 2 5′-TGAG CAAG ACCC AGTG GGAG A-3′ and 5′-GATG TACC ACAG CCAC TTGG TG-3′. AID 5′-ATCT CAGA CTGG GACC TGGA C-3′ and 5′-GGAA CCAG AAGT GTCT TCA-3′.
RESULTS AND DISCUSSION
To determine the rate of mutation of genes by direct sequencing we took advantage of a B cell line in which the rate of mutation at several loci is known. This line of B cells, referred to as 18.81 cells, was generated by transforming murine BALB/c bone marrow cells with Abelson virus (20,21). The 18.81.A3.43 subclone from one transformation undergoes 1.1–4.2 × 10−5 mut/bp per cell generation at the immunoglobulin loci (1) and mutates reporter genes at many other loci at similar rates (19). Consistent with its ability to mutate Ig genes, 18.81 cells express the Ig mutator, the activation-induced cytidine deaminase (AID).
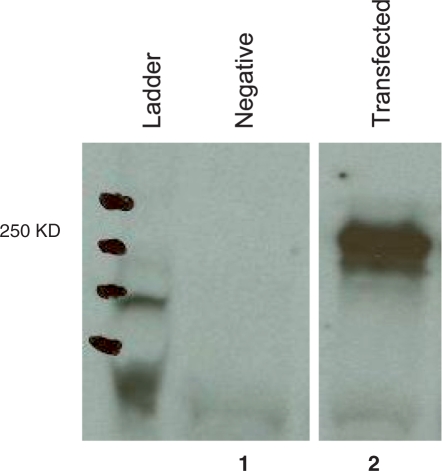
To determine how fast a gene mutates, we first ascertained the gene sequence in the founder cell. Toward this, we transfected the 18.81.A3.43 subclone with the human immuno-deficiency virus-1 envelope gene (gp140), a well-characterized gene (22). To facilitate detecting proteins translated from gp140 variants, the gene was tagged with three copies of murine C3d. Transfected cells expressed both RNA of the gp140 gene (not shown) and the gp140C3d chimeric protein (Figure 1).
Figure 1.
GP140 is expressed in B cells. The figure shows a western blot analysis of 18.81 cells transfected (lane 2) or not (lane 1) with the gp140-C3d vector. Protein extracts were obtained from 30 × 106 cells and separated by 7.5% SDS–PAGE. Proteins were blotted onto PVDF membranes and gp140 was revealed by IgG pooled and purified from human serum of HIV positive patients (HIVIg) at a 1 : 5000 dilution followed by goat anti-human IgG HRP-conjugated at a dilution of 1 : 2000. The gp140-C3d chimera has the approximate molecular weight of 250 kDa.
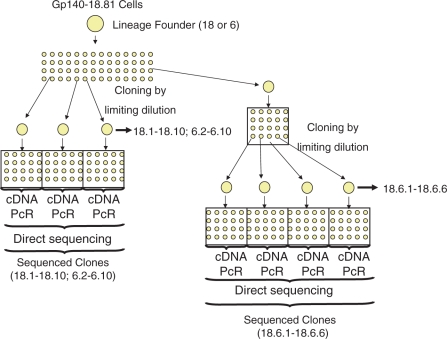
To determine the rate of mutation, we next performed fluctuation analysis. Transfected cells were cloned by limiting dilution to generate lineages of cells (Figure 2). After each cloning step RNA was extracted from more than half of the cells in the clone and cDNA generated by reverse transcription, the gp140 gene was amplified by PCR and sequenced directly. The consensus sequence of the gp140 gene reflected the sequence of the majority of the cells in a clone and thus the sequence in the lineage founder. By successive limiting dilutions, we obtained the sequence of the gp140 gene in 24 daughter cells from two independent lineages. We identified mutations in each clone by comparing the gp140 sequences in the daughter cells with the gp140 sequence in the lineage founder.
Figure 2.
Cloning by limiting dilution. Transfected 18.81 cells were subjected to successive rounds of limiting dilution to obtain wells seeded with single cells. From single cells, clones were grown for 13 days. Each number identifies a clone in which the first digits refer to the lineage founder and the following digits represent subclones. For example, 18.6.1 refers to a clone obtained from lineage founder 18 which was subcloned twice generating first clone 18.6 and after 18.6.1. Sequences were obtained directly, without cloning, from PCR products obtained from cDNA.
To determine the rate of mutation, we first determined the probability of mutating at each base pair according to the Poisson process. We assumed that each mutation reflects an independent event that has a small probability of occurring and has an infinitesimal probability of occurring twice at the same position. Thus each base pair could be considered an independent Bernoulli trial with an output of either mutated or not mutated and the average number of mutations in a cell could be calculated by the zero order of the Poisson process. The average frequency of mutations (N0/N where N0 is the number of unaltered positions and N the total number of bases sequenced) thus equaled e−λ. Alternatively, for large numbers of N, the average number of mutations per base pair can be obtained by 1−N0/N. This is because according to the Taylor series eλ = 1 + λ when λ is small. Thus e−λ = 1 − λ = N0/N, which is much easier to calculate than the zero order of the Poisson distribution. Using the Taylor series to estimate the average frequency of mutations allows easy determination of its accuracy by calculating the standard error of the estimate, which equals the square root of the average frequency of mutations divided by the square root of N. The number of base pairs to sequence to obtain an accurate estimate of the average frequency of mutations can be deduced from the standard error.
In one example (Table 1), we sequenced 18 000 bp (trials) and found seven mutated base pairs and 17 993 non-mutated base pairs. Because the Ig mutator in this particular line targets only G/C base pairs (5), we calculated the rate considering only the number of G/C pairs. Therefore, N0 = 11 016, N = 11 025 and e − λ = 11 016/11 025. λ = −ln 0.9991836 = 8.1666 × 10−4. This number represents the average number of mutations per base pair (i.e. frequency of mutation). According to the Taylor series the average frequency is 8.164 × 10−4.
Table 1.
The number of mutations in progeny of clone 18
| Clone 18 | Mutation no. |
|---|---|
| 18.1 | 1 |
| 18.3 | 0 |
| 18.4 | 0 |
| 18.5 | 0 |
| 18.6 | 3 |
| 18.7 | 0 |
| 18.8 | 1 |
| 18.9 | 2 |
| 18.10 | 0 |
Each clone was grown for 22.97 cell generations from founder. Nine clones were analyzed, 2000 bp containing 1225 G or C nucleotides were sequenced, or a total of 11 025 G or C base pairs. Estimation of the average frequency of mutations per base pair was done according to the Poisson distribution, the zero order value was − ln(11 016/11 025) = − ln 0.9991836 = 8.1666 × 10−4. The mutation rate was 3.55 × 10−5 mut/bp per cell generation. According to the Taylor series, the average frequency was 8.164 × 10−4 which yielded the same mutation rate.
The mutation rate was calculated by dividing the average frequency of mutation per base pair by the number of cell generations. The clone represented in Table 1 grew from a single cell to 8.24 × 106 cells prior to sequencing. The average number of cell generations that passed from the clone founder to any one of the nine cells sequenced was 22.97 (because 2n = 8.24 × 106) assuming no cells died. Therefore, the mutation rate was 3.55 × 10−5 mut/bp per cell generation.
We calculated the rate of mutation in two other lineages of cells generated from the same founder population. Results shown in Table 2 indicated a rate of 4.65 × 10−5 mut/bp per cell generation deduced from the zero order of the Poisson distribution and a rate of 4.69 × 10−5 mut/bp per cell generation deduced from the Taylor series. Results shown in Table 3 indicated a mutation rate of 1.26 × 10−5 mut/bp per cell generation deduced from the zero order of the Poisson distribution and a rate of 1.39 × 10−5 mut/bp per cell generation according to the Taylor series.
Table 2.
The number of mutations in subclone 18.6
| Subclone 18.6 | Mutation no. |
|---|---|
| 18.6.1 | 2 |
| 18.6.2 | 0 |
| 18.6.3 | 3 |
| 18.6.4 | 1 |
| 18.6.5 | 2 |
| 18.6.6 | 0 |
Each clone was grown for 23.41 cell generations from founder. Six clones were analyzed, 2000 bp containing 1225 G or C nucleotides were sequenced, or a total of 7350 G or C base pairs. Estimation of the average frequency of mutations per base pair was done according to the Poisson distribution, the zero order value was − ln(7342/7350) = − ln 0.9989 = 1.089 × 10−4. The mutation rate was 4.65 × 10−5 mut/bp per cell generation. According to the Taylor series, the average frequency was 1.1 × 10−3 which yielded a mutation rate of 4.69 × 10−5 mut/bp per cell generation.
Table 3.
The number of mutations in progeny of clone 6
| Clone 6 | Mutation no. |
|---|---|
| 6.2 | 0 |
| 6.3 | 2 |
| 6.4 | 0 |
| 6.5 | 0 |
| 6.6 | 0 |
| 6.7 | 1 |
| 6.8 | 0 |
| 6.9 | 0 |
| 6.10 | 1 |
Each clone was grown for 28.71 cell generations from founder. Nine clones were analyzed, 2000 bp containing 1225 G or C nucleotides were sequenced, or a total of 11 025 G or C base pairs. Estimation of the average frequency of mutations per base pair was done according to the Poisson distribution, the zero-order value was − ln(11 021/11 025) = −ln 0.9996 = 3.63 × 10−4. The mutation rate was 1.26 × 10−5 mut/bp per cell generation. According to the Taylor series, the average frequency was 0.4 × 10−3 which yielded a mutation rate of 1.39 × 10−5 mut/bp per cell generation.
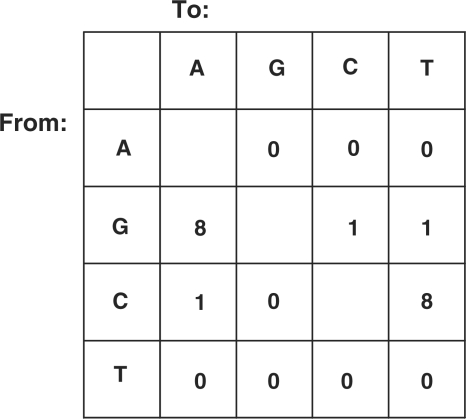
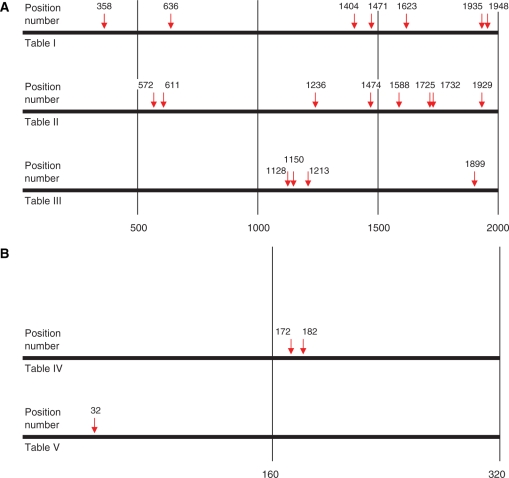
The mutations in the gp140 gene targeted only C and G base pairs and 84% were transitions (Figure 3) consistent with the properties of the immunoglobulin mutator in this cell line (19). However, mutations in gp140 were dispersed throughout the 2000 bp gene length (Figure 4). This distribution is in contrast with the typical distribution of mutations at the immunoglobulin loci which generally targets the first few hundred base pairs, but extending as far as 1.5 kb downstream (4). In agreement, the three mutations in the immunoglobulin VH81X exon were found within the first 200 bp out of the 300 sequenced. Dispersion of mutations throughout the length of the gp140 gene suggests that the clustering of mutations towards the transcription initiation site associated with the immunoglobulin mutator on the immunoglobulin locus may not hold for other loci.
Figure 3.
Mutation table depicting the type of mutations found in the gp140 gene. Results pooled from three independent experiments.
Figure 4.
Schematic representation of mutations in the gp140 and VH81X genes. Shown is the relative position and exact base pair number of each mutation which occurred in either the gp140 (A) or VH81X (B) genes in all of the clones sequenced.
To calculate the rate of mutation by sequencing, we assumed that only one copy of the gene of interest (gp140 in our case) was being expressed in each clone analyzed. For a typical cellular gene this is a reasonable assumption to make. However, multiple copies of a gene might integrate into the genome during transformation or transduction. However, so long as only one copy of the gene is expressed, and expression is stable, then sequencing the cDNA will accurately estimate the rate of mutation. This is because since we measure the rate of mutation by directly sequencing amplified gene fragments obtained from RNA, the sequence reflects the consensus of the majority and avoids errors generated during reverse transcription, amplification or sequencing. In the case of transgenes with only one copy expressed, the sequence obtained in this way reflects the gene sequence. In the case of genes that are expressed from two copies, as is the case of most cellular genes, a mutation in one of the copies but not in the other will originate an ambiguous reading at that position which could be resolved by cloning. Alternatively selective amplification of one of the copies (by taking advantage of known polymorphisms) would allow unambiguous detection of mutations by direct sequencing.
To validate our method, we measured the rate of mutation of the immunoglobulin heavy chain variable region gene in 18.81 cells and compared the result with previous estimates (1). We sequenced 300 bp of the VH81X immunoglobulin variable region from each clone comprehending 176 G or C containing nucleotides, and identified mutations as described. We found no mutations in nine daughter cells from lineage 18, two mutations in six daughter cells from lineage 18.6 (Table 4 and Figure 4B) and one mutation in nine daughter cells from lineage 6 (Table 5 and Figure 4B). The rate of mutation was 8.1 × 10−5 mut/bp per cell generation or 8.54 × 10−5 mut/bp per cell generation, according to the Poisson process or the Taylor series, respectively. The rate of mutation in lineage 6 (Table 5 and Figure 4B) was 2.2 × 10−5 mut/bp per cell generation or 3.5 × 10−5 mut/bp per cell generation, according to the Poisson process or the Taylor series, respectively. Therefore the rate of mutation at the immunoglobulin heavy chain locus is in the same order of magnitude as the rate calculated for the HIV-1 envelope gene.
Table 4.
The number of mutations in the progeny of a subclone of clone 18
| Subclone 18.6 | Mutation no. |
|---|---|
| 18.6.1 | 0 |
| 18.6.2 | 0 |
| 18.6.3 | 0 |
| 18.6.4 | 0 |
| 18.6.5 | 2 |
| 18.6.6 | 0 |
Each clone was grown for 23.41 cell generations from founder. Six clones were analyzed, 300 bp sequenced comprehending 176 G or C containing nucleotides, or a total of 1056 G or C base pairs. Estimation of the average frequency of mutations per base pair was done according to the Poisson distribution, the zero-order value was − ln(1054/1056) = −ln 0.998 = 1.90 × 10−3. The mutation rate was 8.1 × 10−5 mut/bp per cell generation. According to the Taylor series, the average frequency was 2 × 10−3 which yielded a mutation rate of 8.54 × 10−5 mut/bp per cell generation.
Table 5.
The number of mutations in progeny of clone 6
| Clone 6 | Mutation no. |
|---|---|
| 6.2 | 0 |
| 6.3 | 0 |
| 6.4 | 0 |
| 6.5 | 0 |
| 6.6 | 0 |
| 6.7 | 1 |
| 6.8 | 0 |
| 6.9 | 0 |
| 6.10 | 0 |
Each clone was grown for 28.71 cell generations from founder. Nine clones were analyzed, 300 bp, 176 G or C containing nucleotides, were sequenced per clone, or a total of 1584 G or C base pairs. Estimation of the average frequency of mutations per base pair was done according to the Poisson distribution, the zero-order value was − ln(1583/1584) = −ln 0.999 = 6.32 × 10−4. The mutation rate was 2.2 × 10−5 mut/bp per cell generation. According to the Taylor series, the average frequency was 1 × 10−3 which yielded a mutation rate of 3.5 × 10−5 mut/bp per cell generation.
We confirmed the rate of mutation in the 18.81.A3.43 subclone by performing a classic fluctuation analysis. We used immunofluorescence to identify clones with cells expressing immunoglobulin µ heavy chain and clones without cells expressing µ heavy chain. Because reversion of an amber STOP codon by a single nucleotide mutation allows expression of immunoglobulin µ heavy chain, the rate of reversion equals the rate of mutation. On average, we found 1.442 revertants per clone by the zero order of the Poisson distribution. The rate of reversion was calculated by dividing the average frequency of reversion by the average number of cells in each clone and by the total number of cell generations in the history of the clone (26). The reversion rate was 7.2 × 10−6 mut/bp per cell generation and at least one order of magnitude lower than the rate of mutation measured by sequencing. Because mutants were identified by expression of µ heavy chain, detection can be impaired by phenotypic lag that is a delay between occurrence of the mutation and expression of the protein.
To determine if conserved genes also mutate in 18.81 cells, we sequenced the β2 microglobulin gene in each of the clones found to mutate the gp140 gene. The β2 microglobulin gene is highly conserved (27) and is not known to mutate in B cells. We found no mutations in 800 bp of the β2 microglobulin gene in any of the clones studied (data not shown). These results indicate that sequencing did not artificially generate mutations and that the β2 microglobulin gene mutates at a rate that is lower than 2.6 × 10−6 mut/bp per cell generation (the approximate rate of mutation if just one mutation was found). This rate is lower than the rates found for the gp140 gene and for the immunoglobulin gene (1) indicating that in agreement with the conclusions of others (28), somatic hypermutation does not target all loci in the B cell genome.
Here, we show that the rate of mutation of a gene can be measured by direct sequencing independently from phenotypic expression of a reporter gene. This approach can be applied to any area of the genome, and even to different areas within one gene, and can be completed in a relatively short time. We show that the rate of mutation of the HIV-1 envelope protein integrated into the genome of a mutating line of B cells is between 1.26 × 10−5 and 3.55 × 10−5 mut/bp per cell generation and within the same range as the rate of mutation of the immunoglobulin gene (1).
FUNDING
NIH (HL079067); Bill and Melinda Gates Foundation (52090). Funding for open access charge: NIH and Bill and Melinda Gates Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wabl M, Burrows PD, von Gabain A, Steinberg C. Hypermutation at the immunoglobulin heavy chain locus in a pre-B-cell line. Proc. Natl Acad. Sci. USA. 1985;82:479–482. doi: 10.1073/pnas.82.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang CL, Wabl M. Hypermutation rate normalized by chronological time. J. Immunol. 2005;174:5650–5654. doi: 10.4049/jimmunol.174.9.5650. [DOI] [PubMed] [Google Scholar]
- 3.Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nature Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 4.Wabl M, Cascalho M, Steinberg C. Hypermutation in antibody affinity maturation. Curr. Opin. Immunol. 1999;11:186–189. doi: 10.1016/s0952-7915(99)80031-4. [DOI] [PubMed] [Google Scholar]
- 5.Bachl J, Wabl M. An immunoglobulin mutator that targets G.C. base pairs. Proc. Natl Acad. Sci. USA. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buettner VL, Hill KA, Scaringe WA, Sommer SS. Evidence that proximal multiple mutations in Big Blue transgenic mice are dependent events. Mutat. Res. 2000;452:219–229. doi: 10.1016/s0027-5107(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 7.Hill KA, Wang J, Farwell KD, Scaringe WA, Sommer SS. Spontaneous multiple mutations show both proximal spacing consistent with chronocoordinate events and alterations with p53-deficiency. Mutat. Res. 2004;554:223–240. doi: 10.1016/j.mrfmmm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557; . doi: 10.1158/0008-5472.CAN-07-5835. discussion 3557. [DOI] [PubMed] [Google Scholar]
- 9.Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lea D, Coulson C. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, Sandri GV, Sarkar S. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J. Appl. Probab. 1992;29:255–267. [Google Scholar]
- 12.Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Q. Statistical and algorithmic methods for fluctuation analysis with SALVADOR as an implementation. Math. Biosci. 2002;176:237–252. doi: 10.1016/s0025-5564(02)00087-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhu M, Rabinowitz JL, Green NS, Kobrin BJ, Scharff MD. A well-differentiated B-cell line is permissive for somatic mutation of a transfected immunoglobulin heavy-chain gene. Proc. Natl Acad. Sci. USA. 1995;92:2810–2814. doi: 10.1073/pnas.92.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 16.Gearhart PJ, Johnson ND, Douglas R, Hood L. IgG antibodies to phosphorylcholine exhibit more diversity than their IgM counterparts. Nature. 1981;291:29–34. doi: 10.1038/291029a0. [DOI] [PubMed] [Google Scholar]
- 17.Reddy M, Gowrishankar J. A genetic strategy to demonstrate the occurrence of spontaneous mutations in nondividing cells within colonies of Escherichia coli. Genetics. 1997;147:991–1001. doi: 10.1093/genetics/147.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachl J, Dessing M, Olsson C, von Borstel RC, Steinberg C. An experimental solution for the Luria-Delbruck fluctuation problem in measuring hypermutation rates. Proc. Natl Acad. Sci. USA. 1999;96:6847–6849. doi: 10.1073/pnas.96.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CL, Harper RA, Wabl M. Genome-wide somatic hypermutation. Proc. Natl Acad. Sci. USA. 2004;101:7352–7356. doi: 10.1073/pnas.0402009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg N. In vitro transformation of lymphoid cells by Abelson murine leukemia virus. Proc. Natl Acad. Sci. USA. 1975;72:1932. doi: 10.1073/pnas.72.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siden E. Immunoglobulin synthesis by lymphoid cells transformed in Vitro by Abelson murine leukemia virus. Cell. 1979;16:389–396. doi: 10.1016/0092-8674(79)90014-x. [DOI] [PubMed] [Google Scholar]
- 22.Balin SJ, Ross TM, Platt JL, Cascalho M. HIV genes diversify in B cells. Curr. HIV Res. 2008;6:10–18. doi: 10.2174/157016208783571919. [DOI] [PubMed] [Google Scholar]
- 23.Bower J, Yang X, Sodroski J, Ross T. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 2004;78:4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 25.Young F, Ardman B, Shinkai Y, Lansford R, Blackwell TK, Mendelsohn M, Rolink A, Melchers F, Alt FW. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Develop. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- 26.Von Borstel RC, Cain KT, Steinberg CM. Inheritance of spontaneous mutability in yeast. Genetics. 1971;69:17–27. doi: 10.1093/genetics/69.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colombani J. Conserved and variable structures in HLA class I molecules: a review. Tissue Antigens. 1990;35:103–113. doi: 10.1111/j.1399-0039.1990.tb01765.x. [DOI] [PubMed] [Google Scholar]
- 28.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti R, Klein U, Kuppers R, Rajewsky K, et al. BCL-6 mutations in normal germinal center B cells: Evidence of somatic hypermutation acting outside Ig loci. Proc. Natl Acad. Sci. USA. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]