Abstract
The article describes a new technology for real-time polymerase chain reaction (PCR) detection of nucleic acids. Similar to Taqman, this new method, named Snake, utilizes the 5′-nuclease activity of Thermus aquaticus (Taq) DNA polymerase that cleaves dual-labeled Förster resonance energy transfer (FRET) probes and generates a fluorescent signal during PCR. However, the mechanism of the probe cleavage in Snake is different. In this assay, PCR amplicons fold into stem–loop secondary structures. Hybridization of FRET probes to one of these structures leads to the formation of optimal substrates for the 5′-nuclease activity of Taq. The stem–loop structures in the Snake amplicons are introduced by the unique design of one of the PCR primers, which carries a special 5′-flap sequence. It was found that at a certain length of these 5′-flap sequences the folded Snake amplicons have very little, if any, effect on PCR yield but benefit many aspects of the detection process, particularly the signal productivity. Unlike Taqman, the Snake system favors the use of short FRET probes with improved fluorescence background. The head-to-head comparison study of Snake and Taqman revealed that these two technologies have more differences than similarities with respect to their responses to changes in PCR protocol, e.g. the variations in primer concentration, annealing time, PCR asymmetry. The optimal PCR protocol for Snake has been identified. The technology’s real-time performance was compared to a number of conventional assays including Taqman, 3′-MGB-Taqman, Molecular Beacon and Scorpion primers. The test trial showed that Snake supersedes the conventional assays in the signal productivity and detection of sequence variations as small as single nucleotide polymorphisms. Due to the assay’s cost-effectiveness and simplicity of design, the technology is anticipated to quickly replace all known conventional methods currently used for real-time nucleic acid detection.
INTRODUCTION
Polymerase chain reaction (PCR) has revolutionized the detection of nucleic acids. This method is the most commonly used laboratory technique for DNA amplification and is capable of detecting as little as a single copy of DNA or RNA. Fluorescent labels can be detected at nanomolar concentrations, and this is well within the range of PCR productivity. Fluorimetric detection of PCR products has simplified readout and made possible ‘real-time’ techniques that allow amplification to be monitored continuously (1,2). The original techniques employed ethidium bromide, which in turn was replaced by more sensitive dyes like SYBR Green (3). This is an inexpensive way to track the PCR reaction but has the drawback of detecting both specific and nonspecific products of amplification. Real-time systems were improved by labeled oligonucleotide probe detection. Fluorescent probes are oligonucleotides designed to bind exclusively to a target amplicon. These probes are usually synthesized with both a reporter fluorescent dye and a quencher dye that are in Förster resonance energy transfer (FRET) interaction (4). When FRET occurs, emission of the reporter dye is extinguished by the quencher. In the case of random dipole orientation and a good overlap between the emission spectrum of the reporter and the absorption spectrum of the quencher, the efficiency of FRET is dependent on the inverse sixth power of the intermolecular separation (4,5). Disruption of FRET by the dye separation results in a fluorescent signal, and this is widely used in probe designs for nucleic acid detection (6). Regardless of design, all FRET probes function by one of two strategies.
Hybridization-triggered FRET probes
In this strategy, the magnitude of FRET is based on a change in distance between the reporter and quencher dyes as the result of sequence-specific hybridization between a target nucleic acid and a fluorescent oligonucleotide probe. For example, when the fluorescent reporter and the quencher moieties are conjugated to opposite ends of the same probe, the quencher moiety is sufficiently close to the reporter dye due to random oligonucleotide coiling. Once the probe is hybridized to a complementary polynucleotide, the quencher and reporter moieties are separated, thus enabling the reporter dye to fluoresce. In reality this approach (7) has not found practical application due to a profoundly inefficient FRET effect in the unhybridized probe, which leads to an elevated fluorescence background. The background problem can be solved by conjugation of a 5′-minor groove binding (MGB) moiety in Eclipse probes (8), synthesizing the oligomers with a flexible PNA backbone (9) or using a hairpin-shaped Molecular Beacon in which the FRET dyes are brought in close proximity by intramolecular stem formation (10). Covalent linking of a molecular beacon probe to one of the PCR primers is a unique property of yet another hybridization-triggered technology named Scorpion primers (11).
Cleavable FRET probes
The best strategy to abolish FRET is based on cleavage of an oligonucleotide probe upon its binding to a target nucleic acid. When cleavage takes place anywhere between the conjugated dyes, the result is a complete and irreversible disruption of FRET. Although a number of ways to achieve the probe cleavage have been explored (12–14), the Taqman technology was the first developed (15) and it remains widely used for real-time nucleic acid detection in PCR (16). The method utilizes the 5′-nuclease activity of Thermus aquaticus (Taq) DNA polymerase. A dual labeled FRET probe is designed to anneal to a target sequence located between two PCR primer binding sites. During strand elongation, Taq polymerase cleaves the probe that is hybridized downstream from a primer site, releasing the reporter dye from the quencher. Unlike in the hybridization-triggered assays, the Taqman probe signal generated at a given PCR cycle is a sum of the signals generated at that particular cycle plus all previous ones. However, the elevated fluorescence background of the Taqman probes overshadows the signal advantage. Conjugation with a 3′-MGB-moiety (17) allows substantial reduction in the probe length, effectively resolving the background issue. MGB-Taqman probes were also shown to have improved single nucleotide polymorphism (SNP) discrimination properties (18).
The present article describes a new Snake system designed for real-time nucleic acids detection which, similar to Taqman, utilizes 5′-nuclease cleavable FRET probes but, unlike Taqman, does not directly depend on the PCR primer extension due to a unique mechanism of action. Similar to the Scorpion technology (11), the Snake system benefits from amplicons folding into a stem–loop structure but, unlike Scorpion, the Snake amplicons do not incorporate FRET components and participate in detection by promoting the 5′-nuclease cleavage of the FRET probes. A preliminary study and evaluation of this new detection technology has been conducted and the technology has been directly compared with Taqman and other conventional techniques.
MATERIALS AND METHODS
PCR reaction components
JumpStartTM Taq DNA polymerase, an antibody-inactivated ‘hot start’ enzyme, and dNTPs were purchased from Sigma-Aldrich. Primers and FRET probes for Snake, TaqMan, Molecular Beacon and Scorpion assays were prepared by Cepheid (Sunnyvale, CA) using reagents from Glen Research (Sterling, VA). The 3′-MGB Taqman probes were purchased from Applied Biosystems Inc (Foster City, CA). The sequences of oligonucleotides used in this study are shown in Figures 1 and 10. FAM in FRET probes is 6-fluorescein and BHQ1 is a Black Hole Quencher from Biosearch Technologies. The oligonucleotide 2 μM stock solutions were prepared and stored refrigerated in 5 mM sodium cacodylate. EvaGreenTM fluorescent dye was purchased from Biotium, Inc. (Hayward, CA).
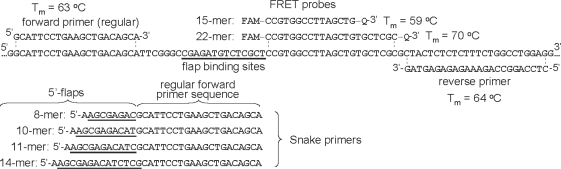
Figure 1.
The scheme shows the sequences of two FRET probes and two PCR primers (forward and reverse) used in the Taqman assays aligned with a detected sequence of 96-mer target oligodeoxyribonucleotide. Also shown are the forward primers used in the Snake assays. These primers comprise the regular primer sequence and 8–14-mer 5′-flap sequences. The segments of the flaps that participate in the intramolecular stem formation of the folded Snake amplicons and their binding site within the target sequence are underlined. FAM is 6-fluorescein and Q is a BHQ1 quencher.
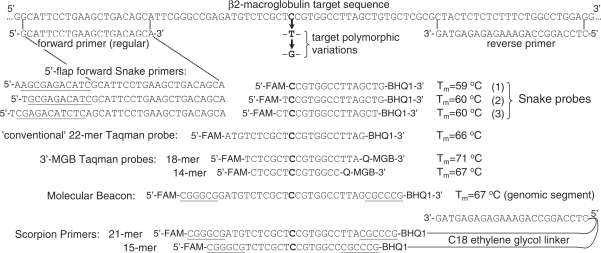
Figure 10.
The scheme shows the sequences of PCR primers and FRET probes used in a study to compare Snake with the other FRET assays in its capability to discriminate single nucleotide variations (SNP) in target nucleic acids. Three otherwise identical 96-mer DNA oligonucleotides, each with a different base at the highlighted nucleotide, were used in this study. Wherever it is possible, the oligonucleotides are aligned with the detected target sequence. Molecular Beacon, Taqman probes and Scorpion Primers were designed to provide the best SNP discrimination with the polymorphic nucleotide (highlighted) located in the middle of the probes. The Molecular Beacon probe and Scorpion Primers incorporated 6-G/C-nucleotide ‘sticky’ ends (underlined). The Scorpion primers were made by coupling the 5′-end of the reverse primer with the 3′-ends of Molecular Beacon-like FRET probes via a C18 polyethylene glycol linker. Three FRET probes were used in the Snake study with the SNP discriminating nucleotide located at the first, second and third position with respect to the 5′-end. Each of these probes was used with the reverse primer and a corresponding 5′-flap forward Snake primer shown on the left side from each of the probes. These 5′-flap primers were designed to provide the optimal cleavage structures shown in Figure 2E. The first (top) combination of a 5′-flap primer and FRET probe represents the 11-mer flap primer and 15-mer probe (Figure 1) used in the Snake assay optimization study. All other assays employed the regular forward and reverse primers in combination with the respective FRET probes. FAM is 6-fluorescein and BHQ1 is a Black Hole Quencher from Biosearch Technologies. MGB is a minor groove binder and the 3′-MGB Taqman probes incorporate its own 3′-quencher (Q).
Physical measurements and calculations
Oligonucleotide extinction coefficients were calculated using CalcExt2.8 software (Cepheid). Melting temperatures (Tm values) of unmodified primers and probes were calculated using the ‘nearest-neighbor’ approach (19,20) for perfect-match duplexes at 200 nM with adjustment for the PCR buffer used in this study. The effect of dyes on duplex stability was disregarded in calculations.
Each real-time curve shown here represents an average of at least 3–5 independent experiments. The fluorescent curve threshold value (Ct) was determined as the cycle number at which the semi-log of fluorescence of the curve reached or was pointing to 0.5. The reported Ct values were averaged based on 3–5 experiments and rounded to an integer. In the case of more accurate Ct measurements, confidence intervals were calculated and reported for 95% confidence level based on 5–8 independent experiments. A relative change in PCR yield ΔY = 100 × (1 – x)% was calculated based on an equation 2N = (1 + x)n, wherein n and N are respectively the Ct value discussed and the reference reaction.
DNA target
A 96-mer synthetic DNA oligonucleotide (Sigma-Genosys) containing a portion of the β2-Macroglobulin gene (GenBank accession #NM004048) was used as a target for PCR. The sequence shown in Figure 1 is a fragment of the target between 5′-CG … and … CTA-3′ ends. Two additional 96-mer DNA oligonucleotides (Figure 10) that were otherwise identical to the natural sequence but different at one nucleotide were used in the SNP discrimination studies. In order to prevent adsorption of the target DNA to plastic tubes at subnanomolar concentrations, it was diluted in 100 nM (dA)18 conjugated at the 3′-end with propane diol. Human genomic DNA (20 ng per 25 μL reaction) was used in quantitative PCR (qPCR) experiments to compare the efficiencies of the Snake and Taqman assays in the detection of the β2-Macroglobulin gene sequence in natural samples.
Real-time PCR (standard protocol)
Unless otherwise indicated in the text, PCR reaction mixtures were conducted in 25 µL volumes and contained target DNA—104 molecules per reaction; forward and reverse PCR primers—200 nM each; FRET probe—200 nM; dNTPs—200 μM each; JumpStart DNA polymerase (Sigma)—0.04 U/μL in 50 mM KCl, 2 mM MgCl2, 20 mM Tris–HCl (pH 8.0). All real-time PCR experiments were performed on SmartCycler (Cepheid). Background fluorescence was subtracted using instrument software and the data (fluorescence versus PCR cycle) were transferred to an Excel format (Microsoft Office) for further processing. PCR was performed in two steps with the annealing and extension stages combined into a single step referred to throughout the text as ‘annealing’. Unless otherwise noted, the reaction mixtures were incubated at 95°C for 2 min to activate the antibody-blocked enzyme followed by 55 PCR cycles consisting of 10 s at 95°C for denaturation and 45 s at 64°C for annealing, i.e. (95°C for 2 min) → (95°C for 10 s → 64°C for 45 s)55. Fluorescence was measured during the annealing stage.
RESULTS
Snake system design and the mechanism of action in real-time PCR
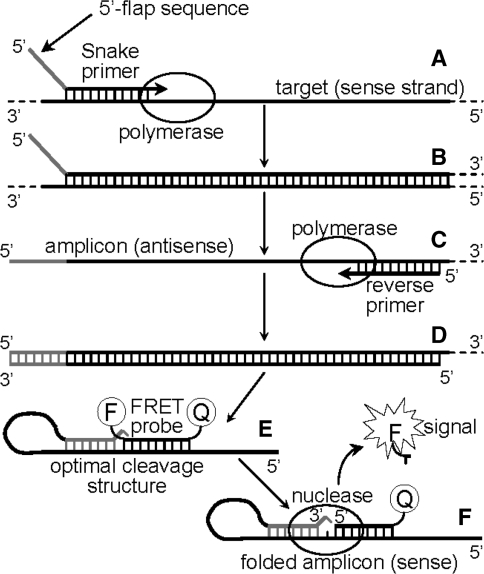
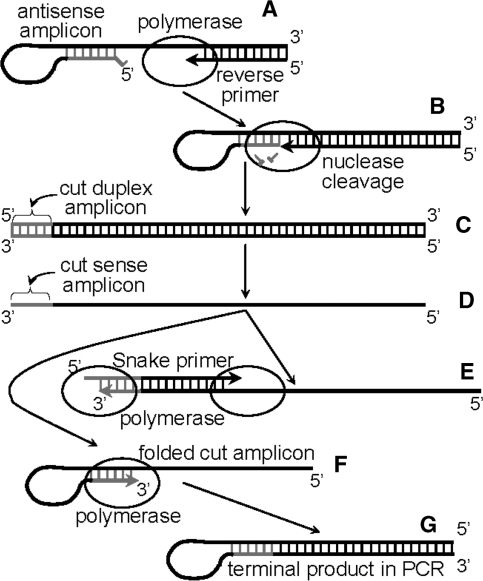
In the Snake assay one of the PCR amplicons not only supplies a detectable target template but also actively participates in FRET probe cleavage by virtue of folding into a hairpin-like secondary structure, wherein the stem of the structure is adjacent to the FRET probe binding site. Hybridization of the probe to this folded amplicon results in an optimal cleavage structure for duplex-specific 5′-flap endonucleases (21) and the 5′-nuclease domain of Taq polymerase (22). Figure 2 illustrates how the Snake system functions. A forward primer contains a 5′-flap sequence (shown in gray) which is complementary, save for the 5′-terminal base, to a target site located downstream from the primer binding site. Extension of this forward primer in stage A results in the synthesis of an antisense strand providing a double stranded amplicon (stage B). After strand separation (95°C), a reverse primer hybridizes to the antisense strand, and DNA polymerase extends the complex (stage C) resulting in yet another double stranded amplicon (stage D). Since the 5′-flap of the forward primer functions as a template for DNA synthesis, a complement to this sequence appears at the 3′-end of a sense amplicon strand (also shown in gray). After another round of strand separation, the sense amplicon (synthesized in stage D) folds into a secondary structure in which the 3′-terminal nucleotide remains mismatched (stage E). A FRET probe hybridizes to the sense strand of the amplicon such that an optimal cleavage structure for 5′-nuclease is created. A 5′-nuclease recognizes the structure, cleaves the probe and releases a detectable fluorescent signal in stage F.
Figure 2.
The diagram shows the key stages of the real-time Snake assay. The mechanism is discussed in the text.
Snake and Taqman detection technologies in real-time PCR
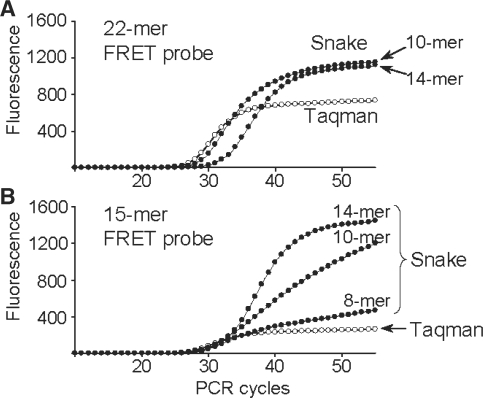
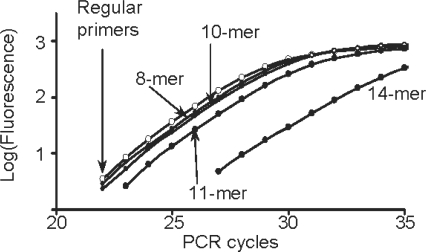
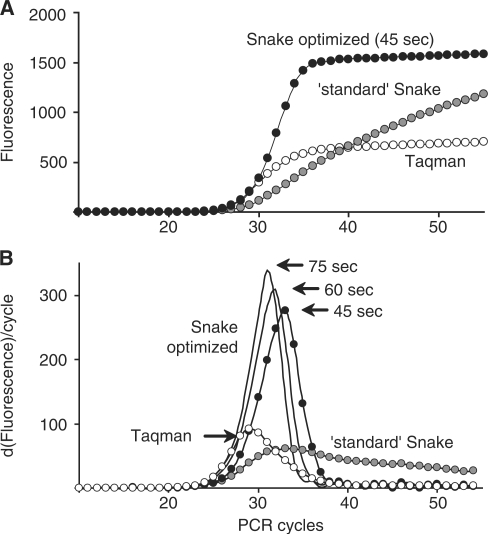
The Taqman and Snake assays were directly compared in real-time detection of the β2-Macroglobulin target sequence, and the results are shown in Figure 3. The long 22-mer FRET probe was designed according to Taqman rules to have elevated hybridization properties (Tm = 70°C) in order to perform at the annealing temperature of 64°C. The respective assay (Taqman, Figure 3A) showed good signal productivity with a plateau at ∼700 fluorescent units (FU). Despite the relatively strong signal, this only accounts for ∼65% of the probe cleavage while ∼35% of the probe remained intact after PCR. In contrast to Taqman, complete digestion of this 22-mer probe was achieved in Snake assays applying either 10-mer or 14-mer 5′-flap forward primers (Snake, Figure 3A).
Figure 3.
Real-time PCR detection of the β2-Macroglobulin target sequence in Taqman (open circle) and Snake (filled circle) assays using 22-mer (A) and 15-mer (B) FRET probes. Regular forward and reverse primers were used in Taqman, whereas the Snake system utilized its own forward primers identified in the charts by the length of 5′-flap sequences, i.e. 8-, 10- and 14-mers. The structures of the primers and FRET probes used in these experiments are shown in Figure 1. The experiments were performed with the standard reaction composition, component concentrations and PCR time/temperature profile described in ‘Materials and Methods’ section.
The 15-mer FRET probe was not designed for the Taqman assay. This probe’s melting temperature (Tm = 59°C) is 5°C below the PCR annealing temperature (64°C). As a result, it showed very low signal productivity (∼250 FU) with the cleavage yield <20% (Taqman, Figure 3B). Nonetheless, this short 15-mer probe was completely cleaved in Snake. Although the shortest 8-mer Snake primer provided a moderate positive effect, the signal strength escalated rapidly with the flap length increase, resulting in complete probe cleavage for 14-mer primer. Note that the increase in the signal plateau (∼300 FU) observed for this primer in Figure 3B versus the same in Figure 3A is due to improvement in fluorescence background of the shorter 15-mer FRET probe versus the longer 22-mer one.
The probe cleavage in Snake takes place ‘before’ the 5′-flap primer anneals to the folded target amplicon, destroying the optimal cleavage structure. At the beginning of detection, when the primers are in excess over the amplicons, the lifetime of the optimal cleavage structure is anticipated to be short. This may explain the 1 cycle delay in real-time curve threshold observed for the 10-mer primer (Ct10-mer = 24.4 ± 0.3 versus Ctreg = 23.4 ± 0.1; Figure 3A) but it may not explain the >3 cycles delay observed for the 14-mer primer (Ct14-mer = 26.7 ± 0.5). Negative effect on PCR due to the amplicon folding in Snake had always been anticipated to emerge at a certain level of secondary structure stability. Interestingly, use of the short 15-mer probe with the same 14-mer primer led to a smaller threshold delay (Ct14-mer = 25.6 ± 0.1; Figure 3B).
Effect of folded PCR amplicons on PCR productivity in Snake assays
There are two different mechanisms by which the Snake amplicons, both of which are folded, can negatively affect PCR yield. First, annealing of a 5′-flap primer to the stem–loop structure of the sense amplicon (e.g. shown in Figure 2E and F) is a slower process compared to that in conventional PCR. Second, the folded antisense amplicons can affect PCR efficiency according to yet another mechanism that is shown in Figure 4. A study was conducted to investigate the issue and the results are shown in Figure 5.
Figure 4.
The scheme illustrates key stages of a reaction pathway that occur in Snake assays if the stem–loop structure of the folded amplicons is too stable. During the extension of a reverse primer hybridized to a folded antisense amplicon (stage A), DNA polymerase reaches the duplex of the folded amplicon and partially cleaves its 5′-end (shown in gray, stage B) while displacing the rest of the sequence and completing the replication. This results in a cut double-stranded amplicon (stage C) that, after strand separation at 95°C, provides a 3′-excised single-stranded sense amplicon (stage D). If a 5′-flap forward primer hybridizes to this amplicon, then the original sequence integrity of the sense amplicon can be restored along with the forward primer extension (stage E). However, this truncated amplicon can also fold into a secondary structure that, unlike normal Snake amplicons (Figure 2E), forms a self-priming intramolecular duplex with no 3′-terminal mismatched nucleotide (stage F). Extension of such a duplex by DNA polymerase leads to a long hairpin-like structure (stage G) that does not participate in PCR due to it exceptional thermal stability.
Figure 5.
Effect of secondary structure in folded Snake amplicons on PCR efficiency. The experiments were performed according to the standard PCR protocol described in ‘Materials and Methods’ section, but the FRET probe was omitted and PCR amplicons were detected in the presence of EvaGreen fluorescent dye (0.625 μl of 20× provider stock solution per 25 μl reaction). Reverse primer was applied with regular forward primer (open circle) or one of the 5′-flap primers (closed circle) as indicated for each curve. Fluorescent signal versus PCR cycle number is presented in a semi-log plot. Structures of the primers are shown in Figure 1.
FRET probes may affect PCR and, in this study (Figure 5), amplicons were detected in real time using EvaGreen fluorescent dye. PCR efficiency of the reactions performed with various Snake primers was compared to the system using regular primers (Ctreg = 21.9±0.2). It was found that PCR efficiency declines with the increase in length of the 5′-flap sequences, as indicated by the respective threshold values calculated for 8-mer (22.0±0.2), 10-mer (22.3±0.1), 11-mer (23.3±0.2) and 14-mer (26.6 ± 0.2) 5′-flap primers. However, this negative effect was fairly small (ΔCt=0.1–1.4 cycle) for 8–11-mer primers, whereas use of the longest 14-mer Snake primer resulted in as many as ∼5 cycle delay (ΔCt = 4.7). Estimates showed that such a Ct delay accounts for ∼23% reduction in PCR cycle yield compared to the regular primer pair. If the slow hybridization of 14-mer primer is the reason of this PCR deficiency, then increase in PCR annealing time should return the real-time threshold back to 22 cycles, the Ct value obtained for the regular primers. In a respective set of experiments, the annealing time of the 14-mer reaction was increased to 60, 75, 90 and 105 s providing Ct values of 25.4 ± 0.1, 25.0 ± 0.3, 24.1 ± 0.3, 24.1 ± 0.2, respectively. Although the extension of annealing time was helpful in improving the PCR efficiency, the threshold value did not reach 22 cycles, and it was stabilized at Ct = 24.1. The respective yield calculations led to the conclusion that about half of the PCR yield reduction (∼12%) is caused by the slow kinetics of 14-mer primer while another half (∼11%) is likely due to the mechanism described in Figure 4.
Dependence of Taqman and Snake systems on PCR primers concentration
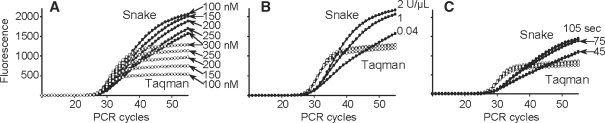
Signal generation in Taqman is directly linked to the PCR amplification process because cleavage of FRET probes takes place during the primer extension. Thus, the signal strength in Taqman is in a direct proportion to the PCR primer concentration. This is illustrated in Figure 6A. The lower the PCR primer concentration, the lower the signal plateau is in Taqman. Although in the Snake assay the mechanism of probe cleavage is different, this technology also relies on an amount of target amplicon, and a trend somewhat similar to Taqman was anticipated. Nevertheless, the same experiments performed in the Snake format showed a very unusual adverse effect on PCR primer concentration (Figure 6A). Decrease in the primer concentration led to an increase in the assay signal. There is apparently yet another reaction component that regulates the signal productivity in Snake. The Taq DNA polymerase was suspected. The same enzyme performs the target amplification and detection in both assays, Taqman and Snake. However, unlike in Taqman, in Snake these two processes are separated in time and in space, i.e. occur in different complexes. The duplex-specific 5′-nuclease is an associated activity of Taq polymerase, the main function of which is to replicate DNA. Therefore, it is reasonable to suggest that the enzyme substrate affinity is not in favor of the optimal cleavage structure. Moreover, during the detectable stage of PCR, i.e. >24–25 cycles, DNA polymerase becomes a reaction-limiting factor. For example, estimates based on the additional information provided by Sigma-Aldrich about the JumpStartTM Taq DNA polymerase used in this study show that the 0.04 U/μl of the enzyme applied in the standard PCR protocol corresponds to ∼0.3–0.8 nM enzyme concentration. During the detectable stage of the amplification, the concentration of the extension complexes, i.e. primers hybridized to PCR amplicons, can reach, in theory, up to ∼100–200 nM if the primers are used at 200 nM concentrations. This means that, at certain PCR cycles, one molecule of the enzyme may need to accomplish the synthesis of up to ∼50–100 or more amplicon strands to hold a high reaction yield. In this vein, the preferential involvement of Taq in the amplification process that, in turn, is stimulated by the increase in primer amounts may negatively affect the Snake signal productivity, and this may explain the phenomenon observed in the experiments of Figure 6A. The experiments shown in Figure 6B support this hypothesis. As judged by the signal strength, the Snake assay positively responded on increase of the enzyme concentration from 0.04 to 1 and 2 U/μL. In contrast to Snake, the same change applied in Taqman had no detectable effect and all three curves overlapped. These results in particular point to the Snake system’s deficiency in the 5′-nuclease function and the benefit of using elevated amounts of Taq polymerase.
Figure 6.
Dependence of Taqman (open circle) and Snake (closed circle) assays on change in concentration of PCR primers (A) and Taq DNA polymerase (B). (C) The effect of annealing time on the assays. All experiments were carried out using regular primers and a 22-mer FRET probe in Taqman; and an 11-mer 5′-flap forward primer, reverse primer and a 15-mer FRET probe in Snake. The structures of these oligonucleotides are shown in Figure 1. The following changes were applied to the standard PCR protocol described in ‘Materials and Methods’ section: in (A), the FRET probes—300 nM and concentrations of both primers were equal and used in the amount indicated for each real-time curve; in (B), the FRET probes and primers—300 nM and DNA polymerase were used at concentrations as indicated; in (C), FRET probes and primers—200 nM (standard) but PCR time/temperature profile was changed to 95°C for 2 min → (95°C for 10 s → 64°C for 45 s)20 → (95°C for 10 s → 64°C for X s)35 wherein X was the applied annealing time of 45 (standard), 75 and 105 s, respectively. In panels B and C, the Taqman real-time curves are shown but not assigned to the protocol changes because of the close curve overlap. All diagrams are shown in the same fluorescence scale.
Dependence of Taqman and Snake systems on increase of PCR annealing time
The folded sense amplicon (Figure 2E and F) is the key element of Snake detection technology. The longer it remains unhybridized to a respective primer, the more signal the Snake system can generate. The lifetime of this amplicon in PCR could increase, especially by the time when PCR amplification is completing and primer concentrations are substantially reduced. Accordingly, increase in annealing time near or after the PCR completion could positively affect the Snake signal. This was investigated in the experiments shown in Figure 6C. The change in PCR annealing time from 45 to 105 s had no effect on Taqman. In contrast to Taqman, the Snake assay produced more signal when the annealing time was increased from 45 to 75 s. However, further increase in time to 105 s had almost no effect.
Consumption of primers in PCR is followed by a proportional increase in amplicon concentrations. The amplicons can reanneal, and this is a factor affecting the lifetime of the signal-producing sense amplicon in Snake. A PCR product was prepared according to the standard protocol, using the reverse primer in combination with an 11-mer 5′-flap primer but with the probe omitted. The amplicon strand reannealing was studied at 64°C in the presence of EvaGreen fluorescent dye, detecting duplex DNAs. SmartCycler is not designed to study very fast kinetics. Nevertheless, analysis of the fluorescence curves showed that ∼88 and ∼99% of the Snake amplicons are reannealed in ∼2–7 and ∼37–42 s, respectively. By the time of ∼70–80 s, the reannealing reaction is 100% complete and this is in good agreement with the results of Figure 6C obtained for Snake.
Asymmetric PCR in Taqman and Snake
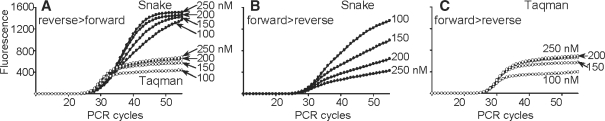
The problem of rapid amplicon reannealing can be resolved using asymmetric PCR (23), wherein forward and reverse primers are applied in unequal molar amounts. In asymmetric PCR, one of the strands is preferentially amplified and the effect of this method on Taqman and Snake systems was investigated (Figure 7). Extension of the regular forward primer (Figure 1) in Taqman leads to the probe cleavage, and a positive response of this system to the disproportionate increase of this primer was anticipated. Indeed, the FRET signal observed in these experiments (Figure 7C) was roughly in a direct proportion to the primer excess. Unexpectedly, the same trend was observed when this primer was proportionately less abundant with respect to the reverse primer (Figure 7A).
Figure 7.
Effect of asymmetric PCR on Taqman (open circle) and Snake (closed circle) assays. Snake assays were performed using a 15-mer FRET probe (200 nM) and an 11-mer 5′-flap forward primer. Regular forward and reverse primers and a 22-mer FRET probe (200 nM) were used in Taqman. The concentration of the excess primer was ranged from 100 to 250 nM as indicated for each real-time curve. The deficient primer concentration in all experiments was 100 nM. Other reaction conditions were the same as described in ‘Materials and Methods’ section. (A) A set of experiments wherein the reverse primer was in excess (reverse > forward). (B and C) illustrate an opposite scenario (forward > reverse). The curves marked with 100 nM represent normal, symmetric PCR wherein primers were used at equal (100 nM) concentrations. All diagrams are shown in the same fluorescence scale.
In contrast to Taqman, the Snake system’s response to the primer asymmetry brought no surprises. Increase in the sense amplicon concentration versus antisense stimulated by the excess of the reverse primer was followed by a steady growth of the real-time signal (Figure 7A). An inverse effect was anticipated in the Snake assay, wherein the antisense amplicon was stimulated by a reverse primer asymmetry (forward >reverse). As predicted, the signal declined in the experiments shown in Figure 7B.
Snake system performance with optimized reaction conditions
The meticulous study of the Snake assay led to the discovery of the particular reaction conditions that would be optimal for this technology. Figure 8A shows the considerable changes in the real-time Snake performance when the optimized reaction conditions, such as an asymmetric PCR format (reverse > forward primer, 300/70 nM) and 5-fold increase in the Taq polymerase amount (from 0.04 to 0.2 U/μL), were implemented. As can be better seen in Figure 8B, the main consequence of this reaction modification was a ∼3-fold increase in the probe cleavage rate. Increasing the time of the detection system to digest the probe at the annealing stage kept increasing the cleavage rate (‘60’ and ‘75’ s real-time curves in Figure 8B). This also improved the assay’s sensitivity. For example, compared to the standard Snake assay (Ct = 24.1 ± 0.2), the optimized Snake conditions reduced the threshold values to 22.8 ± 0.3, 22.3 ± 0.1 and 21.5 ± 0.2 when annealing times of 45, 60 and 75 s were respectively used. However, these positive changes may not be significant for routine use in Snake, especially taking into account the associated increase in the overall assay time (up to ∼30–55%). Moreover, with 45 s of annealing time (standard in this study), the optimized Snake assay remarkably outperformed Taqman in signal productivity while matching the competitor in the detection sensitivity (Snake ‘45 sec’ Ct = 22.8 ± 0.3 versus Taqman Ct = 23.0 ± 0.3).
Figure 8.
The effect of optimized reaction conditions and PCR protocol on the Snake system performance in real time. (B) the first derivatives of the real-time curves for the Taqman (open circle) and Snake (closed circle) assays shown in (A). Snake assays were carried out using a 15-mer FRET probe and an 11-mer 5′-flap forward primer. Regular forward and reverse primers and a 22-mer FRET probe were applied in Taqman. The structures of these oligonucleotides are shown in Figure 1. The Taqman and ‘standard’ Snake assays were performed using the reaction conditions and PCR time/temperature profile described in ‘Materials and Methods’ section. The reaction mixtures in the ‘optimized’ Snake comprised target DNA—104 molecules per reaction; 11-mer 5′-flap forward primer—70 nM; reverse primer—300 nM; 15-mer FRET probe—200 nM; dNTPs—200 μM each; JumpStart DNA polymerase (Sigma)—0.2 U/μl in 50 mM KCl, 2 mM MgCl2, 20 mM Tris–HCl (pH 8.0). The optimized Snake PCR time/temperature profile was 95°C for 2 min → (95°C for 10 s → 64°C for X s)55 wherein the annealing time X was 45 (standard), 60 or 75 s as indicated for each curve in both diagrams.
Validation of the Snake technology for qPCR analysis
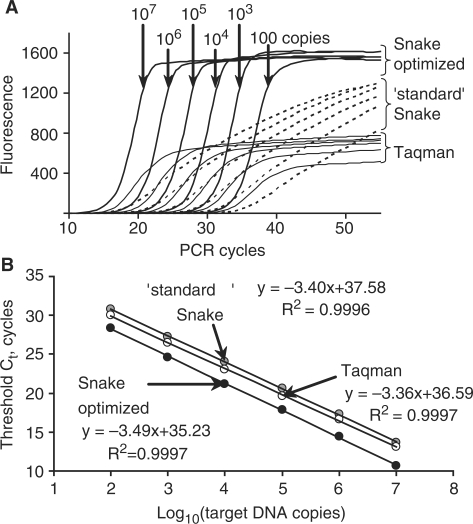
The results of a target-titration study shown in Figure 9 prove the applicability of the Snake assay for quantitative assessments of nucleic acids in test samples. Regardless of the reaction compositions, i.e. performed in standard or optimized formats, the Snake assay showed a linear dependence of the threshold values on the logarithm of the target amounts (copies) used in PCR. All three lines shown in Figure 9B for the Taqman and Snake assays are essentially parallel with the ‘least squares’ factor R2 close to the theoretical maximum (>0.999). The linear slope coefficient was varied insignificantly and close to the theoretical value of –3.322. Deviations of the linear slope coefficient provide average PCR yields of 96.8, 98.4 and 93.4% for the standard Snake, Taqman and Snake optimized assays, respectively. The relatively reduced PCR efficiency in the case of the Snake optimized format is at least in part due to asymmetric PCR and, in particular, low forward primer concentration (70 nM). In contrast, use of symmetric PCR in the standard Snake format with both primer concentrations at 200 nM improves the PCR yield. Notwithstanding the 5% lower amplification yield compared to Taqman, the linear trendline of the Snake optimized assay is positioned below the Taqman trendline, indicating ∼1.4 cycle earlier target detection. The standard Snake assay is less sensitive than Taqman in the target detection, and its trendline is shifted up by one PCR cycle. Based on the linear trendlines of Figure 9B, the copy numbers of the β2-macroglobulin gene were measured in 20 ng of a human genomic DNA sample using all three qPCR methods. The standard Snake, Taqman and Snake optimized assays provided close values of 469 ± 70, 523±37 and 428±42 gene sequence copies, respectively.
Figure 9.
Use of the Snake assay for quantitative measurements of target nucleic acids. (A) The real-time fluorescence curves for the detection of variable amounts of the β2-macroglobulin target sequence by the Snake assay performed in optimized and standard reaction conditions and conventional Taqman. As indicated, the amounts of the target sequence in the reactions ranged from 100 to 107 copies with an order of magnitude increment. The reaction compositions and PCR protocols used for each of the methods are described in Figure 8. The optimized Snake assay was carried out using 75 s of annealing time. The threshold values were determined for each of the curves and plotted versus the logarithm of the target DNA amounts used in the detection experiments. The results are shown in (B). Also shown are the linear trendline equations and coefficients of determination (R2) calculated for each of the assays.
Snake technology in the detection of target polymorphic variations
Detection of target polymorphic variations, commonly as small as SNPs, is yet another application of the probe-based real-time PCR assays. The Snake approach was validated in the discrimination of two unnatural SNPs introduced into the β2-Macroglobulin target sequence. The SNP discrimination study was expanded to include, in addition to the conventional Taqman, its enhanced version based on 3′-conjugated minor groove binder (18), Molecular Beacon (10) and Scorpion Primers (11). The target polymorphic variations and the probe and primer designs corresponding to each individual technology are shown in Figure 10. The Scorpion primers, Taqman and Molecular Beacon probes were designed according to the guidance provided for each technology in the above cited articles. One design principle common to all of the conventional assays is that the SNP discriminating nucleotide is placed in the middle of the probe. The Snake technology is different in this regard in that the SNP location is preferably located at or near the probe 5′-end. As shown in Figure 10, three Snake probes were prepared to have equal hybridization properties (Tm values) with the SNP discriminating nucleotide at the first, second and third positions with respect to the 5′-end.
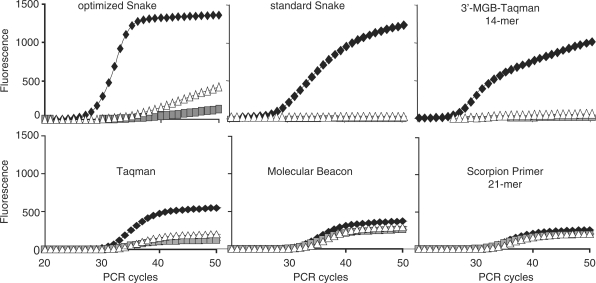
All conventional assays closely track the amplification process, and the SNP discrimination in Taqman, Molecular Beacon and Scorpion does not appreciably change within the detectable stage of PCR. Once again, the Snake system does not follow the conventional trend. In Snake, the SNP discrimination improves if you measure it in an earlier PCR cycle. Nonetheless, to insure the direct technology comparison, all assays including Snake (standard Snake) were performed using the conventional symmetric PCR protocol, in which the primers and probes are used in equal amounts (200 nM), and the SNP discrimination factor was determined at the PCR cycle 50, when the signal in the conventional assays reaches its maximum. The details of this standard protocol are described in ‘Materials and Methods’ section. The optimized Snake assay cannot be directly compared to the other systems because of the changes to the PCR protocol and, as discussed above, the considerable variations in SNP discrimination factor versus PCR cycle. However, the exemplary real-time curves and summary of the SNP discrimination study are also provided for this assay (see Figure 11 and Table 1). The SNP discrimination factor of the optimized Snake format was measured at the PCR cycle 32, when half of the probe is cleaved and the generated signal roughly matches the maximum signal productivity of the conventional Taqman assay.
Figure 11.
Detection of SNP variations in the β2-macroglobulin target sequence by Snake and other FRET probe detection technologies. The primers and probes used in each individual assay are shown in Figure 10. A second set of a FRET probe and a corresponding 5′-flap forward primer was used in the Snake assays shown here. All assays except optimized Snake were performed using equal concentrations of primers and probe (200 nM, symmetric PCR) according to the standard reaction composition and PCR time/temperature profile described in ‘Materials and Methods’ section. The optimized Snake assay was performed using the reaction compositions and PCR protocol (45 s annealing) described in Figure 8. The real-time curves for the match target are shown by black rectangles whereas white triangles and gray squares are used to show the mismatch target signals. The fluorescence scale is identical for all diagrams. The results of this technology assessment study are summarized in Table 1.
Table 1.
Characteristics of PCR performance of various FRET technologies in the detection of SNP variations in the β2-macroglobulin gene
| Assay type | Fluorescence background | Signal of match target at cycle 50 (background subtracted) | SNP discrimination factorsa |
|
|---|---|---|---|---|
| C/C mismatch | C/A mismatch | |||
| Optimized Snake: | ||||
| First position probe | 93 ± 4 | 1600 ± 39 | 0.46 ± 0.02 | 0.59 ± 0.02 |
| Second position probe | 130 ± 5 | 1358 ± 45 | 1.00 ± 0.00 | 0.97 ± 0.00 |
| Third position probe | 143 ± 5 | 1335 ± 38 | 0.95 ± 0.01 | 0.93 ± 0.02 |
| Standard Snake: | ||||
| First position probe | 100 ± 3 | 1220 ± 60 | 0.96 ± 0.01 | 0.92 ± 0.02 |
| Second position probe | 130 ± 5 | 1210 ± 115 | 1.00 ± 0.00 | 0.99 ± 0.01 |
| Third position probe | 150 ± 6 | 1370 ± 50 | 0.99 ± 0.00 | 0.99 ± 0.01 |
| 3′-MGB-Taqman: | ||||
| 14-mer | 120 ± 7 | 1010 ± 180 | 1.00 ± 0.00 | 0.94 ± 0.00 |
| 18-mer | 270 ± 9 | 790 ± 90 | 0.87 ± 0.01 | 0.81 ± 0.01 |
| Taqman 22-mer | 200 ± 13 | 550 ± 60 | 0.78 ± 0.01 | 0.63 ± 0.04 |
| Molecular Beacon | 110 ± 6 | 370 ± 30 | 0.31 ± 0.05 | 0.20 ± 0.10 |
| Scorpion Primers: | ||||
| 21-mer | 250 ± 10 | 260 ± 30 | 0.24 ± 0.02 | 0.20 ± 0.14 |
| 15-mer | 190 ± 10 | 150 ± 10 | 0.93 ± 0.01 | 0.83 ± 0.04 |
aThe SNP discrimination factors for the individual technologies were calculated as 1-(Signal of mismatch/Signal of match), wherein the signal was measured at cycle 50 for all assays but optimized Snake. In optimized Snake, the SNP discrimination was measured at cycle 32.
Each of the FRET probes listed in Figure 10 forms either matched C/G or mismatched C/C and C/A duplexes with one of the target DNA strands during PCR amplification. The results of the best performing probes for each of the tested assays are shown in Figure 11. The assays’ fluorescence backgrounds, signal strengths and SNP discriminating capabilities are summarized in Table 1. The results show that the Snake and 3′-MGB-TaqMan technologies outperformed the other assays in both signal generation and specificity. Snake generated twice as much signal as the conventional TaqMan assay. The relatively short probes used in these assays have a low background of ∼90–150 FU. The Molecular Beacon probe has half of the fluorescence background (∼110 FU) of the TaqMan probe but generates a lower signal, likely due to ‘residual’ quenching in the probe–target hybrid (24). Interestingly, the 21-mer Scorpion primer has an elevated fluorescence background of ∼250 FU despite having the same ‘sticky’ ends used in the design of the Molecular Beacon probe. Unlike the Molecular Beacon probe, the Scorpion probe is covalently linked to the reverse primer and partial complementarity with it may elevate the level of background fluorescence.
Two factors, thermodynamic and enzymatic, contribute to the specificity of any detection technology used in SNP detection. Thermodynamically, shorter probes are more sensitive to a mismatch in the probe–target hybrid. Enzymatically, the 5′-nuclease of Taq polymerase is very sensitive to structural abnormalities within or near its cleavage site (25). In this regard, the Snake assay is similar to the Endo IV assay (14) in which Escherichia coli endonuclease IV has a very low affinity for duplexes that contain a mismatch at its binding site. Placing the base pair mismatch at the 5′-end of the FRET probes in Snake increases the contribution of both factors, the nucleic acids thermodynamics and 5′-nuclease structural specificity. The second 5′-position appeared to be the best, providing in Snake nearly absolute SNP discrimination power regardless of the assay format, standard or optimized (0.97–1, Table 1). Conventional hybridization assays rely exclusively on the ‘thermodynamic’ factor, which explains their relatively weak discrimination between perfect-match and mismatched targets. Conjugation to a MGB-moiety (17) is an effective solution for the TaqMan technology (18) because it allows considerable reduction in the probe length with no sacrifice in signal. The 3′-MGB Taqman appears to be the only technology tested that can effectively compete with Snake in SNP discrimination. However, it requires precise design of probes that are as short as possible. For example, compared to the 14-mer 3′-MGB-Taqman probe, the homologous 18-mer probe showed an increased fluorescence background and a noticeable decrease in the signal strength and SNP discriminating capabilities.
Molecular Beacons probes have been reported to have enhanced specificity (26), but in this study the one probe of this type performed poorly. Note that the discriminating power of this type of probe can be improved by reducing its length, but only with a concurrent decrease in signal strength. For example, this effect was observed when a 21-mer genomic segment of a Scorpion primer was shortened to 15-mer (Table 1). The SNP discrimination values were remarkably increased from 0.20–0.24 to 0.83–0.93. Also improved was the fluorescence background, which dropped from ∼250 to ∼190 FU. These improvements, however, were accompanied by a loss in overall signal from 260 to 150 FU.
DISCUSSION
It has been shown that the progression of several types of DNA polymerases, from prokaryotes, phages and eukaryotes, is impeded by stable secondary structure in the template strand (27–29). Furthermore, it is well established that secondary structures within oligonucleotide binding sites negatively affect the hybridization (30,31). For these reasons, amplification of self-complementary sequences is avoided in PCR whenever it is possible. However, the presently described Snake detection technology, one of three recently discovered (32), is deliberately based on fabrication of PCR amplicons that fold into stem–loop structures. The study described here shows that up to a certain level of stability these secondary structures do not interfere with the amplification process but effectively catalyze the FRET probe cleavage. Unstable structures have little effect on PCR but they do not contribute to the real-time signal, whereas over-stabilization leads to reduction in PCR yield according to the two mechanisms discussed. Nevertheless, for any specific PCR condition, there is an apparent gap in thermodynamic stability of the folded amplicons that has essentially no effect on PCR but considerably benefits the 5′-nuclease assay signal productivity. Although the reported data indicate that this thermodynamic gap is sufficiently broad, a number of potential improvements to the Snake technology have already been sought. For example, preliminary results point to an advantage of using base-modified duplex-stabilizing dNTPs (33). Along with the anticipated benefit to the hybridization properties of PCR primers and probes, this approach would lead to a disproportional stabilization of the key sense amplicon in comparison to the problematic antisense one. On the other hand, the ‘undesired’ stem–loop structure in the antisense amplicons could be destabilized by applying duplex-destabilizing nucleoside analogs in the design of the 5′-flap sequences, e.g. deoxyuridine and deoxyinosine replacing deoxythymidine and deoxyguanosine, respectively. Moreover, the hybridization properties of the 5′-flap primers might be improved, for example, by elongation of the loop segment in the sense amplicons or incorporation of duplex-stabilizing base modifications (34) within the regular, but not flap, primer sequence. Applied individually or in combination, these approaches are aimed at eliminating a negative effect of the Snake amplicon folding on PCR. It is needless to point out that the future development of software designated for the primer and probe design in Snake will be very helpful for the adoption of this new technology in scientific research and exploring all the benefits that it offers. In the meantime, those researchers who would like to test the technology can follow the general guidance provided in the present study.
The 5′-terminal adenosine in the flap sequences of Snake primers shown in Figure 1 is not complementary to the target DNA. Thus, the sense amplicons in Snake inherit a corresponding 3′-mismatched thymidine moiety. This is an important element of the Snake system design because it prevents self-extension of the sense amplicons in PCR. This single 3′-unpaired nucleoside is also a key element of the optimal cleavage structure (21,22). According to Ichihara and Kurosawa (35), more than half of the products of PCR contain an extra adenosine residue at the 3′ end incorporated by Taq DNA polymerase that has no 3′-exonuclease ‘proofreading’ activity. If this is correct, then most of the sense amplicons in Snake incorporate not 3′-mono but 3′-dinucleotide mismatched termini and this does not support the 5′-nuclease cleavage. It remains unclear whether such a suboptimal cleavage structure contributes to the assay fluorescence or the signal is exclusively generated by the smaller fraction of the ‘optimal’ amplicons. In either case, the data obtained so far prove the assay’s superiority in signal productivity over Taqman and other real-time technologies.
Probe cleavage in Snake can potentially cycle, providing more than one cleaved probe per target template molecule during a given PCR cycle. If cleavage is indeed cycling, that would explain the strong signal in Snake PCR, especially when the assay is performed under optimized conditions (Figure 8). However, initial analysis of the data does not indicate that Snake belongs to the class of cycling probe technologies (36,37). Calculations show that the Snake system can digest up to 25% of the probe at a PCR cycle when the maximum cleavage rate is reached (Figure 8). This corresponds to 1.25 pmole of the probe cleaved. On the other hand, asymmetric PCR, when it enters into the linear amplification stage, can produce up to 1.75 pmole per cycle of newly synthesized target amplicon, in addition to what has already been produced in all previous cycles. These estimates do not support the idea of cycling of the probe cleavage in Snake but point to the most likely reason of the assay’s signal superiority: the unique mechanism, wherein the probe cleavage takes place not only ‘during’ but also ‘after’ the active state of amplification.
For the optimal Snake performance, the two catalytic activities, 5′-nuclease and DNA polymerase, need to be balanced. However, this is hard to achieve when both activities are provided by the same enzyme, i.e. Taq DNA polymerase. Use of elevated amounts of the enzyme to eliminate the assay’s 5′-nuclease deficiency may negatively affect the detection sensitivity by stimulating undesired reactions like primer-dimer formation. An alternative and likely better approach is to supplement the Snake reaction with one of the thermostable 5′-flap endonucleases (21,22). This actually opens the opportunity to use other DNA polymerases, particularly those with no 5′-nuclease activity. Note, however, that the use of DNA polymerases with 3′-nuclease proofreading activity has to be avoided in Snake. These enzymes can remove the 3′-uncomplementary nucleotide in folded sense amplicons (Figure 2E and F) and extend the remaining self-priming duplex with all of the negative consequences on PCR illustrated in Figure 4F and G. An example of DNA polymerase that has neither 5′- nor 3′-nuclease activity is VentR® (exo−) DNA polymerase from New England Biolabs, Inc.
Evidently the Snake technology has bright prospects to become an indispensable molecular tool for genetic analysis. It supersedes conventional assays in the detection of sequence variations as small as SNPs. The technology can be applied for qPCR at least as effectively as all other known real-time detection techniques. The inexpensive dual-labeled FRET probes used in Snake are the same as those that are currently applied in conventional Taqman, only shorter. Addition of a short 5′-sequence to one of the two primers appears to solve all major issues identified so far in real-time PCR. Moreover, the solutions come in an absolute and ultimate fashion leaving little, if any, opportunity for further improvement. In this regard, it is really difficult to imagine another next-generation technology that would be able to compete with Snake in terms of the key factors such as signal productivity, SNP discrimination power and cost-effectiveness.
FUNDING
National Institute of Health (SBIR grant 1R43GM079018-01A1). Additional and substantial project support was provided by Cepheid. Funding for open access charges: Perpetual Genomics funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Access to the necessary facilities and instrumentation was provided by Cepheid. The author would like to thank Cepheid’s employees and managers particularly John Bishop, David Persing, Humberto Reyes, Peter Dailey, John Smith, Alexander Gall, Sergey Lokhov, Mikhail Podyminogin and Ekaterina Viazovkina for interest, technical help and support. The assistance of Vassily Kutyavin (Haverford College, Haverford, PA) in the editing and revising of the manuscript is gratefully acknowledged.
REFERENCES
- 1.Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR: Real time monitoring of DNA amplification reactions. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 3.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechnologies. 1998;24:954–958. [PubMed] [Google Scholar]
- 4.Förster T. Delocalized excitation and excitation transfer. In: Sinanoglu O, editor. Modern Quantum Chemistry, Istanbul Lectures, part III. New York: Academic Press; 1965. pp. 93–137. [Google Scholar]
- 5.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 6.Didenko VV. DNA probes using fluorescence resonance energy transfer (FRET): design and application. BioTechniques. 2001;31:1106–1121. doi: 10.2144/01315rv02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livak KJ, Flood SJA, Marmaro J, Mullah KB. Self-quenching fluorescent probe. US Patent. 1998;5:723. 591. [Google Scholar]
- 8.Afonina IA, Reed MW, Lusby E, Shishkina IG, Belousov YS. Minor groove binder-conjugated DNA probes for Quantitative DNA detection by hybridization-triggered fluorescence. BioTechniques. 2002;32:940–949. doi: 10.2144/02324pf01. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz E, Estrada G, Lizardi PM. PNA molecular beacons for rapid detection of PCR amplicons. Mol. Cell. Probes. 1998;12:219–226. doi: 10.1006/mcpr.1998.0175. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi S, Kramer FR. Molecular beacons-probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 11.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 12.Duck P, Bender R, Crosby W, Robertson JG. Nucleic acid compositions with scissile linkage useful for detecting nucleic acid sequences. US Patent. 1989;4:876. 187. [Google Scholar]
- 13.Harvey JJ, Lee SP, Chan K, Kim JH, Hwang E-S, Cha C-Y, Knutson JR, Han MK. Characterization and application of CataCleave probe in real-time detection assays. Anal. Biochem. 2004;333:246–255. doi: 10.1016/j.ab.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 14.Kutyavin IV, Milesi D, Belousov Y, Podyminogin M, Vorobiev A, Gorn V, Lukhtanov EA, Vermeulen NMJ, Mahoney W. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelfand DH, Holland PM, Saiki RK, Watson RM. Homogeneous assay system using the nuclease activity of a nucleic acid polymerase. US Patent. 1993;5:210. 015. [Google Scholar]
- 16.Lie YS, Petropoulos CJ. Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotech. 1998;9:43–48. doi: 10.1016/s0958-1669(98)80082-7. [DOI] [PubMed] [Google Scholar]
- 17.Kutyavin IV, Lukhtanov EA, Gamper HB, Meyer RB. Oligonucleotides with conjugated dihydropyrroloindole tripeptides: base composition and backbone effects on hybridization. Nucleic Acids Res. 1997;25:3718–3723. doi: 10.1093/nar/25.18.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allawi HT, Kaiser MW, Onufriev AV, Ma WP, Brogaard AE, Case DA, Neri BP, Lyamichev VI. Modeling of flap endonuclease interactions with DNA substrate. J. Mol. Biol. 2003;328:537–554. doi: 10.1016/s0022-2836(03)00351-6. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser MW, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev VI. A comparison of Eubacterial and Archaeal structure-specific 5’-exonucleases. J. Biol. Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 23.Gyllensten UB, Erlich HA. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc. Natl Acad. Sci. USA. 1988;85:7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Detection of nucleic acid hybridization by nonradioactive fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1988;85:8790–8804. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyamichev V, Mast A, Hall J, Prudent J, Kaiser M, Takova T, Kwiatkowski R, Sander T, de Arruda M, Arco D, et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- 26.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaDuca RJ, Fay PJ, Chuang C, McHenry CS, Bambara RA. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983;22:5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- 28.Bedinger P, Munn M, Alberts BM. Sequence-specific pausing during in vitro DNA replication on double- stranded DNA templates. J. Biol. Chem. 1989;264:16880–16886. [PubMed] [Google Scholar]
- 29.Bierne H, Michel B. When replication forks stop. Mol. Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 30.Uhlenbeck OC. Complementary oligonucleotide binding to transfer RNA. J. Mol. Biol. 1972;65:25–41. doi: 10.1016/0022-2836(72)90489-5. [DOI] [PubMed] [Google Scholar]
- 31.Lima WF, Monia BP, Ecker DJ, Freier SM. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- 32.Kutyavin I. PCT patent publication; 2007. Use of products of PCR amplification carrying elements of secondary structure to improve PCR-based nucleic acid detection. WO/2007/127999. [Google Scholar]
- 33.Kutyavin IV. Use of base-modified duplex-stabilizing deoxynucleoside 5’-triphosphates to enhance the hybridization properties of primers and probes in detection polymerase chain reaction. Biochemistry. 2008;47:13666–13673. doi: 10.1021/bi8017784. [DOI] [PubMed] [Google Scholar]
- 34.Lebedev Y, Akopyans N, Azhikina T, Shevchenko Y, Potapov V, Stecenko D, Berg D, Sverdlov E. Oligonucleotides containing 2-aminoadenine and 5-methylcytosine are more effective as primers for PCR amplification than their nonmodified counterparts. Genet. Anal. 1996;13:15–21. doi: 10.1016/1050-3862(96)00139-8. [DOI] [PubMed] [Google Scholar]
- 35.Ichihara Y, Kurosawa Y. Construction of new T vectors for direct cloning of PCR products. Gene. 1993;130:153–154. doi: 10.1016/0378-1119(93)90361-6. [DOI] [PubMed] [Google Scholar]
- 36.Walder JA, Walder RY. Nucleic acid hybridization and amplification method for detection of specific sequences in which a complementary labeled nucleic acid probe is cleaved. US patent. 1995;5:403. 711. [Google Scholar]
- 37.Duck P, McNevin J. Cycling probe cleavage detection of nucleic acid sequences. US patent. 1997;5:660. 988. [Google Scholar]