Abstract
The non-essential VTS1 gene of Saccharomyces cerevisiae is highly conserved in eukaryotes and encodes a sequence- and structure-specific RNA-binding protein. The Vts1 protein has been implicated in post-transcriptional regulation of a specific set of mRNAs that contains its-binding site at their 3′-untranslated region. In this study, we identified VTS1 as a multi-copy suppressor of dna2-K1080E, a lethal mutant allele of DNA2 that lacks DNA helicase activity. The suppression was allele-specific, since overexpression of Vts1 did not suppress the temperature-dependent growth defects of dna2Δ405N devoid of the N-terminal 405-amino-acid residues. Purified recombinant Vts1 stimulated the endonuclease activity of wild-type Dna2, but not the endonuclease activity of Dna2Δ405N, indicating that the activation requires the N-terminal domain of Dna2. Stimulation of Dna2 endonuclease activity by Vts1 appeared to be the direct cause of suppression, since the multi-copy expression of Dna2-K1080E suppressed the lethality observed with its single-copy expression. We found that vts1Δ dna2Δ405N and vts1Δdna2-7 double mutant cells displayed synergistic growth defects, in support of a functional interaction between two genes. Our results provide both in vivo and in vitro evidence that Vts1 is involved in lagging strand synthesis by modulating the Dna2 endonuclease activity that plays an essential role in Okazaki fragment processing.
INTRODUCTION
Chromosomal DNA replication in eukaryotes requires the coordinated action of a large number of replication factors (1–5). Unlike leading strand DNA synthesis, lagging strand DNA synthesis proceeds discontinuously via the generation of Okazaki fragments. DNA polymerase (pol) α-primase first synthesizes ∼10 nucleotides (nt) of RNA, followed by the addition of short DNA stretches (6,7), which are designated ‘RNA–DNA primers’. These are different from the RNA-only primers observed in prokaryotes. The second DNA polymerase, pol δ, extends the RNA–DNA primers to form new Okazaki fragments with the aid of proliferating cell nuclear antigen (PCNA) and replication factor C (RFC). Pol δ not only extends the newly synthesized Okazaki fragments but it also creates flap structures by displacing the 5′-end region of downstream Okazaki fragments (8,9). Flaps can be processed by nucleases to form ligatable nicks, which are sealed by DNA ligase I to produce continuous double-stranded (ds) DNA. Short flap regions can be removed by the activity of flap endonuclease 1 (Fen1), which is a structure-specific endonuclease (10–12). However, when the flap region is relatively long (>20 nt), flap cleavage requires Dna2, an endonuclease/helicase (8,13). Replication protein A (RPA) can bind efficiently to DNA flaps longer than 27 nt in length, which inhibits Fen1 activity. In contrast to Fen1, RPA-bound flaps are efficiently cleaved by Dna2 because its endonuclease activity is markedly stimulated (13). Thus, RPA can act as a nuclease switch for the sequential processing of long flaps that are processed first by Dna2, and then by Fen1 to form a nick.
Although the majority of Okazaki fragments appear to be removed in vitro via generation of short flaps in a Fen1-dependent manner, a substantial fraction of flaps generated in vitro are long (11), and their processing requires both Fen1 and Dna2. Therefore, both pathways are essential for processing of all flaps. In vivo, however, the level of long and/or short flaps formed is not known. The production of long flaps can be influenced by the failure of cleavage at the early stages in strand displacement reaction due to inadequate Fen1 activity, enhanced strand displacement by pol δ (14) or the failure of 3′→5′ proofreading function of pol δ (11,15). Alternatively, there may be sequences that can be more readily displaced by pol δ. All of these factors can lead to elevate the levels of long flaps.
In order to gain more insights into the processing of long flaps by Dna2, we searched for genetic suppressors that could rescue a particular defect caused by a mutation in DNA2. This approach identified the non-essential VTS1 gene of S. cerevisiae as a suppressor of the dna2-K1080E mutant (glutamate substitution for lysine at position 1080, the ATP-binding motif), which lacks ATPase/helicase activities (16–18). VTS1 was originally identified in S. cerevisiae as a multi-copy (and low-copy) suppressor of vti1–2 mutant cells that displayed defects in growth and vacuole transport (19). However, it is not clear whether Vts1 is directly involved in vesicle trafficking (19). One notable structural motif present in Vts1 is the sterile-α motif (SAM) domain. This domain is highly abundant in eukaryotes and known to mediate protein–protein interactions for transcription regulation, signal-transduction cascades and nucleic acid binding (20–22). A subclass of SAM domain can bind RNA (22–25). For example, both Vts1 of S. cerevisiae and Smaug of Drosophila melanogaster recognize and bind RNAs possessing a specific structure (named SRE, SMAUG recognition element) (22). This structural element consists of a RNA stem and a pentanucleotide loop (pentaloop; CNGGN, N for any nucleotide) (22,26,27). Disruption of the stem or a single base change in any of the three conserved nucleotides in the pentaloop significantly reduced the RNA-binding activity of Smaug or Vts1 (22,28). Conversely, a single amino acid substitution in the SAM domain of Smaug or Vts1 abolished its ability to bind the RNA-stem-loop structure. Consistent with the role of Smaug in Drosophila, S. cerevisiae Vts1 regulated the stability of artificial mRNAs containing an SRE at their 3′-untranslated region (22). However, target mRNAs that are regulated by Vts1 have not been identified yet.
The SAM domain has also been implicated in DNA binding. For example, the RuvA subunit, a component of the prokaryotic RuvABC complex that processes recombination intermediates, contains this domain that is required for binding Holliday junction structures (29). Since VTS1 was isolated as a suppressor of defects in Okazaki fragment processing, we investigated its biochemical interactions with Dna2 and Fen1. In this study, we present both in vitro and in vivo evidence that Vts1 is involved in Okazaki fragment processing by modulating the endonuclease activity of Dna2.
MATERIALS AND METHODS
Chemicals, nucleotides, enzymes and DNA
The oligonucleotides (listed in Table 1) used to construct various DNA substrates were synthesized commercially from Genotech (Daejeon, Korea) and gel-purified prior to use. Nucleoside triphosphates and DNase I were obtained from Sigma-Aldrich. [γ-32P] ATP (>5000 Ci/mmol) and [α-32P] dCTP (>5000 Ci/mmol) were purchased from IZOTOP (Budapest, Hungary). Restriction endonucleases, PCR polymerases, Klenow Fragment (3′→5′ exo−) were purchased from either New England Biolabs (Beverly, MA) or EnzynomicsTM (Daejeon, Korea). The pRS plasmids were purchased from New England Biolabs. The pET28 vectors used for protein expression in Escherichia coli were from Novagen (Darmstadt, Germany). Isopropyl β-d-1-thiogalactopyranoside (IPTG) and X-gal were from ElpisBiotech, Inc. (Daejeon, Korea). Imidazole was from Acros Organics (Geel, Belgium), and 5-fluorooritic acid (5-FOA) was obtained from Duchefa Biochemie (Haarlem, the Netherlands).
Table 1.
Oligonucleotides used for construction of substrates
| No. | Nucleotide sequences (length in nt) | Name |
|---|---|---|
| 1 | 5′-GAAAACATTATTAATGGCGTCGAGCTAGGCACAAGGCGAACTGCTAACGG-3′ (50) | 5TY-1 |
| 2 | 5′-GGAAAACATTATTAATGGCGTCGAGCTAGGCACAAGGCGAACTGCTAACGG-3′ (51) | 5TY |
| 3 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTAGCTCGACGCCATTAATAATGTTTTC-3′ (50) | ANTI-5TY-1 |
| 4 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTA-3′ (25) | 5TB |
| 5 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTAG-3′ (26) | 5TBG |
| 6 | 5′-CGCTCGACGCCATTAATAATGTTTTC-3′ (26) | 729-FLAP-1 |
| 7 | 5′-CGAACAATTCAGCGGCTTTAACCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (52) | 729 |
| 8 | 5′-GCTCGACGCCATTAATAATGTTTTC-3′ (25) | 5TA |
| 9 | 5′-CTCGACGCCATTAATAATGTTTTC-3′ (24) | 5TA-1 |
| 10 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (60) | T60 |
| 11 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTT-3′ (59) | T60-CH-1 |
| 12b | 5′-GCUCGAGGCUCUGGCAGCCUCGAGC-3′ (25)a | SRE-RNA |
aUnderlined are inverted repeat sequences that form hairpin or stem structure.
bRibooligonucleotide.
Dna2, Dna2Δ405N and Dna2(1–405) (an N-terminal 405-amino-acid fragment of Dna2) were purified as described previously (9). Rad27 (yeast Fen1) was purified as described (13).
Preparation of the substrates
The position of radio-isotopic label in the substrates and substrate structures are indicated in each figure. Oligonucleotide labeling at its 5′-end by incorporating [γ-32P]ATP with T4 polynucleotide kinase was described previously (30). To label at 3′-end, downstream oligonucleotide was first annealed to the template and upstream oligonucleotides in a molar ratio of 1:1.2:1.4 (downstream/template/upstream oligonucleotides, respectively). The annealing reaction was performed by using PCR machine (95°C, 5 min; 65°C, 30 min; −0.5°C/min). Note that downstream oligonucleotide is two nucleotide shorter when annealed with template oligonucleotide (Table 1). Annealed substrate was then labeled at 3′-end of downstream oligonucleotide by incorporating [α-32P] dCTP with Klenow Fragment (3′→5′ exo−), and then chased with excess amount of cold dCTP. The resulting flap substrate was gel-purified prior to use.
Construction of yeast strains
S. cerevisiae strains used in this study were as follows: YPH499 (MATa ade2-101 ura3-52 lys2-801 trp1-63 his3-200 leu2-Δ1 GAL+), YJA2 (MATa ade2-101 ura3-52 lys2-801 trp1-63 his3-200 leu2-Δ1 GAL+ dna2Δ405N), YJA1B (MATa ade2-101 ura3-52 lys2-801 trp1-63 his3-200 leu2-Δ1 GAL+ dna2::HIS pRS316-DNA2), 4053-5-2 (MATa trp1 leu2 ura3 his7 can1) and 7720-x (MATa trp1 leu2 ura3 his7 can1 dna2-x) (31). The VTS1 gene was disrupted from YPH499 and YJA2 by replacing it with the HIS3 gene. In order to perform one-step gene disruption, two primers were used: VH-F (5′-GTT AAT GGT TAT ACA GCT GCC ATT TGA CCG TGC ACC ACA GCA AAT TAT Cct gag agt gca cca taa ttc-3′) and VH-R (5′-GTT AAG TAA ATA GTT TTA TGC CAT TAA ACG GTG CAA AAA CAG GTG TAA TCg cat atg atc cgt cga gtt c-3′). The sequences in capital letters are homologous to the flanking sequences of the VTS1 open reading frame (ORF), while sequences in small letters are homologous to HIS3. Using these two primers and pRS303 (with His3+ as a selection marker) as the template, the HIS3 gene was amplified. The amplified fragments thus contained the HIS3 gene flanked by upstream and downstream sequences of VTS1 and were transformed into YPH499 and YJA2. Transformants that grew in the histidine-dropout medium were analyzed for correct disruption.
The VTS1 gene was also disrupted in 7720-x strains, which possess different point-mutations in DNA2, by replacing it with the LEU2 gene. Two primers, VL-1 (5′-GCa agc ttG ACT TTC TCA TCG ATG TTG TGC CTA GAA G-3′) and VL-2 (5′-GCg gat ccG ATT TCT TTG CTG ACA ATT ACT TTA TAT G-3′), were used to amplify the upstream region of VTS1 ORF. VL-3 (5′-GCc tcg agG AAC ATA TTT CAT TTA TTT TGT AAT TTT AAC-3′) and VL-4 (5′-GCa agc ttC ATT TAC TAC TAG TAT ATT TCC TTT TGT GAA G-3′) were used to amplify the downstream region of VTS1 ORF. Small letters in these sequences indicate restriction enzyme sites for ligation into the pRS305 vector. The PCR products (each, 350 bp) were cloned into pRS305 (with Leu2+ as a marker) and digested with HindIII. The resulting linearized DNA was transformed into each of the 7720-x dna2 mutant strains. Transformants were then spread on leucine-dropout media and analyzed by PCR and Southern blotting.
YPH499 (VTS1-FLAG) is a strain in which the endogenous VTS1 gene was 5×FLAG-tagged at its C-terminus. The 5×FLAG sequence with the KanMX gene was from the pKL258 vector. Two primers, VFLAG-1 (5′-GAA TCG TTA TTG ATT GAT TAC AAA GAA CGT GAT TTA ATT GAT AGA TCT GCT TAT cgt acg ctg cag gtc gac-3′) and VFLAG-2 (5′-CAA AGT AAA TAA AAA TTA AAT ATT TAT GCA ACG TCA AGA CAA TCA Act cga tga att cga gct cg-3′), were used to amplify the DNA fragment using pKL258 as a template. The sequences in capital letters in VFLAG-1 and VFLAG-2 are homologous to the upstream and downstream sequences, respectively, of the stop codon in the VTS1 ORF. The sequences in small letters in both primers are homologous to flanking sequences of the 5×FLAG plus KanMX gene in pKL258 vector. Thus, the amplified fragments contained the 5×FLAG and KanMX gene flanked by sequences upstream and downstream of the VTS1 stop codon and were used for integration into the C-terminus of VTS1 ORF by homologous recombination. Transformants that grew in YPD containing G418 were selected and analyzed by PCR and western blotting.
Measurement of growth rate
The rate of cell growth was measured by counting cell numbers using a hematocytometer. Cells were first inoculated in 2 ml of YPD liquid media and grown at 25°C until saturation. The cells were then diluted in 30 ml of fresh YPD media to a concentration of 1×106 cells/ml (∼0.1 of A600) and incubated at 32°C in a shaking incubator. Aliquots containing the same number of cells of each strain were withdrawn at indicated times and subjected to brief sonication to disperse clumped cells, and then vortexed for 30 s to generate homogeneous suspension.
Cell growth defects were examined by the drop-dilution assay. Cells were inoculated in 2 ml YPD liquid media and grown at 25°C until saturation. The cells were then diluted to a concentration of 1×107 cells/ml (∼1.0 of A600) and spotted in 10-fold serial dilutions (105, 104, 103 and 102 cells) on YPD plates. Plates were incubated at different temperatures (25°C and 30°C) for 3 days.
Purification of recombinant Vts1
The VTS1 ORF was inserted into pET28 vector that expresses the ORF as an N-terminal 6 × His-tagged protein. The pET28-Vts1 plasmid was transformed into E. coli BL21 (CodonPlus), and cells (1 l) were grown at 25°C until the A600 value reached 0.5. At this point, cells were induced with 0.1 mM IPTG (final concentration) for 4 h. Cells were harvested by centrifugation and the cell pellet was resuspended in 30 ml of buffer T500 [50 mM Tris–HCl/pH 8.0, 500 mM NaCl, 10% glycerol, 0.1% Nonidet P-40 (NP-40), 1 mM phenylmethlysulfonyl fluoride (PMSF), 0.1 mM benzamidine, 1.25 µg/ml leupeptin and 0.625 µg/ml pepstatin A]. The subscript number in T500 indicates the concentration of NaCl in mM. Then, cells were sonicated for four cycles with a 1-min interval on ice. The supernatant was adjusted to 5 mM imidazole (IDZ, final concentration). A Ni2+-NTA agarose (1.5 ml, Qiagen; Valencia, CA) column was pre-equilibrated with T500 plus 5 mM IDZ. In all subsequent steps, the final concentration of NP-40 in T500 was reduced to 0.02%. The supernatant was loaded onto a Ni2+-NTA agarose column. The column was then washed with 5-column volumes of T500 plus 70 mM IDZ, and eluted with T500 plus 480 mM IDZ. The eluate was subjected to glycerol gradient sedimentation (5 ml, 15–35% glycerol in buffer T500) at 48 000 r.p.m. for 24 h in a SW55 Ti rotor (Beckman). Fractions (250 µl each) were collected from the bottom of the tube. Peak fractions were stored at −80°C. The Vts1-A498Q mutant and Vts1(1∼406) were purified using the same procedure as described above. Vts1SAM was also purified similarly, but most of proteins were eluted in the 70 mM IDZ wash from the Ni2+-NTA agarose column.
Nuclease assays
Standard nuclease assays were performed in reaction mixtures (20 µl) containing 50 mM Tris–HCl/pH 7.8, 2 mM MgCl2, 2 mM DTT, 0.25 mg/ml BSA, 62.5 mM NaCl and 15 fmol of DNA substrate. Reactions were incubated at 37°C for 15 min, followed by the addition of 4 µl of a 6 × stop solution (60 mM EDTA/pH 8.0, 40% sucrose, 0.6% SDS, 0.25% BPB, 0.25% xylene cyanol). The products were subjected to electrophoresis for 30 min at 150 V in 0.5 × TBE (45 mM Tris, 45 mM boric acid and 1 mM EDTA). The gels were dried on a DEAE-cellulose paper and autoradiographed. Labeled DNA products were quantified with the use of a Phosphor Imager (BAS-1500, Fujifilm).
Electrophoretic mobility shift assay
Reaction mixtures (20 µl) containing 50 mM Tris-HCl/pH 7.8, 5% glycerol, 2 mM DTT, 0.25 mg/ml BSA, 37.5 mM NaCl, 5 fmol of labeled DNA substrate and indicated amounts of Vts1 or its derivatives were preincubated on ice for 10 min, followed by incubation at 37°C for 5 min. Complexes formed were resolved in a 5% polyacrylamide gel in 0.5× TBE at 4°C, and the amount of complex formed was measured using a Phosphor Imager.
Alkaline protein extraction from S. cerevisiae
The expression level of proteins in yeast was determined after alkaline protein extraction. YJA1B harboring pRS314-dna2-K1080E, pRS316-DNA2 and the pRS325-suppressor gene were grown to saturation. Expression of the suppressor gene cloned in pRS325 was driven by a constitutive ADH1 promoter or its native promoter. Cells (5×107, total) were harvested and resuspended in the lysis buffer (150 μl; 1.85 M NaOH, 7.5% β-mercaptoethanol). Then, the mixture was incubated on ice for 10 min. TCA (20% v/v, 150 μl) was added and incubated on ice for an additional 10 min. Samples were then centrifuged at 4°C for 5 min at 20 000 g in an Eppendorf centrifuge. Supernatants were discarded, and 1× SDS sample buffer was added to the precipitated materials. The samples were neutralized with 1–2 μl of 2 M Tris/pH 11.0, and resuspended by rigorous vortexing after 30 min of incubation at 60°C, followed by boiling for 10 min. The samples were then subjected to electrophoresis in a 10% SDS–PAGE gel. Gels were Coomassie-stained and analyzed by western blotting.
Enzyme-linked immunosorbent assay
ELISA was used to detect the physical interaction between Dna2 and Vts1 as described previously (32) with some modifications. Vts1 was diluted to a concentration of 10 nM in the carbonate buffer (16 mM Na2CO3/34 mM NaHCO3, pH 9.6). Vts1 was then added to appropriate wells of a 96-well ELISA plate (100 µl/well), and the plate was incubated for 2 h at 25°C. As control, BSA was substituted for Vts1 in the coating step. Wells were aspirated and washed three times with the wash buffer (50 mM Tris–HCl/pH 7.5, 200 mM NaCl, 2.5 mM KCl, 0.05% Tween 20). The blocking buffer (50 mM Tris–HCl/pH 7.5, 150 mM NaCl, 2.5 mM KCl, 0.05% Tween 20 and 3% BSA) was added and the mixture was incubated for 2 h at 25°C. Wells were aspirated and washed three times with the blocking buffer. Preparations of Dna2, Dna2Δ405N and Dna2(1–405) were serially diluted (as indicated) in the binding buffer (50 mM Tris–HCl/pH 7.5, 100 µg/ml BSA, 200 mM NaCl and 0.02% NP-40), and then added to Vts1-coated wells (100 µl/well). The plates were incubated for 30 min at 25°C. Wells were aspirated and washed four times with the binding buffer. Primary antibodies (rabbit polyclonal antibodies against Dna2) were diluted 2000-fold in the blocking buffer, added to appropriate wells. The mixtures were then incubated for 1 h at 25°C. Wells were aspirated and washed five times with the blocking buffer. Secondary antibodies, diluted 10 000-fold in the conjugate buffer (50 mM Tris–HCl/pH 7.5, 150 mM NaCl, 0.05% Tween 20, 1% BSA), were added to appropriate wells and the mixtures were incubated for 30 min at 25°C. Wells were aspirated and washed five times with the conjugate buffer. Dna2–Vts1 complexes were detected using the TMB substrate (Sigma). Absorbance readings were corrected by subtracting background absorbance generated with BSA-coated control wells.
DNA footprinting
Reaction mixtures (20 μl) contained 50 mM Tris–HCl/pH 7.8, 2 mM DTT, 5% glycerol, 0.25 mg/ml BSA, 62.5 mM NaCl, 20 fmol of DNA substrate and indicated amounts of Vts1. Reaction mixtures were preincubated at 4°C for 10 min, followed by further incubation at 37°C for 5 min to allow Vts1 to bind DNA substrates. In order to cleave unprotected regions in the DNA substrate, DNase I was added as indicated in each experiment with 2 μl of 10 × Ca2+/Mg2+ buffer (5 mM CaCl2, 25 mM MgCl2). The mixtures were then incubated at 4°C for 10 min, treated with 2 × stop buffer (23 μl; 20 mM EDTA/pH 8.0, 95% formamide, 0.25% xylene cyanol) and boiled for 5 min. Samples were subjected to electrophoresis for 1.5 h at 30 W in 1× TBE (90 mM Tris, 90 mM boric acid and 2 mM EDTA) through a 15% denaturing gel containing 7 M urea.
Isolation of yeast nuclei
Cells (25 ml) were grown in YPD to A600 = 1.5, harvested and resuspended in 1 ml of 50 mM Tris/pH 7.5 and 30 mM DTT. After incubation at 30°C for 15 min, cells were centrifuged for 3 min in 1700 g (Sorvall SS-34 rotor), and pellets were resuspended in 667 μl of YPD/S (YPD with 1 M sorbitol). Then, 333 μl of 2 M sorbitol was added, followed by the addition of 10 µl of 4 mg/ml lyticase (Sigma). Samples were then incubated for 45 min at 30°C; this treatment resulted in efficient formation of spheroplasts (∼90%). The spheroplasts were collected by centrifugation, resuspended in 1 ml of YPD/S and incubated at 30°C for 15 min to allow cells to recover. After one additional wash cycle at 4°C, cells were resuspended in 800 μl of buffer N (25 mM K2SO4, 30 mM HEPES-NaOH/pH 7.5, 5 mM MgSO4, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 3 mM DTT and protease inhibitors). Cells were homogenized with 15–20 gentle strokes and stored at 4°C to permit cell debris to settle. The supernatant (nuclei + cytoplasmic components) was centrifuged in 3800 g (Sorvall SS-34 rotor) in order to pellet nuclei. The supernatant, designated ‘cytoplasmic fraction’, was used in this experiment. The pellet fractions containing nuclei and/or cell debris were resuspended in the same volume of buffer N as described for the isolation of the cytoplasmic fraction. Equal volumes of each fraction were then loaded on a 10% SDS–PAGE gel for western blotting.
RESULTS
Vts1 is an allele-specific suppressor of dna2 mutations
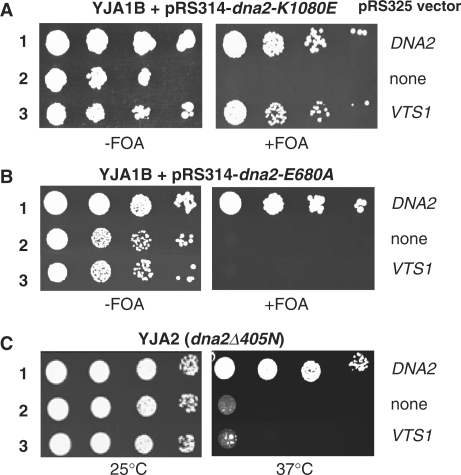
To better understand mechanistic aspects of Dna2 during processing of Okazaki fragments, we searched for novel proteins that function jointly with Dna2 endonuclease/helicase. This screen identified VTS1 as a multi-copy suppressor of dna2-K1080E, a mutant allele devoid of helicase activity. Subsequent subcloning confirmed that the multi-copy expression of VTS1 alone was sufficient to suppress the Dna2 helicase-deficient mutant. As shown in Figure 1A, pRS325 empty vector, pRS325-DNA2 and pRS325-VTS1 were separately transformed into the dna2Δ YJA1B strain that harbors both pRS314-dna2-K1080E and pRS316-DNA2. Plasmids pRS314, pRS316, and pRS325 contained Trp1+, Ura3+ and Leu2+, respectively, as a selection marker. All transformants were able to grow in the absence of 5-FOA (Figure 1A, left). When cells were grown in the presence of 5-FOA, only cells that had lost Ura3+-pRS316 (hence wild-type DNA2) could grow. In the presence of pRS325-VTS1, the YJA1B strain could grow with pRS314-dna2-K1080E only and did not require pRS316-DNA2 any longer (Figure 1A, right), indicating that multi-copy expression of VTS1 suppressed the growth defects of dna2-K1080E. In contrast, VTS1 expression failed to restore growth of either dna2-E680A or dna2Δ405N mutant cells (Figure 1B and C), which lack endonuclease activity and the N-terminal 405-amino-acid domain, respectively. These results indicate that suppression by Vts1 requires both the endonuclease activity and the N-terminal domain of Dna2.
Figure 1.
VTS1 is a multi-copy suppressor specific for Dna2-K1080E, a Dna2 helicase-negative mutant. (A) Overexpression of VTS1 suppresses the dna2 helicase-negative mutant. The dna2Δ strain YJA1B harboring both pRS314-dna2-K1080E and pRS316-DNA2 plasmids was transformed with each of the three plasmids indicated at the right of the figure. Transformants were grown in liquid media and cells were spotted in 10-fold serial dilutions (105, 104, 103 and 102 cells) onto plates without (−FOA) or with (+FOA) 5-FOA, followed by incubation for 3 days at 30°C, as shown. (B) Overexpression of Vts1 does not suppress the dna2 endonuclease-deficient mutant. The same plasmids used in panel A were transformed into dna2Δ strain YJA1B harboring pRS314-dna2-E680A and pRS316-DNA2 plasmids. Cells were grown as shown in panel A. (C) Overexpression of Vts1 does not suppress dna2Δ405N that lacks the N-terminal 405-amino-acid domain. The same plasmids used in panel A were transformed into YJA2 (dna2Δ405N). Transformants were grown in liquid media and the cells were spotted in 10-fold serial dilutions (105, 104, 103 and 102 cells) onto SD-Leu plates. The plates were incubated for 3 days at 25°C or 37°C as indicated.
The vts1Δ dna2 double mutant displays synergistic growth defects
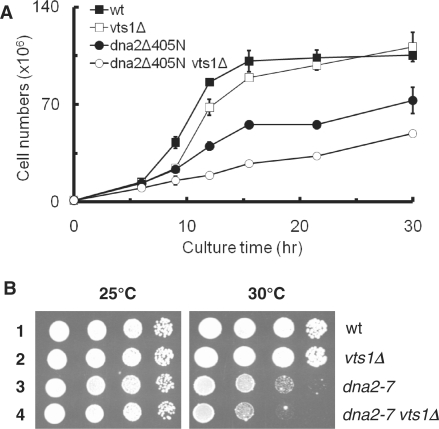
We expected that vts1Δ dna2 double mutation would lead to more severe growth defects than that caused by a single mutation alone. Since both dna2-K1080E and dna2-E680A cells are not viable, this analysis was not feasible (Figure 1B and C). Instead, we examined the growth of vts1Δ dna2Δ405N (Figure 2A) and vts1Δ dna2-7 (Figure 2B) under semipermissive conditions. The dna2-7 allele contains an aspartic acid for glycine substitution at position 913 that renders the dna2-7 mutant cells both temperature- and MMS-sensitive (31). When grown at 32°C (a semipermissive temperature for dna2Δ405N), vts1Δ mutants exhibited a longer lag phase period, but they doubled in cell numbers with a doubling time of ∼3 h (Figure 2A, open square) similar to wild type (closed square) at the later stage (9 h after incubation). In contrast, dna2Δ405N grew more slowly (doubling time, ∼6 h) (Figure 2A, closed circle) than either wild-type or vts1Δ mutant cells. The vts1Δ dna2Δ405N double mutant cells, however, grew significantly slower with a doubling time of ∼9 h (Figure 2A, open circle) than cells harboring vts1Δ or dna2Δ405N alone.
Figure 2.
Synergistic growth defects associated with dna2 mutations and vts1Δ. (A) Cells were incubated in 32°C and the cell numbers were measured at the time point indicated using a hematocytometer. (B) Cells were spotted in serial 10-fold dilutions on YPD media and plates were incubated for 3 days at the temperatures indicated.
We also tested dna2-7 alone and dna2-7 vts1Δ. The vts1Δ dna2-7 double mutant cells displayed synergistic growth defects at 30°C, a semipermissive temperature for dna2-7 (Figure 2B). The vts1Δ dna2-7 double mutant cells grew more poorly (∼10 fold) than cells with vts1Δ or dna2-7 alone. These results, together with genetic suppression data shown above, suggest that VTS1 plays a role in a process that requires the function of DNA2.
Vts1 stimulates endonuclease activity of Dna2 in vitro
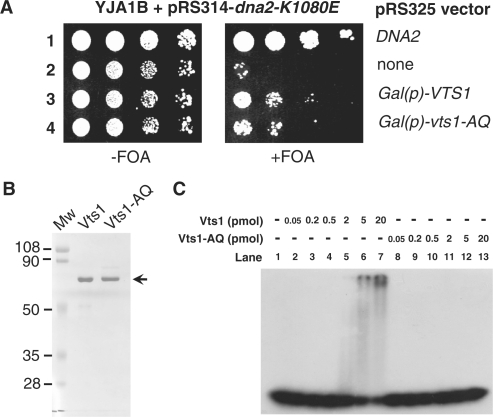
The SAM domain of Vts1 is critical for the in vivo posttranscriptional regulation of target mRNAs. The vts1-A498Q mutation, containing the single substitution (Ala into Gln) at position 498 amino acid in the SAM domain, abolished the binding of Vts1 to SRE-RNA containing a specific stem-loop structure, resulting in defects in the posttranscriptional regulation of the target mRNAs (22). In order to examine whether this structure-specific RNA-binding activity of Vts1 is required for the suppression of dna2-K1080E, we prepared the vts1-A498Q mutant allele by site-directed mutagenesis and cloned into pRS325. The resulting plasmid, pRS325-vts1-A498Q, was introduced into dna2Δ strain YJA1B harboring both pRS314-dna2-K1080E and pRS316-DNA2. Multi-copy expression of this mutant protein failed to restore the growth of dna2-K1080E mutation unlike wild-type VTS1 (data not shown). This effect was probably due to the poor expression of the Vts1-A498Q mutant protein, because the Vts1-A498Q protein was hardly detected (data not shown). For this reason, the native promoter of Vts1 was replaced with a GAL1 promoter to generate pRS325-GAL(p)-VTS1 and pRS325-GAL(p)-vts1-A498Q. When expression of Vts1-A498Q was induced by galactose, both Vts1 and Vts1-A498Q suppressed dna2-K1080E to similar extents (Figure 3A), indicating that the SAM domain of Vts1 is not critical for its functional interaction with Dna2.
Figure 3.
Purification of Vts1 and Vts1-A498Q. (A) The structure-specific RNA-binding activity of Vts1 is dispensable for suppression of the dna2-K1080E mutant. Plasmids containing genes indicated at the right of figure were introduced into dna2Δ strain YJA1B harboring pRS314-dna2-K1080E and pRS316-DNA2 plasmids. Cells were grown in SG-His-Trp-Leu (2% galactose without glucose) and 10-fold serial dilutions (105, 104, 103 and 102 cells) were spotted onto SG-His-Trp-Leu plates without (−FOA) or with (+FOA) 5-FOA. The plates were incubated for 5 days at 30°C as shown. (B) An SDS–PAGE analysis of wild-type Vts1 and mutant Vts1-A498Q (Vts1-AQ) proteins. Both proteins were purified as described in ‘Material and Methods’ section. Of each protein, 1 µg was subjected to electrophoresis in a 10% SDS–PAGE gel, which was then stained with Coomassie Blue G-250. (C) Vts1 binds to the SRE-RNA substrate, while Vts1-A498Q does not. Electrophoretic mobility shift assays were performed as described in ‘Materials and Methods’ section. The SRE-RNA substrate (5 fmol) was incubated in the presence of increasing amounts (0.5–20 pmol) of Vts1 or Vts1-A498Q at 4°C for 10 min, followed by additional 5-min incubation at 37°C. After incubation, reaction mixtures were subjected to electrophoresis in a 6% native polyacrylamide gel.
In order to confirm our in vivo observations in vitro, both wild-type Vts1 and mutant Vts1-A498Q proteins were purified from E. coli to near homogeneity (Figure 3B), as described in ‘Materials and Methods’ section. Both proteins contained a hexahistidine epitope tag in their N-terminal region. We confirmed that the epitope-tagged proteins retained the ability to suppress dna2-K1080E (data not shown). Our preparations of Vts1 and Vts1-A498Q were devoid of any detectable helicase (data not shown) or nuclease activities (Figure 4A, lanes 3 and 4). Using purified proteins, we first carried out electrophoretic mobility shift assays to confirm that purified Vts1 possesses SRE-RNA-binding activity and Vts1-A498Q does not. For this purpose, the labeled SRE-RNA substrate was prepared (see Table 1 for their nucleotide sequence) and incubated with increasing levels (0.05–20 pmol) of either Vts1 or Vts1-A498Q (Figure 3C). Consistent with the previous report (22), Vts1, but not Vts1-A498Q, bound to the SRE-RNA substrate in electrophoretic mobility shift assays (Figure 3C).
Figure 4.
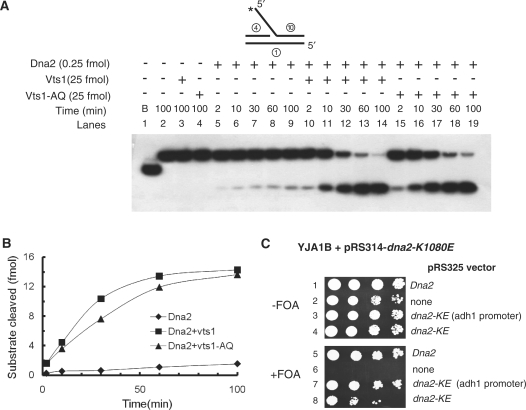
Vts1 stimulates the endonuclease activity of Dna2 in vitro. (A) Both Vts1 and Vts1-A498Q stimulate Dna2 endonuclease activity in vitro. Three sets of reaction mixtures were assembled on ice; one with Dna2 (0.25 fmol) only (lanes 5–9), the second with Dna2 (0.25 fmol) and Vts1 (25 fmol) and the third with Dna2 (0.25 fmol) and Vts1-A498Q (25 fmol). The reaction mixtures were then incubated at 37°C for varying periods of time (0, 2, 10, 30, 60 and 100 min). Cleavage products were analyzed on a 10% denaturing polyacrylamide gel as described in ‘Materials and Methods’ section. ‘B’ in lane 1 represents boiled. (B) The amount of product formed from reactions in panel A was plotted against the time of incubation. (C) Overexpression of Dna2-K1080E (Dna2-KE) suppresses its own growth defects. The experimental procedures are as described in Fig. 1A. Note that the expression of Dna2-K1080E was driven either by the ADH1 promoter or by its own promoter.
Next, we examined the influence of Vts1 and Vts1-A498Q on the Dna2 endonuclease activity. For this purpose, time-course experiments were carried out with a low level (0.25 fmol) of Dna2 in the presence or absence of Vts1 or Vts1-A498Q (each at 25 fmol). As shown in Figure 4A (lanes 5–9), Dna2-catalyzed cleavage products slowly increased with time (up to 100 min), indicating that Dna2 was stable over this incubation time (Figure 4A, lanes 5–9; Figure 4B, diamond). However, the rate of cleavage by Dna2 was markedly (∼8- to 10-fold) stimulated by the presence of either Vts1 (Figure 4A, lanes 10–14; Figure 4B, square) or Vts1-A498Q (Figure 4A, lanes 15–19; Figure 4B, triangle) and plateaued after 60 min of incubation. These findings indicate that both Vts1 and Vts1-A498Q stimulate the endonuclease activity of Dna2 with almost identical efficiency, confirming that the structure-specific RNA-binding activity of Vts1 was not required for the stimulation of the Dna2 endonuclease activity.
Increased levels of Dna2 endonuclease activity are sufficient to suppress the dna2-K1080E mutation in vivo
Since both Vts1 and Vts1-A498Q stimulated the Dna2 endonuclease activity and suppressed the dna2-K1080E mutation, we determined whether increased Dna2 endonuclease activity could rescue the growth defects of dna2-K1080E. This question can be addressed directly by increasing the copy number of the dna2-K1080E mutant allele. We constructed two pRS325 plasmids in which the dna2-K1080E mutant allele was differentially expressed, one plasmid in which expression of dna2-K1080E was driven by its native promoter and the other by the constitutive strong ADH1 promoter. The two plasmids were introduced into the dna2Δ strain YJA1B harboring both pRS314-dna2-K1080E and pRS316-DNA2, and the extent of suppression was examined. Expression of dna2-K1080E in both plasmids resulted in suppression of dna2-K1080E mutation (Figure 4C). In fact, the expression of dna2-K1080E driven by the ADH1 promoter suppressed the growth defects of dna2-K1080E more efficiently than its native promoter, indicating that the increased endonuclease activity of Dna2 could be the cause of suppression. Based on this result, we concluded that elevated levels of the Vts1 protein increase the endonuclease activity of Dna2 and suppress the lethal phenotype of dna2-K1080E.
Stimulation of Dna2 endonuclease by Vts1 requires a direct protein–protein interaction
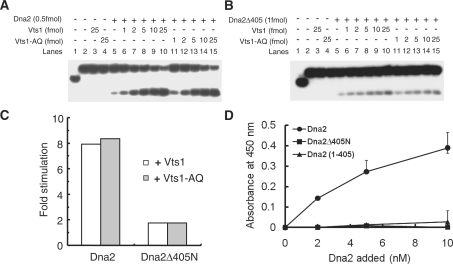
As shown in Figure 1C, overexpression of VTS1 failed to suppress the temperature-sensitive growth defects of dna2Δ405N. This mutant enzyme, lacking the N-terminal 405-amino-acid domain, possesses the same endonuclease activity as the wild-type enzyme (13). In order to further substantiate our notion that the stimulation of Dna2 endonuclease activity by Vts1 is responsible for the suppression observed in vivo, we examined whether increasing amount of Vts1 or Vts1-A498Q affected the endonuclease activities of wild-type Dna2 (Figure 5A) and Dna2Δ405N (Figure 5B). The endonuclease activity of wild-type Dna2 was markedly stimulated (∼8-fold) in response to increasing levels of both Vts1 and Vts1-A498Q (Figure 5A and C), while the stimulation of Dna2Δ405N was marginal (<2-fold) (Figure 5B and C). This is in keeping with the results obtained from the time-course experiments described above (Figure 4A). In conclusion, our findings demonstrate that the stimulation of the Dna2 endonuclease activity by Vts1 requires the N-terminal domain of Dna2 and that suppression of dna2-K1080E involves the stimulation of Dna2 endonuclease activity.
Figure 5.
Vts1 stimulates Dna2 endonuclease via interaction with the DNA-binding domain of Dna2. (A) Vts1 stimulates Dna2 endonuclease in vitro. Reaction mixtures containing indicated amounts of Dna2 and either Vts1 wild-type or Vts1-A498Q (Vts1-AQ) were incubated for 15 min in 37°C. Products were analyzed on 10% denaturing polyacrylamide gel. (B) Vts1 does not stimulate Dna2Δ405N endonuclease in vitro. Experiments with Dna2Δ405N were done same as described in panel A. (C) The stimulation folds of Dna2 endonuclease activity by Vts1 (or Vts1-AQ) in reactions described in A and B were plotted. (D) Vts1-coated wells were incubated with increasing concentrations (2, 5 and 10 nM) of Dna2 derivatives indicated at the top of the figure. Wells were aspirated, washed three times and bound Dna2, Dna2Δ405N and Dna2(1–405) were detected by ELISA using rabbit polyclonal antibodies against Dna2. Absorbance readings at each point were corrected for background absorbance generated with BSA-coated wells.
Experiments using ELISA assays revealed that highly purified Dna2 and Vts1 proteins directly interacted with each other. Vts1 was first adsorbed to a microtiter plate that was incubated in the presence of increasing levels (2–10 nM) of purified wild-type Dna2, Dna2Δ405N and Dna2(1–405) (the N-terminal 405-amino-acid fragment). After washing, the amount of Dna2 and its derivatives adsorbed to the wells was detected with polyclonal antibodies specific to Dna2. The polyclonal antibodies were appropriately diluted in order to detect each Dna2 derivative to the same extent (data not shown). As shown in Figure 5D, a specific interaction between purified Vts1 and Dna2 was observed, which increased as the level of wild-type Dna2 was increased (Figure 5D, circle). However, Dna2Δ405 and Dna2(1–405) failed to interact with Vts1 (Figure 5D, square and triangle, respectively). We also carried out the reciprocal experiment; Dna2 was first adsorbed to the wells of the microtiter plate, and Vts1 protein was then added. Detection was done with polyclonal antibodies specific for Vts1. Consistent with the result described above (Figure 5D), Vts1 bound to the wells coated with wild-type Dna2, neither Dna2Δ405 nor Dna2(1–405) (data not shown). These findings suggest that the interaction of Vts1 with Dna2 requires both the N-terminal domain and some other region of Dna2. Taken together, our in vitro results are in keeping with in vivo observations that VTS1 fails to suppress the temperature-sensitive growth defects of dna2Δ405N, further supporting the notion that the stimulation of the Dna2 endonuclease activity by Vts1 is the direct cause of the suppression of dna2-K1080E and that this effect is dependent on a specific protein–protein interaction.
Vts1 is a flap-specific DNA-binding protein
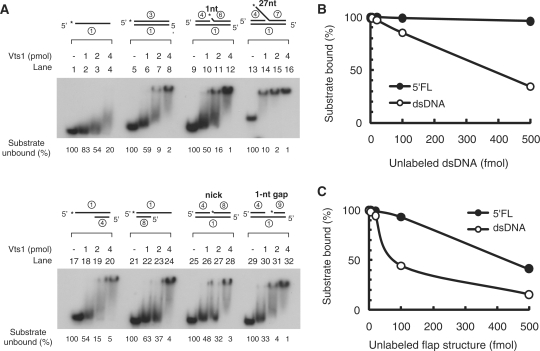
Vts1 could stimulate Dna2 endonuclease activity by recruiting Dna2 more efficiently to a DNA substrate. Since this mechanism would likely require the binding of Vts1 to DNA, we examined this possibility with different structures using electrophoretic mobility shift assays. The substrates tested included single-stranded (ssDNA, 50-nt), double-stranded (dsDNA, 50-bp), 1-nt 5′-flap, 27-nt 5′-flap (5′FL), 5′-overhang, 3′-overhang, nicked duplex and 1-nt gapped duplex DNAs. All substrates were constructed with a common 50-nt oligonucleotide (Table 1, oligonucleotide 1). The interaction of Vts1 with ssDNA yielded a smeared electrophoretic mobility shift (Figure 6A, lanes 1–4) at relatively high protein levels (>2 pmol), suggesting that binding, if it occurred, was not stable. In contrast, all other substrates containing a duplex DNA region were more efficiently complexed than the ssDNA substrate. Among them, Vts1 bound the 27-nt 5′-flap DNA with the greatest affinity and stability, since it formed the highest level of a discrete nucleoprotein complex (Figure 6A, lanes 13–16). The binding of Vts1 to the 1-nt 5′-flap DNA (Figure 6A, lanes 9–12) was similar to that observed with the other partial duplex DNA substrates. Thus, Vts1 appears to bind preferentially to a duplex containing a long 5′-ssDNA flap region.
Figure 6.
Vts1 is a flap DNA-specific-binding protein. (A) Electrophoretic mobility shift assays were performed as described in ‘Materials and Methods’ section. Each substrate (15 fmol), shown at the top of the gel, was preincubated with increasing amounts (1, 2 and 4 pmol) of Vts1 at 4°C for 10 min, followed by an additional 5 min of incubation at 37°C. After incubation, samples were loaded on a 6% native polyacrylamide gel and the Vts1–DNA complexes formed were analyzed by autoradiography as described in ‘Materials and Methods’ section. The amount of substrate left was measured and the values are presented at the bottom of the gel. (B and C) Substrate challenge assays. The Vts1–DNA complex was first formed by incubating 5 fmol of labeled substrate (indicated at the top of the gel) with excess Vts1 (4 pmol) as described in panel A. Increasing amounts (5, 20, 100 and 500 fmol) of unlabeled competitor DNA were added to reactions containing the Vts1/labeled substrate complex. The amount of labeled substrate left was plotted against the amount of unlableled competitor DNA used.
In order to confirm that Vts1 binds more stably to the 5′-flap structure than duplex DNAs, substrate-challenge experiments were carried out. We first incubated 5 fmol of labeled 27-nt 5′-flap (5′FL) or simple duplex DNA (dsDNA) with a fixed amount (4 pmol) of Vts1 to allow formation of the nucleoprotein complex. As shown in Figure 6A, the level of Vts1 (4 pmol) used shifted nearly all of the 27-nt 5′FL or dsDNA substrate added. The complexes formed were then challenged with increasing amounts of unlabeled competitor DNAs. The addition of increasing levels (up to 500 fmol, ∼100-fold molar excess) of unlabeled dsDNA as competitor had no effect on the Vts1-5′FL complex (Figure 6B, closed circle), while the Vts1-dsDNA complex was linearly reduced (up to ∼60%) by the addition of unlabeled dsDNA (Figure 6B, open circle). In contrast, the Vts1–dsDNA complex (open circle) was affected to a greater extent than the Vts1–5′FL complex (closed circle) by increasing concentrations of unlabeled competitor 5′FL DNA (Figure 6C). These data confirm that Vts1 is more stably bound to 5′-flap DNA than dsDNA without a flap.
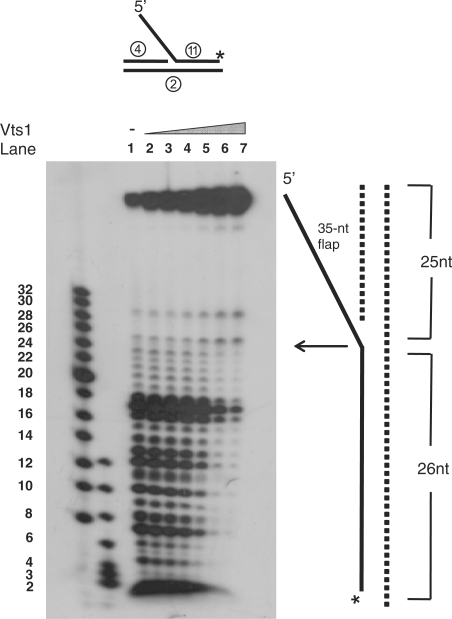
Since the efficient binding of Vts1 to DNA appeared to require the presence of a flap structure, DNA footprinting analyses were carried out with the Vts1/35-nt 5′FL DNA complex. We first determined the optimal concentration of DNase I that partially cleaved the 35-nt 5′FL DNA labeled at the 3′-end of the flap strand. The amount of DNase I required for optimal partial cleavage was 0.1U per reaction (data not shown). The 35-nt 5′FL DNA was first complexed with increasing levels (0.5–16 pmol) of Vts1 and then treated with DNase I (0.1U) to cleave unprotected regions in the DNA. Two changes in the protection pattern were detected. At high Vts1 concentrations (4–16 pmol), the double-stranded region of the flap-containing strand (Figure 7, lanes 5–7) was effectively protected from DNase I digestion. This protection was more pronounced near the duplex end and progressed toward the ssDNA–dsDNA junction with increasing concentrations of Vts1 (Figure 7, compare lanes 5–7). We noted that Vts1 had an intrinsic property to bind to duplex DNA ends. These results as well as those described above suggest that Vts1 binds to the single-strand flap first, and then subsequently binds to the duplex region.
Figure 7.
DNase I protection analysis of Vts1 bound to 5′-flap DNA. DNase I protection assays were carried out with the DNA substrate indicated above the gel. The 3′-end of the strand indicated above the gel was 32P-labeled. Increasing amounts of Vts1 (0.5, 1, 2, 4, 8 and 16 pmol) were preincubated at 4°C for 10 min, followed by additional 5-min incubation at 37°C. DNase I (0.1U/reaction) was then added to the reaction mixtures, followed by 10-min incubation at 4°C. The reaction products were analyzed in a 15% high-resolution sequencing gel containing 7 M urea.
Stimulation of Dna2 endonuclease by Vts1 requires its DNA-binding activity
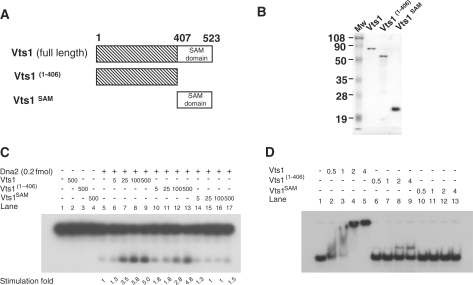
To investigate whether the DNA-binding activity of Vts1 is critical for its stimulation of Dna2 endonuclease activity, we purified two truncated fragments of Vts1, Vts1(1–406) (a 44-kDa N-terminal fragment, amino acid 1-406), and Vts1SAM (a 13-kDa protein containing the SAM domain, amino acid 407–523) (Figure 8A and B). Vts1(1–406) stimulated Dna2, but less efficiently than wild-type Vts1 (Figure 8C, compare lanes 5–9 and 10–13), while Vts1SAM failed to stimulate Dna2 endonuclease (Figure 8C, lanes 14–17). DNA-binding experiments revealed that Vts1(1–406) was capable of binding to the 5′-flap DNA, while Vts1SAM was not (Figure 8D). The DNA-binding activity of Vts1(1–406) was much weaker than wild-type Vts1 (Figure 8D, compare lanes 2–5 and 6–9). These results suggest that the stimulation of Dna2 endonuclease activity by Vts1 depends on its ability to bind DNA. It is worthwhile to note that Vts1(1–406) formed a unique and distinct DNA complex different from the smeared high-molecular weight complex formed by wild-type Vts1. Thus, we conclude that Vts1(1–406) contains DNA-binding activity and the SAM domain is required for the formation of higher order complexes with DNA substrates.
Figure 8.
DNA-binding activity of Vts1 is required for the stimulation of Dna2 endonuclease activity. (A) A schematic diagram of Vts1 and its truncated fragments used in this experiment. Note that the C-terminal 117-amino-acid fragment contains the intact SAM domain. (B) Purified Vts1 (full-length), Vts1(1–406) and Vts1SAM proteins (0.5 µg each) were subjected to electrophoresis on a 12% SDS–PAGE. The gel was then Coomassie-stained. (C) The N-terminal fragment of Vts1 is able to stimulate Dna2 endonuclease activity. The reaction mixtures containing indicated amounts of Dna2 and Vts1 (or fragmented derivatives) were incubated for 15 min at 37°C. Cleavage products were analyzed in a 10% denaturing polyacrylamide gel. (D) The N-terminal fragment of Vts1 and Vts1 are able to bind 5′-flap DNA. Electrophoretic mobility shift assays were carried out as described in ‘Materials and Methods’ section. The 27-nt 5′-flap substrate (5 fmol) was incubated at 4°C for 10 min in the presence of increasing amounts (0.5, 1, 2 and 4 fmol) of Vts1, followed by additional 5 min of incubation at 37°C.
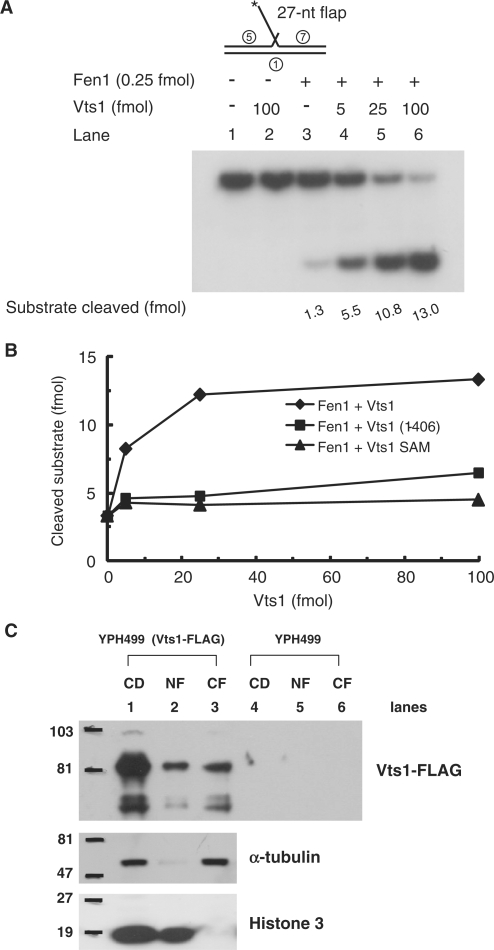
Vts1 stimulates yeast Fen1 endonuclease
Yeast Fen1, Rad27, is a critical enzyme that cleaves flap structures to generate ligatable nicks. Since Vts1 is a flap-specific binding protein, we tested whether it also stimulates the nuclease activity of Fen1. As shown in Figure 9A, Vts1 increased (∼10 fold) the Fen1 endonuclease activity. Unlike Dna2, both Vts1(1–406) and Vts1SAM failed to significantly stimulate Fen1 (Figure 9B). Thus, full-length Vts1 is required to stimulate Fen1. As discussed above, RPA is a potent inhibitor of Fen1, and this inhibition was unaffected by the addition of Vts1 (data not shown).
Figure 9.
Vts1 stimulates yeast Fen1 endonuclease activity. (A) The reaction mixtures containing indicated amounts of Fen1 and wild-type Vts1 were incubated with 5′-flap substrate (15 fmol) for 15 min in 37°C. Cleavage products were analyzed in a 10% denaturing polyacrylamide gel. (B) The same reactions as panel A were repeated with Vts1 and its derivatives as indicated. The amount of product formed was plotted against the amount of Vts1 or its derivates added. (C) Cellular localization of Vts1 in S. cerevisiae. As described in ‘Materials and Methods’ section, nuclear and cytoplasmic fractions were prepared from spheroplasts of two strains, YPH499 (negative control) and YPH499 (Vts1-FLAG), that expresses endogenous FLAG-tagged Vts1. CD, cell debris fraction; NF, nuclear fraction; CF, cytoplasmic fraction. The fractions were subjected to western-blot analyses in a 10% SDS–PAGE with specific monoclonal antibodies against α-tubulin, histone 3 and the FLAG epitope. Proteins detected were as indicated at the right of each blot.
Vts1 is present in nuclei
Our in vivo and in vitro findings support that Vts1 is involved in the processing of Okazaki fragments in yeast. If Vts1 were to play this role in vivo, we would expect it to be present in nuclei. Based on cell images available from the Organelle DB of S. cerevisiae Genome Database (http://organelledb.lsi.umich.edu/gene.php?sys_name=YOR359W), Vts1 appears to be distributed throughout the whole cell, obscuring its detection in nuclei. In order to address this issue, we determined whether Vts1 is also present in the nucleus. For this purpose, yeast nuclei were isolated from spheroplasts and three different cellular fractions were isolated that included cell debris (CD), nuclear fraction (NF) and cytoplasmic fraction (CF). Each fraction was subjected to western blotting with specific monoclonal antibodies to detect the presence of FLAG-epitope tagged Vts1 (Vts1-FLAG) and two marker proteins, α tubulin (cytoplasmic) and histone 3 (nuclear), were used as controls. As expected, cell debris derived from spheroplasts contained both cytoplasmic and nuclear proteins, because both α-tubulin and histone 3 were present (Figure 9C, lane 1). In contrast, histone 3 and α-tubulin were detected only in nuclear (Figure 9C, lane 2) and cytoplasmic (Figure 9C, lane 3) fractions, respectively, demonstrating that our nuclear and cytoplasmic protein preparations were not cross-contaminated. Vts1 was detected in both cytoplasmic and nuclear fractions (Figure 9C, lanes 2 and 3), demonstrating that Vts1 is also present in nuclei. This finding is in keeping with our notion that Vts1 can function with Dna2 in nuclei.
DISCUSSION
Though dna2 helicase-deficient mutant yeast cells fail to grow in media containing glucose, they grow slowly in the presence of poor carbon sources such as lactate and glycerol (32). These cells are viable in the presence of low levels of hydroxyurea that do not affect the growth of wild-type cells (data not shown). These findings suggest that the helicase activity of Dna2 is not essential and is dispensable under certain growth conditions unlike its endonuclease activity. Thus, it is likely that the in vivo function of the Dna2 helicase activity can be replaced by other helicases or enzymes. These considerations prompted us to screen for the genes that suppress the growth defects of the dna2-K1080E helicase-deficient mutant to find a new helicase involved in Okazaki fragment processing in eukaryotes. During the course of this work, we identified Vts1 as a genetic suppressor of helicase-negative dna2-K1080E. Contrary to our expectation, we found that Vts1 is devoid of helicase activity and has no effect on the Dna2 helicase activity (data not shown). Although Vts1 was originally reported to play a role in cytoplasmic events such as posttranscriptional regulation, our findings suggested that Vts1 affects nuclear events as well, particularly in Okazaki fragment processing in yeast. This conclusion is supported by a substantial number of results presented here. These include the following: (i) The double mutants such as vts1Δ dna2Δ405N and vts1Δ dna2-7 grew more slowly than single mutants alone. (ii) The stimulation of Dna2 endonuclease activity by Vts1 required specific protein–protein interactions between the two proteins. Not only the physical interaction between Vts1 and Dna2 but also the stimulation of the Dna2 endonuclease activity by Vts1 required the N-terminal 405-amino-acid domain of Dna2. (iii) Vts1 was shown to be a flap-structure-specific DNA-binding protein. (iv) Vts1 stimulated yeast Fen1, another endonuclease essential for processing of Okazaki fragments in eukaryotes. (v) Finally, we showed that Vts1 was present in yeast nuclei, consistent with its function in Okazaki fragment processing.
Our biochemical and genetic data indicate that the stimulation of Dna2 endonuclease activity by Vts1 is a major factor contributing to its suppression of the dna2-K1080E mutation. This is supported by our findings that the suppression of dna2-K1080E was observed when the mutant dna2-K1080E allele was provided in a multi-copy plasmid. In addition, the extent of suppression depended on the levels of Dna2-K1080E expression; suppression was more pronounced when Dna2-K1080E expression was driven by the strong ADH1 promoter (Figure 4C). Thus, elevated levels of Dna2 endonuclease activity are required to overcome the helicase deficiency caused by this mutation. Since the stimulation of Dna2 endonuclease activity by Vts1 requires its DNA-binding activity, it is possible that the binding of Vts1 to DNA substrates could alter the DNA structure and render it more susceptible to Dna2 cleavage. However, this notion is unlikely because the stimulation was allele-specific. Vts1 stimulated wild-type Dna2, but not Dna2Δ405N. In addition, Vts1 interacted with wild-type Dna2, but not Dna2Δ405N in ELISA assays. These results indicate that the stimulation of Dna2 endonuclease by Vts1 requires its DNA-binding activity and its ability to interact with Dna2. For these reasons, we conclude that the specific and stable binding of Vts1 to flap DNA substrates contributes to efficient Okazaki fragment processing by targeting Dna2 efficiently to flap DNA.
Although Vts1 significantly stimulated yeast Fen1, it was unable to reverse the RPA inhibition of the endonuclease activity of Fen1 (data not shown). This finding suggests that Vts1 cannot displace RPA from RPA-coated flaps. Thus, it is possible that Vts1 binds to the flap structure less strongly, individually or cooperatively, than RPA. In the presence of both RPA and Vts1, Vts1 stimulated the degradation of flaps shorter than 20 nt, at which RPA binds poorly (data not shown). Though Vts1 robustly stimulated Fen1 activity in vitro, it is unlikely that this effect contributes to the suppression of the dna2-K1080E mutation since Vts1 in a multi-copy plasmid failed to suppress dna2Δ405N. If suppression of the dna2-K1080E mutation occurred solely by the stimulation of Fen1, we would expect that overexpression of Vts1 could suppress both dna2-K1080E and dna2Δ405N because overexpression of Fen1 suppressed both dna2-K1080E (data not shown) and dna2Δ405N (33). Therefore, it appears that the stimulation of Fen1 by Vts1 in vivo may not be sufficient to suppress dna2Δ405N. However, it is possible that the stimulation of Fen1 by Vts1 could provide an explanation for more severe growth defects observed with the double mutants, vts1Δ dna2Δ405N and vts1Δ dna2-7. The following scenario would be possible; in vts1Δ cells, the deletion of VTS1 may reduce the function of Fen1 compared to that found in wild-type cells. Despite the normal protein levels of Fen1, the absence of Vts1 (and lack of Fen1 stimulation) could lead to a decrease of Fen1 activity in vivo. This scenario could account for the more severe growth defects observed with vts1Δ dna2Δ405N or vts1Δ dna2-7 compared to the single mutation.
It is noteworthy that Vts1 can bind to duplex ends efficiently. This property was especially evident in the presence of high levels of Vts1 and with substrates containing a 1-nt overhang at its 3′-end (data not shown). If this were the case, the DNase I protection observed near the duplex ends would not be physiologically relevant during Okazaki fragment processing since duplex ends are not available for the binding. However, it is possible that the duplex end binding activity of Vts1 could function in other DNA transactions.
In summary, we suggest that Vts1 is involved in the processing of Okazaki fragments, and possibly other DNA events such as repair and recombination. During repair and recombination, flap-like structures can be formed and they can be efficiently removed by Fen1 and Dna2 in the presence of Vts1.
FUNDING
Korea Science and Engineering Foundation Grants funded by the ministry of Education, Science and Technology. Funding for open access charge: Grant number N01090092.
Conflict of interest statement. None declared.
ACKNOWLEDEGEMENT
The authors thank Dr Jerard Hurwitz for critical reading of the manuscript.
REFERENCES
- 1.MacNeill SA. DNA replication: partners in the Okazaki two-step. Curr. Biol. 2001;11:R842–R844. doi: 10.1016/s0960-9822(01)00500-0. [DOI] [PubMed] [Google Scholar]
- 2.Hübscher U, Seo YS. Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells. 2001;12:149–157. [PubMed] [Google Scholar]
- 3.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 4.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 5.Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 2006;106:453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- 6.Denis D, Bullock PA. Primer-DNA formation during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 1993;13:2882–2890. doi: 10.1128/mcb.13.5.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro: II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J. Biol. Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 8.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 9.Bae SH, Kim JA, Choi E, Lee KH, Kang HY, Kim HD, Kim JH, Bae KH, Cho Y, Park C, et al. Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 2001b;29:3069–3079. doi: 10.1093/nar/29.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast: I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 11.Rossi ML, Bambara RA. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J. Biol. Chem. 2006;281:26051–26061. doi: 10.1074/jbc.M604805200. [DOI] [PubMed] [Google Scholar]
- 12.Harrington JJ, Lieber MR. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 13.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 14.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast II: cooperation between the polymerase δ and 3′ to 5′ exonuclease activities of pol δ in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 16.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 17.Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 18.Bae SH, Kim DW, Kim J, Kim JH, Kim DH, Kim HD, Kang HY, Seo YS. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 2002;277:26632–26641. doi: 10.1074/jbc.M111026200. [DOI] [PubMed] [Google Scholar]
- 19.Dilcher M, Kohler B, von Mollard GF. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 2001;276:34537–34544. doi: 10.1074/jbc.M101551200. [DOI] [PubMed] [Google Scholar]
- 20.Hall TM. SAM breaks its sterotype. Nat. Struct. Biol. 2003;10:677–679. doi: 10.1038/nsb0903-677. [DOI] [PubMed] [Google Scholar]
- 21.Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 22.Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- 23.Green JB, Gardner CD, Wharton RP, Aggarwal AK. RNA recognition via the SAM domain of Smaug. Mol. Cell. 2003;11:1537–1548. doi: 10.1016/s1097-2765(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 24.Edwards TA, Butterwick JA, Zeng L, Gupta YK, Wang X, Wharton RP, Palmer AG, 3rd, Aggarwal AK. Solution structure of the Vts1 SAM domain in the presence of RNA. J. Mol. Biol. 2006;356:1065–1072. doi: 10.1016/j.jmb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Johnson PE, Donaldson LW. RNA recognition by the Vts1p SAM domain. Nat. Struct. Mol. Biol. 2006;13:177–178. doi: 10.1038/nsmb1039. [DOI] [PubMed] [Google Scholar]
- 26.Aviv T, Amborski AN, Zhao XS, Kwan JJ, Johnson PE, Sicheri F, Donaldson LW. The NMR and X-ray structures of the Saccharomyces cerevisiae Vts1 SAM domain define a surface for the recognition of RNA hairpins. J. Mol. Biol. 2006;356:274–279. doi: 10.1016/j.jmb.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 27.Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat. Struct. Mol. Biol. 2006;13:160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 28.Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 2006;13:168–176. doi: 10.1038/nsmb1053. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, West SC, Egelman EH. Structure and subunit composition of the RuvAB-Holliday junction complex. J. Mol. Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 30.Cho IT, Kim DH, Kang YH, Lee CH, Amangyelid T, Nguyen TA, Hurwitz J, Seo YS. Human replication factor C stimulates flap endonuclease 1. J. Biol. Chem. 2009;284:10387–10399. doi: 10.1074/jbc.M808893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Formosa T, Nittis T. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae KH, Kim HS, Bae SH, Kang HY, Brill S, Seo YS. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JH, Kang YH, Kang HJ, Kim DH, Ryu GH, Kang MJ, Seo YS. In vivo and in vitro studies of Mgs1 suggest a link between genome instability and Okazaki fragment processing. Nucleic Acids Res. 2005;33:6137–6150. doi: 10.1093/nar/gki900. [DOI] [PMC free article] [PubMed] [Google Scholar]