Abstract
Mitochondrial DNA encodes a set of 13 polypeptides and is subjected to constant oxidative stress due to ROS production within the organelle. It has been shown that DNA repair in the mitochondrion proceeds through both short- and long-patch base excision repair (BER). In the present article, we have used the natural competence of mammalian mitochondria to import DNA and study the sub-mitochondrial localization of the repair system in organello. Results demonstrate that sequences corresponding to the mtDNA non-coding region interact with the inner membrane in a rapid and saturable fashion. We show that uracil containing import substrates are taken into the mitochondrion and are used as templates for damage driven DNA synthesis. After further sub-fractionation, we show that the length of the repair synthesis patch differs in the soluble and the particulate fraction. Bona fide long patch BER synthesis occurs on the DNA associated with the particulate fraction, whereas a nick driven DNA synthesis occurs when the uracil containing DNA accesses the soluble fraction. Our results suggest that coordinate interactions of the different partners needed for BER is only found at sites where the DNA is associated with the membrane.
INTRODUCTION
The mammalian mitochondrial genome (mtDNA) is a small (∼16.5 kb) circular genome found in multiple copies in all nucleated cells. It encodes 22 mt-tRNAs, 2 mt-rRNAs and 13 proteins which are integral components of the complexes that couple oxidative phosphorylation. As a direct consequence of its close proximity to the mitochondrial electron transfer chain and a lack of protective histones, mtDNA is considered as being more damage prone than its nuclear counterpart (1). As mtDNA mutations are known to cause a variety of mainly neurodegenerative disorders and have been implicated in the ageing process itself (2–4), it is important that such lesions are efficiently repaired.
The topic of DNA repair has received substantial interest (5,6). Whilst the nucleus possesses multiple well-characterized DNA repair mechanisms, the molecular components of only the base excision repair (BER) pathway have been described in mammalian mitochondria, although a mismatch repair and non-homologous end-joining pathway may also exist (7,8). This BER mechanism is divided in two sub-pathways, the short-patch BER (spBER) and the long-patch BER (lpBER) (9), whose main difference lies in the DNA repair synthesis step. The existence of the latter in mitochondria has been recently demonstrated by several groups (10,11) whilst the former was first described in the organelle >10 years ago (12).
In more detail, the first step in both cases is the recognition and cleavage of the damaged base by a specific (broad range) DNA glycosylase. The state of the resulting abasic (AP) lesion then normally determines whether spBER or lpBER is needed to complete the repair reaction. After cleavage by a monofunctional DNA glycosylase such as uracil DNA glycosylase (UDG), a synthesis-compatible 3′-hydroxyl end is generated by the AP endonuclease 1 (APE1), leaving a 5′-deoxyribose phosphate (dRp). When a bi-functional glycosylase such as OGG1 is involved, the base removal is followed by a lyase activity resulting in a terminal 5′-phosphate. The lesion is further resolved by APE1 which produces a 3′-OH. In spBER, mitochondrial DNA polymerase γ, the sole mitochondrial replicative and repair polymerase, adds the missing nucleotide before ligation. In cases where the 3′-terminus is non-ligatable, repair synthesis stalls after addition of one nucleotide. The lpBER facilitates the rescue of such stalled complexes by allowing strand displacement to occur. Recent reports have shown that both Fen1 and the DNA2 helicase/nuclease are involved in mtDNA strand displacement and cleavage (13). Synthesis of 2–6 nt occurs, displacing the original DNA strand, which is then recognized by Fen1/DNA2 and cleaved, leaving a 5′-phosphate that can be ligated by the mitochondrial DNA ligase III. Normally, simple abasic sites are repaired by spBER. However, it has recently been shown that sites generated by uracil-N-glycosylase activity in mitochondria can also act as substrates for lpBER (14).
Although evidence for the two forms of BER in mammalian mitochondria is compelling, the physical organization of these processes in the mitochondrion is unclear and potentially very elaborate. It is generally accepted that mtDNA is anchored to the mitochondrial membrane by an uncharacterized mechanism involving mtDNA sequence from the control region (15). DNA glycosylases, DNA polymerase γ and DNA ligase III also seem to interact with an inner membrane-containing particulate fraction, but strikingly this interaction does not appear to be mediated by the mtDNA (16). Moreover, despite their co-localization in the mitochondrial membrane, the different repair components do not seem to stably interact with each other and to build complete repair complexes in mitochondria, at least for the uracil repair pathway (17). Further investigation of these mechanisms has been hindered by the absence of a possibility to analyse the repair of a traceable DNA substrate in a physiological context.
We recently demonstrated that isolated mammalian mitochondria are competent for low level DNA uptake (18). In the present work, we developed in organello DNA import and repair assays based on this mechanism. We showed that exogenous DNA fragments associate with the mitochondrial inner membrane upon import. Using specifically designed synthetic sequences, we demonstrate that both mitochondrial spBER and lpBER function to remove and replace uracil in imported DNA. Finally, we show that bona fide DNA repair synthesis occurs only in association with the membranes in the in organello physiological context, while generic DNA-dependent DNA synthesis predominates in the soluble mitochondrial compartment. We therefore conclude that DNA must be anchored to the membrane to facilitate BER in mammalian mitochondria.
MATERIALS AND METHODS
Preparation of internally labelled standard DNA import substrates
Five DNA amplicons, ranging from 221 to 235 bp were PCR-generated from isolated rat mtDNA using the primers described in Supplementary Table S1. Four corresponded to different sections of the rat mtDNA non-coding region (NCR) (NC1–NC4) and one, ExtNC, was internal to rat MT-CO1. A sixth probe, ctGFP, corresponded to a 390 bp fragment from the GFP coding sequence of pGFPDloopLuc (18). Aliquots (50 ng) of each PCR product were transferred to a new reaction mixture with the same primer pair but with cold dCTP substituted with radiolabelled [α32P]dCTP (50 µCi, 3000 Ci/mmol). One PCR cycle with a 10 min elongation was performed before the addition of 0.2 nmol cold dCTP and another 5 min elongation step. For strand-specific labelling of NC2, a similar single cycle was performed from 50 ng of the PCR product as template but with only one primer. After labelling, the displaced, cold, single DNA strand was digested by Exo-Sap (USB) following the manufacturer's; recommendations. Unincorporated radionucleotide was removed from all products by passing through a Sephadex G50 column followed by ethanol precipitation.
Preparation of end-labelled uracil-containing duplex import substrates
Both 110 nt uracil-containing oligomers were purchased from Sigma-Aldrich and were used directly or further subjected to standard RP-HPLC. Their sequences are given in Supplementary Table S1. Oligomer ct110 is the identical standard oligodeoxynucleotide to U/Arep, with the two uracils substituted with thymine (Figure 3). Prior to annealing with their standard complementary strand, 50 pmol were end-labelled by T4 polynucleotide kinase in the presence of 17 pmol of [γ32P]ATP (10 mCi/ml, 3000 Ci/mmol). Free ATP was eliminated by gel-filtration through Sephadex G50. The DNA was then phenol extracted, ethanol precipitated and resuspended in hybridization buffer (NaCl 1 mM, EDTA 10 mM, Tris–HCl 100 mM pH 7.6). An excess (500 pmol) of complementary strand was added and the temperature was raised to 70°C for 10 min before slowly cooling down to room temperature. The annealed duplex was separated from the remaining single strands by electrophoresis through a 15% (w/v) non-denaturating polyacrylamide gel and the labelled dsDNA was visualized by autoradiography. The matching gel area was cut out and submitted to elution in 3 mM EDTA, 0.5 M ammonium acetate, 10 mM magnesium acetate, 0.1% (w/v) SDS overnight at 4°C.
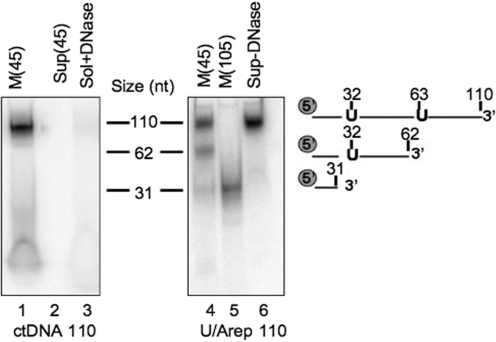
Figure 3.
Imported uracil-containing DNA is cleaved at the uracils. Standard 110 nt oligodeoxynucleotide (ctDNA, lanes 1–3) or containing uracil residues at positions 32 and 63 (U/Arep 110; lanes 4–6) was γ32P-end-labelled, annealed to its complement and incubated with rat liver mitochondria in standard IM as described. Following import for 45 (lanes 1–4 and 6) or 105 min [M (105); lane 5], mitochondria were DNase treated (lanes 1, 2 and 4–6) or solubilized prior to DNase treatment (sol+DNase; lane 3). DNA was then extracted from the mitochondrial pellet (M; lanes 1, 4 and 5), solubilized mitochondria (lane 3) or supernatant (Sup; lanes 2 and 6), precipitated and separated by denaturing polyacrylamide gel electrophoresis before autoradiography. Markers were prepared by end-labelling oligomers of the indicated sizes and corresponded to the migration of the U/Arep DNA cleaved at the incorporated uracils as indicated.
Preparation of randomly incorporated uracil-containing DNA import substrates
A 1060 bp (NCR probe) or a 225 bp (NC2 probe) section of the rat mDNA NCR was PCR-amplified using the primers indicated in Supplementary Table S1. The reactions were run in the presence of 100 µM of the four dNTPs and were supplemented with the following amounts of dUTP: no dUTP (U0 probes), 10 µM (U1 probes), 100 µM (U2 probes) or 1 mM (U3 probes). We observed that dUTP concentrations higher than 1 mM inhibited the PCR reaction. Uracil incorporation was checked using a commercial uracil excison mix (Epicentre) following the manufacturer's; instructions. The PCR products were purified by electrophoresis through 1% (w/v) low melting agarose gels, extracted and precipitated.
Isolation of mitochondria
Mitochondria were prepared from the livers of Wistar rats (200–250 g), dispatched by cervical dislocation and used at most 4 h post-mortem. The liver was transferred directly into 25 ml of ice-cold MSE [220 mM mannitol, 70 mM sucrose, 10 mM HEPES–KOH pH 7.4, 1 mM MgCI2, 1 mM EGTA, 1% (w/v) BSA]. All of the following steps were implemented on ice in a 4°C room. The liver was minced, suspended in 30 ml of MSE with 0.33 mM PMSF and homogenized by 15 strokes of a Potter–Elvehjem, glass–teflon homogenizer. Following centrifugation (400 g, 4 min), the pellet was discarded and the supernatant centrifuged at 8600g for 10 min. The mitochondrial pellet was carefully resuspended in 4.5 ml of MSE and the differential centrifugation was repeated. Mitochondria were finally washed and resuspended in MSE minus BSA prior to estimating the protein concentration by the Bradford assay (BioRad).
Mitochondrial import reactions
Mitochondria (250 µg of protein) were pre-equilibrated in 200 µl import buffer (IB) (220 mM mannitol, 70 mM sucrose, 20 mM Tris–HCl pH 7.4, 1 mM EDTA, 1 mM EGTA, 10 mM glutamate, 1 mM malate) for 20 min at 30°C under mild shaking. Radiolabelled DNA substrate (∼10 ng) was then added and the organelles were further incubated for the indicated time. The suspension was subsequently supplemented with DNase I (200 µg) and MgCl2 (10 mM) to digest the non-imported DNA. After 20 min, the medium was further supplemented with EDTA (40 mM) and EGTA (10 mM) and centrifuged for 10 min at 9000g. The pellet was resuspended in 200 µl IB plus EDTA/EGTA and harvested again before mitochondrial sub-fractionation.
Mitochondrial sub-fractionation
Following DNA import, mitochondrial pellets were frozen and thawed three times in 100 µl IB before centrifugation at 100 000g for 1 h. The supernatant was saved as the soluble fraction and the pellet was resuspended in an equal volume of 20 mM Tris–HCl pH 8.0, 2 mM EDTA, 100 mM NaCl, 1% (w/v) SDS to produce the membrane-rich particulate fraction. To confirm successful separation of particulate and matrix soluble fractions, aliquots were fractionated by SDS-PAGE on 12% (w/v) polyacrylamide gels and transferred onto an immobilon® membrane (millipore) in 25 mM Tris, 192 mM glycine, 0.02% (w/v) SDS, 20% (v/v) methanol, pH 8.3. Blots were probed with the indicated antibodies and visualized using the ECL+ chemiluminescence kit (GE Healthcare) following binding of HRP-conjugated secondary antibodies.
Pre-loading of mitochondria with radionucleotides and DNA repair synthesis
For in organello DNA repair synthesis experiments, mitochondria (250 µg of protein) were incubated at 30°C under mild shaking in 300 µl of RB (75 mM mannitol, 25 mM sucrose, 100 mM KCl, 10 mM Tris–HCl, 10 mM KH2PO4, 0.05 mM EGTA, 10 mM glutamate, 1 mM malate, pH 7.4) supplemented with 60 nM of either [α32P]dCTP or [α32P]dTTP and 250 µM of the three other, unlabelled dNTPs. After 20 min, the mitochondria were harvested by centrifugation for 10 min at 9000g, washed, resuspended in 300 µl of RB and further incubated for 45 min to 2 h as indicated, with 500 ng of unlabelled uracil-containing probe.
Nucleic acid extraction and analysis following mitochondrial import/repair assays
To analyse the import of NC1–NC4, ExtNC and ctGFP probes, mitochondrial sub-fractions were phenol extracted at 55°C. Nucleic acids were ethanol precipitated, fractionated by electrophoresis on either 10% (w/v) polyacrylamide gels or 10% (w/v) polyacrylamide gel with urea 7 M (denaturing conditions; see figure legends) in TBE (90 mM Tris, 90 mM boric acid, 2 mM EDTA, pH 8.0) and visualized by autoradiography. Where necessary, signal intensities were compared using the ImageJ software (http://rsbweb.nih.gov/ij/).
To analyse the import/incision of the end-labelled 110 bp probes ctDNA 110 and U/Arep 110, post-import mitochondrial pellets were phenol extracted at 55°C. Nucleic acids were ethanol precipitated and denatured for 5 min at 95°C in DNA loading buffer [formamide 95% (v/v), EDTA 20 mM, bromphenol blue 0.05% (w/v), xylene cyanol 0.05% (w/v)]. Cleavage products were separated by migration on 15% (w/v) polyacrylamide, 7 M urea gels in TBE. Gels were exposed to a phosphorimager plate (Fuji) and visualized by ImageQuant analysis.
To analyse in organello complete repair of the uracil-containing 1060 bp DNA fragments NCFU0– NCFU3, post-import mitochondrial pellets were phenol extracted at 55°C. Nucleic acids were ethanol precipitated before migration on 1% (w/v) agarose gels in TAE (40 mM Tris–acetate, 1 mM EDTA, pH 8.0), transfer to a nylon membrane and exposure to a phosphorimager plate.
To analyse the localization of the repair reactions with the 225 bp NC2U0–NC2U3 probes or the 110 bp U/Arep and U/Anick probes, nucleic acids were prepared as above from the sub-mitochondrial fractions, denatured for 5 min at 95°C in DNA loading buffer and separated on 15% (w/v) polyacrylamide, 7 M urea gels in TBE. Repair products were visualized by autoradiography.
RESULTS
Exogenous DNA probes show association with the membrane upon mitochondrial import
As has been published previously, limited amounts of naked DNA (∼10 ng DNA per milligram mitochondrial protein) can access the inner compartment of isolated mammalian mitochondria or mitoplasts (18). The imported DNA can be transcribed, implying that it is recruited by organelle enzymes and factors involved in mtDNA metabolism. Previous DNA uptake studies had shown a rapid and saturable association of the imported DNA with the endogenous mtDNA when the imported molecule contained sequences corresponding to the NCR (18). Further, it has been reported that mtDNA, despite being a large polyanion, is anchored to the inner membrane (IM), primarily via interactions with its NCR (15,19). We therefore wished to determine the sub-mitochondrial localization of the exogenous DNA upon import and to assess whether this localization was dependent on sequences present within the NCR.
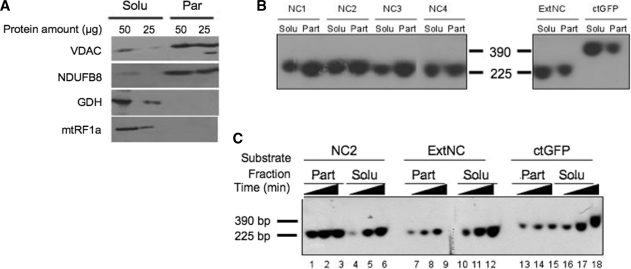
Four DNA fragments ranging from 221 to 235 bp (NC1–NC4), spanning the rat mtDNA NCR was prepared. In addition, two other DNA probes corresponding either to a mtDNA sequence taken outside of the NCR (ExtNC, 224 bp) or to the GFP coding sequence (ctGFP, 390 bp) were generated. Import assays into isolated rat liver mitochondria were then performed with the six probes. After 45-min incubation, a DNase treatment was performed to eliminate all non-imported DNA and the organelles were sub-fractionated into a soluble and particulate fraction. The two fractions were used for western blot analysis and nucleic acid extraction. Western blot analysis confirmed a substantial enrichment of mitochondrial inner membrane components in the particulate fraction and of matrix proteins in the soluble fraction, as shown in Figure 1A and Supplementary Figure S1.
Figure 1.
Sequence-dependent association of imported DNA with the mitochondrial particulate fraction. (A) Mitochondrial sub-fractionation. Isolated rat liver mitochondria were disrupted by repeated freezing and thawing. Soluble (Solu) and particulate (Part) fractions were separated by centrifugation at 100 000g for 1 h and the indicated amount of protein from the different fractions was subjected to western blot analysis. Purity of fractions was assessed by blotting with antibodies against marker proteins. The VDAC and a component of respiratory chain complex I (NDUFB8) are outer and inner membrane markers, respectively; glutamate dehydrogenase (GDH) and the mitochondrial translation release factor (mtRF1a) are matrix soluble markers. (B) NCR corresponding sequences show a strong association with the particulate fraction after import into isolated rat mitochondria. Four probes ranging from 221 to 235 bp (NC1–NC4) spanning most of the NCR, an additional mtDNA probe from outside the NCR (ExtNC) and a 390 bp probe from the GFP coding sequence (ctGFP) were PCR amplified and internally labelled with [α32P]dCTP (primers and locations are given in Supplementary Table S1). Following a 45-min incubation with coupled rat mitochondria, non-imported DNA was DNase I digested, mitochondria sub-fractionated, protected DNA was extracted and subjected to non-denaturing gel electrophoresis prior to autoradiography and visualization as detailed in ‘Materials and Methods’ section. Typical results are shown from three repeats. Solu and Part refer to the soluble and particulate mitochondrial fractions, respectively. Markers refer to labelled probes (NC2 and ctGFP) of the indicated sizes. (C) Imported NCR DNA associates rapidly with the particulate fraction in a saturable process. A representative probe for the NCR, NC2 and the two additional probes ExtNC and ctGFP were internally labelled as in Figure 2B. Incubation of mitochondria with each probe was terminated after 15, 30 or 45 min (indicated as increasing time) before sub-fractionation, DNA extraction and visualization as for Figure 2B. Typical results are shown from three repeats.
Representative results of nucleic acid analysis are displayed in Figure 1B. After import, all probes showed partitioning between the two fractions. However, sequences NC1–NC4 were found mostly associated with the particulate fraction, whereas a major part of ExtNC and ctGFP was recovered in the soluble fraction. Three additional probes generated to other regions of the rat mtDNA outside the NCR were also associated mainly with the soluble fraction (Supplementary Figure S2). These data infer that the imported DNA binds to the membranes and that sharing sequence identity with the mtDNA NCR further promotes such a membrane association.
Association of the imported DNA with the IM is a rapid and saturable process
As mentioned, Figure 1B shows that after a 45-min import experiment, there was an interaction of the imported DNA with the mitochondrial membrane. All probes derived from the mtDNA NCR shared a higher membrane-binding capacity but the bias appeared to be relatively mild, with 30–40% of the NCR probe still being found in the soluble fraction at the end of the experiment. Conversely, for non-NCR sequences, ∼70% of the probe was found in the soluble fraction after 45 min (66 and 72% for ExtNC and ctGFP, respectively).
To characterize the process in more detail, import kinetics were assessed over a 45 min time course, with the mitochondria from each sample being sub-fractionated as above. No obvious selectivity was apparent in the sorting of the NC1–NC4 probes in previous assays (Figure 1B). We thus decided to focus only on NC2 as a representative probe for the NCR, together with ExtNC and ctGFP. As shown in Figure 1C, the association of NC2, ExtNC and ctGFP with the membrane was rapidly saturated, with only a minimal increase between 15 and 45 min (lanes 1–3, 7–9 and 13–15). However, the NC2 sequence had a markedly stronger signal (lane 1 cf. 7, 13). In contrast, the imported DNA accumulated in the soluble fraction in a time-dependent manner (lanes 4–6, 10–12 and 16–18). These data are consistent with the imported DNA first associating with an available membrane-located ‘receptor’ upon import through the IM. Subsequent accumulation of the incoming DNA in the soluble fraction would then depend on the affinity of the probe for the receptor.
The exact nature of the ‘receptor’ tethering the probe to the membrane is unclear. The imported DNA might be recognized by a membrane protein(s). However, all probes derived from the NCR appeared to associate with the membrane with equal avidity, whilst all other probes showed lower level association. This suggests that the NCR has some special affinity for the receptor and is consistent with the view that the mtDNA anchors to the IM primarily via its NCR. In previous experiments, imported DNA containing the NCR sequence was shown to form a higher molecular weight structure with the endogenous mtDNA (18). Another possible explanation would thus be that, besides recognition by a protein receptor, the NC probes can undergo a strand invasion of the endogenous displacement loop, which normally assumes a partially open triple-stranded structure (Figure 2A). The most obvious option in this respect seems to be that the NCR probes would displace the third strand, i.e. the 7S DNA strand. In such a view, the mtDNA itself would be an anchoring ‘receptor’ element.
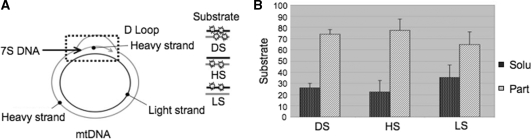
Figure 2.
Sequence selective association of imported DNA with the particulate fraction is not strand specific. (A) Schematic representation of the mitochondrial genome, with emphasis on the triplex displacement (D)-loop region. The short 7S species is indicated. (B) Association of imported NC DNA with the particulate fraction is not strand specific. Three different 225 bp NC2 probes were prepared, carrying radiolabel on both strands (DS), the heavy strand (HS) or the light strand (LS) as illustrated. Import experiments were performed with isolated rat mitochondria for 15 min before DNaseI treatment and sub-fractionation as described in the legend of Figure 2A. Radiolabelled probe from each different fraction was analysed after extraction and electrophoresis in non-denaturing conditions as in Figure 2B. The percentage of the DNA retrieved in each of the two fractions was calculated as described in ‘Materials and Methods’ section and are represented as a histogram. Bars show standard deviations from five independent repeats.
To test the hypothesis of a 7S DNA strand displacement, we determined whether the interaction of the NC2 probe was strand specific. We labelled either strand of duplex NC2 independently as described in the ‘Materials and Methods’ section and mitochondrial import was allowed to proceed for 15 min before sub-fractionation and nucleic acid extraction. Representative autoradiograms are shown as Supplementary Figure S3. The control probe labelled on both strands displayed the same association pattern as previously observed (Figure 2B DS; see NC2, Figure 1B for comparison), the vast majority of the imported DNA being found associated with the membrane fraction. Similarly, duplexes radiolabelled in either the heavy or light strand also accumulated in the particulate fraction after 15 min (Figure 2B; HS, LS). As the 7S DNA is a heavy strand sequence, it was not expected to be displaced by a labelled light strand probe. The data thus suggest that strand invasion is unlikely to explain the sequence-specific membrane association or the interaction with the endogenous mtDNA that has been noted above.
Uracil-containing DNA imported into mitochondria can be cleaved at the level of uracil
Previous studies showed that DNA imported into isolated rat liver mitochondria can also act as a template for DNA synthesis (18). What exactly constitutes this DNA synthesis has not been defined so far and we postulated that it might include DNA repair processes. First, we wished to assess whether uracil residues could be recognized and removed on import into isolated mitochondria. A 110-bp end-labelled dsDNA fragment was either synthesized with only standard deoxyribonucleotides (ctDNA110) or contained two uracil bases (U/Arep 110) at positions 32 and 63 of the radiolabelled strand, as detailed in Figure 3. These two substrates were in turn used for import assays with rat liver mitochondria. After 45 min of import in standard IM followed by DNase treatment, nucleic acids were extracted and subsequently analysed by denaturing gel electrophoresis.
As shown in Figure 3, the control DNA (ctDNA110) of 110 nt was protected after DNase treatment (lane 1) and was lost if the mitochondria were solubilized prior to treatment (lane 3). In contrast, three species were found after import of the U/Arep 110 (lane 4), a 110 nt intact form and two specific cleavage products of 31 and 62 nt. These products corresponded to cleavage occurring specifically at the positions of uracil and thus were presumably mediated by the mitochondrial uracil–DNA glycosylase [UNG, (20,21)] and AP endonuclease(s) (22,23). An import incubation extended to 105 min led to an accumulation of the 31 nt species (lane 5). In standard IM, we did not observe any appreciable ligation of cleaved products in the extended incubation. To rule out the possibility that the specific cleavage may have occurred outside the mitochondrion, the reaction supernatant was removed and analysed just prior to DNase treatment. Crucially, only full-length molecules were present in the post-mitochondrial supernatant after incubation (lane 6), confirming that the uracil excision activities must have occurred within the mitochondrion. These data demonstrate that the imported DNA can indeed recruit DNA repair enzymes and confirms that natural competence can be used to study mitochondrial DNA repair in organello.
Imported uracil-containing DNA can act as a template for DNA synthesis in isolated mitochondria
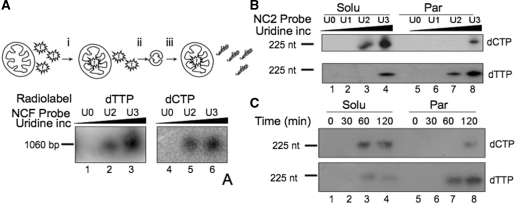
Having shown that imported uracil-containing DNA can recruit the enzymes leading to damage excision, we next assessed the ability of isolated mammalian mitochondria to carry out complete repair, including repair synthesis and potentially religation. For that purpose, we first tested the mitochondrial import and repair-mediated DNA synthesis potential of a uracil-containing 1060 bp DNA fragment (NCF) corresponding to the rat mtDNA NCR. DNA probes NCFU0–NCFU3 were prepared by PCR with increasing concentrations of dUTP, the incorporated uracil pairing with adenine. To assess DNA repair synthesis following the removal of uracil on import into mitochondria, experiments were performed with two independent radionucleotides. Incorporation of [α32P]dTMP should occur irrespective of the mechanism of uracil replacement, as any repair process would start with the incorporation of this nucleotide to substitute the displaced uracil. Similar experiments were also performed with [α32P]dCTP to determine whether repair synthesis was capable of incorporating >1 nt.
Prior to the DNA import and repair assays, isolated rat liver mitochondria were first loaded with free [α32P]dTTP or [α32P]dCTP in DNA repair buffer, harvested and the non-imported free dNTPs removed (see ‘Materials and Methods’ section and Figure 4A). Mitochondria were then incubated for 45 min in DNA repair buffer with DNA probes NCFU0, NCFU2 and NCFU3, followed by nucleic acid extraction and non-denaturing gel electrophoresis. Typical results are shown in Figure 4A. [α32P]dTMP and [α32P]dCMP were both incorporated into probes NCFU2 and NCFU3, with a stronger signal for NCFU3 (lanes 3 and 6, respectively), consistent with the increased amount of uracil in the probe. The absence of incorporation into NCFU0 (lanes 1 and 4) showed that the DNA synthesis observed with NCFU2 and NCFU3 was indeed damage driven and that repair had occurred with the uracil-containing probes. Control experiments using NCFU3 with aphidicolin (500 µM) present during the incubation confirmed that incorporation was not mediated by contaminating aphidicolin-sensitive polymerases (Supplementary Figure S4). Although non-imported free dNTPs had been removed from the loaded mitochondrial preparation, it was crucial to ensure that the damage-driven DNA synthesis did not result from cytosolic contamination but did occur in the mitochondria. We therefore precipitated the DNA probe from the post-import supernatant prior to DNase I treatment. No signal could be demonstrated in the precipitant, consistent with either degradation of DNA outside the mitochondrion or with the absence of repair of non-imported DNA (data not shown).
Figure 4.
DNA repair mechanisms differ dependent on their mitochondrial location. (A) Mitochondria repair uracil-containing DNA on import. In these import and repair experiments the isolated mitochondria were first (i) incubated with the free radionucleotide (dTTP lanes 1–3; dCTP lanes 4–6), (ii) harvested and (iii) washed twice to remove unincorporated radiolabel. Aliquots of 1060 bp probe were prepared with increasing levels of uracil incorporation (U0–U3) as detailed in ‘Materials and Methods’ section and incubated with the pre-loaded mitochondria in DNA repair buffer (RB) for 45 min before DNase I treatment, DNA extraction, separation through non-denaturing gels, transfer to nylon membranes and visualization. (B) Mitochondrial sub-fractions show differing DNA repair mechanisms. Aliquots of probe NC2 were prepared with increasing amounts of uridine prior to incubation with pre-loaded mitochondria in RB for 2 h. Following import, mitochondria were sub-fractionated and incorporation of radiolabel into the probe in each fraction was monitored after migration under denaturing conditions and autoradiography. Upper panel, mitochondria were pre-loaded with dCTP; lower panel, dTTP. (C) Repair driven DNA synthesis as a function of time. A similar experiment was performed with pre-loaded mitochondria incubated for the indicated times with probe NC2U3. Mitochondria were sub-fractionated, DNA isolated and analysed exactly as described in B.
From these assays, we concluded that the imported DNA can recruit not only the glycosylase and AP endonuclease activities but also the DNA polymerase (and potentially the ligase), further validating DNA import into isolated mitochondria as a tool for physiological analysis of repair. As revealed with [α32P]dCTP, more than one deoxyribonucleotide was incorporated into the repair patch, suggesting that the spBER was not the only repair pathway occurring with our uracil-containing probes.
The mechanism of DNA damage-driven synthesis differs between the membrane and soluble compartment
The mammalian mtDNA BER machinery has been found to be at least partially associated with an IM-containing mitochondrial particulate fraction (16). According to the above experiments, DNA imported into isolated rat liver mitochondria also associates with the membrane, with greater affinity if the sequence shares similarity with any part of the NCR. This interaction then becomes saturated and further imported probes accumulate in the soluble matrix compartment. On this basis, we used our in organello approach to determine whether repair occurred predominantly in the membrane-associated DNA fraction or in the matrix-localized fraction. Further, selective repair with radiolabelled dCTP or dTTP enabled us to investigate whether different mechanisms exist in different compartments for DNA damage-driven synthesis. To examine these processes, we first incorporated increasing amounts of uracil into the 225 bp NC2 fragment (see above) and yielded the NC2U0–NC2U3 probes. In organello DNA repair experiments were then performed as described above in Figure 4A except with an increased incubation period to 2 h to promote complete repair, followed by sub-fractionation steps as in Figure 1, DNA extraction and visualization following denaturing gel electrophoresis.
As shown in Figure 4B and C, radiolabelled dCMP and dTMP were incorporated into a complete 225 nt product, both in the membrane-associated DNA fraction and the matrix-localized fraction. DNA synthesis was again dependent on the amount of uracil in the probe and hence, was damage driven. The efficiency was nevertheless quite different between the soluble and membrane fractions. Incorporation of dCMP into a uracil-containing template showed that synthesis of >1 nt at the repair site predominated in the soluble fraction (compare NC2U2 and NC2U3 soluble versus NC2U3 particulate). The mechanism of DNA repair synthesis thus seemed to differ regarding the mitochondrial sub-localization of the probe. dTMP is the first nucleotide to be incorporated in a BER-type DNA repair synthesis but a signal from dTMP incorporation does not necessarily imply that only 1 nt has been added during the repair step. Nevertheless, different patterns were also observed with this radiolabel between NC2U2/NC2U3 soluble and NC2U2/NC2U3 particulate. Strikingly, the strongest signals were obtained for the membrane fraction in this case, the opposite to what was observed with radioactive dCTP.
It cannot be excluded that comparing the signals produced from the incorporation of dCMP and dTMP is biased, as the relative rates of mitochondrial import for these two nucleotides or their overall concentration in the matrix have not been determined. However, if the assumption is made that both are comparable, as the signal for dTMP incorporation was stronger than that for dCMP incorporation in the particulate fraction it suggests that spBER is the main repair process in the membrane-associated DNA fraction.
We next conducted a time course of in organello import and repair of the NC2U3 probe to further elucidate the process (Figure 4C). In the soluble fraction, the earliest and strongest signal came from the incorporation of [α32P]dCMP, while the incorporation of labelled dTMP was again far more efficient in the particulate fraction. This difference further supported the previous observations that the freely soluble DNA mainly incorporated more than one nucleotide, while membrane-associated DNA incorporated only one nucleotide in most cases, a canonical feature of spBER. Nevertheless, dCMP was also significantly incorporated into the particulate fraction, supporting the concept of two mechanisms of DNA repair occurring at the membrane level.
Both spBER and lpBER occur in the membrane-associated DNA fraction
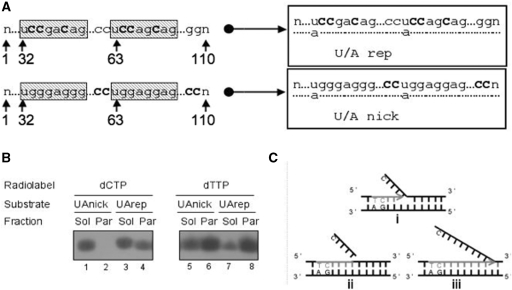
As mentioned, all experiments at that stage suggested that the major DNA repair mechanism at the membrane level mediates the substitution of a single uridine for a thymidine, consistent with spBER. However, as cytidine was also incorporated into membrane-associated DNA, albeit with an apparent lower efficiency, this could be representative either of lpBER or of some generic DNA synthesis driven by a nicked dsDNA template. In order to discriminate between bona fide lpBER and generic, nick-mediated DNA synthesis, an additional probe was designed. This was similar to the above double-stranded 110 bp U/Arep bearing two uracils at positions 32 and 63 but with an appropriately biased sequence (Figure 5 A). In the U/Arep template, three cytidine nucleosides could be incorporated within six residues from each of the original uracil positions, a distance that would be covered by the lpBER pathway. Alternatively, U/Anick had no cytosine in close proximity to the uracil and was therefore suitable to uniquely monitor long-range generic DNA synthesis through radiolabelled dCMP incorporation.
Figure 5.
DNA synthesis in the particulate fraction is limited to the long patch BER range. (A) Design of repair template. Two uracil-containing sequence-bias 110 nt oligonucleotides were designed to evaluate different DNA repair mechanisms in the mitochondrion, end-labelled and annealed to their complementary standard oligomer. Uracils were incorporated at positions 32 and 63 as indicated. To assess the mode of DNA repair, cytosines were added distal to the uracils and well beyond the 6–8 nt long patch region (U/A nick), or specifically within the long patch (U/A rep). (B) BER occurs at the mitochondrial membrane. Isolated mitochondria were preloaded with the indicated radionucleotide before 45 min incubation in RB with either U/Arep or U/Anick and sub-fractionation as described in ‘Materials and Methods’ section. DNA was extracted post-import and subjected to native gel electrophoresis prior to autoradiography. Images are accurate representations from one of three independent repeats. (C) Schematic representation to describe the modes of DNA repair observed. In (i) uracil is removed from the template. In (ii) enzymes coordinate uracil removal and nucleotides incorporation into a patch spanning six residues with the excision of the ‘flapping structure’, consistent with lpBER as seen only with U/Arep and only in the particulate fraction. In (iii) generic nick-directed DNA synthesis occurs on the oligomer, with cytosine replaced throughout the template, as seen with the U/Anick template.
Rat liver mitochondria were pre-loaded with [α32P]dTTP or [α32P]dCTP, as described, subjected to import/repair assays with either U/Arep or U/Anick and finally sub-fractionated into a soluble and particulate fraction. Not surprisingly, thymidine incorporation was observed with both templates (Figure 5B), as repair synthesis will always start with the substitution of uridine for thymidine and would occur with any of the three mechanisms, spBER, lpBER or generic nick-directed DNA synthesis. Cytidine was incorporated into both U/Arep and U/Anick in the soluble DNA fraction, suggesting that nick-directed DNA synthesis could account for at least part of the damage-driven DNA synthesis observed in this fraction. In contrast, cytidine incorporation into the membrane-associated DNA only occurred with U/Arep, which carried cytosines close to the uracils, i.e. within the lpBER range. The absence of [α32P]dCMP labelling for U/Anick, in which cytosines were located much further downstream of the uracils, showed that in the membrane-associated DNA de novo incorporation stopped after a few nucleotides. Hence, repair of uracil in the membrane compartment involved lpBER but not generic nick-directed DNA synthesis. Finally, the incorporation of [α32P]dTMP into the particulate fraction of U/Arep was much greater than that of [α32P]dCMP, although in the sequence there were three cytosines in the lpBER range downstream of each uracil. As the specific activities of the two radionucleotides were similar and provided that their mitochondrial import efficiencies and matrix concentrations were also comparable, this means that in a large sub-population of the molecules, repair synthesis only replaced the uridine, implicating again spBER.
We have therefore observed three different types of DNA repair synthesis proceeding from the cleavage of uracil. In the soluble DNA fraction, the synthesis extends further than 6 nt downstream of the strand incision and could correspond to a form of nick-translation or strand displacement. In the membrane-associated DNA fraction, synthesis following removal of uracil occurs through both spBER and lpBER (Figure 5C).
DISCUSSION
spBER and lpBER activities have been described in mammalian mitochondrial extracts (10,12,14). Selective repair enzymes and the mtDNA itself have been reported as interacting with the inner membrane, but the organization of the repair reactions in physiological conditions was unknown. In this report, we took advantage of the natural DNA import competence of mammalian mitochondria (18) to study the complexity and distribution of the repair reactions in the context of intact organelles. Naked DNA uptake competence has been demonstrated for isolated plant, mammalian and yeast mitochondria (24–26) but the detailed molecular mechanism of the import has yet to be elucidated. Reports implicate the voltage dependent anion channel (VDAC) as the mediator for DNA translocation across the outer membrane. However, the inner membrane DNA carrier or channel remains elusive and to date no effective inhibitor of DNA import across the inner membrane of mammalian mitochondria has been found. Nevertheless, in the absence of in vivo mitochondrial transfection procedures, apart from the unicellular organisms Saccharomyces cerevisiae and Chlamydomonas reinhardtii (27,28), the simplicity of this assay makes it an appealing system for experimental investigation of organellar DNA maintenance processes. Based on the import of appropriate DNA substrates, we demonstrated previously that plant mitochondria possess a spBER pathway (24). We have now been able to exploit the DNA import system to address the mechanisms of DNA repair in mammalian organelles. We show here that bona fide DNA repair reactions take place at sites where the DNA is anchored to the mitochondrial inner membrane. An unrelated, generic nick-directed DNA repair synthesis occurs in the soluble compartment.
A critical aspect of our experiments is the pre-loading of mitochondria with radiolabelled pyrimidine deoxyribonucleotides to ensure that the repair-driven incorporation of radiolabel must occur within the organelle. The mammalian mitochondrial membrane has been proposed to possess a deoxyribonucleotide carrier (DNC, encoded by the human SLC25A19 gene) (29) and a pyrimidine nucleotide carrier (PNC1, homologue of the S. cerevisiae RIM2) (30–32). Both of these might have promoted mitochondrial uptake of [α32P]dCTP and [α32P]dTTP in our assays. On the other hand, assessing the mitochondrial import with low dTTP concentrations (5 nM cf 60 nM for our experiments), Ferraro et al. (33) showed that dTTP was dephosphorylated by enzymes associated with the isolated mitochondria and that dTMP was imported by a further, highly sensitive carrier which remains to be identified. The thymidine monophosphate would then be used as a progenitor of dTTP in the organelles, possibly being sequentially phosphorylated by the recent candidate mitochondrial kinases TMPK2 (34) and NDPK (35). Which of these pathways operated in our experiments has not been defined, as it was not the aim of this report to study the import mechanisms of deoxyribonucleotides but to use these mechanisms for loading. Nevertheless, all our control assays established that the observed repair synthesis reactions occurred inside mitochondria, implying that, in the conditions we used, the organelles gained relevant levels of radiolabelled dCTP and dTTP.
In previous studies, the destination of imported DNA inside mitochondria was not clarified. We show here that, following uptake, the exogenous DNA is distributed between the membrane, i.e. the site of nucleoids and the soluble compartment. This means that, upon import, at least part of the DNA joins the physiological location of the endogenous mtDNA. Although there is no obvious influence of the DNA sequence on the mitochondrial import efficiency (18), we also demonstrate here that sequences corresponding to the NCR will preferentially drive the imported DNA to the membrane association sites. The form of these DNA interactions with the inner membrane remains to be clarified. It has been postulated for some time that the endogenous mtDNA preferentially interacts with the inner membrane through the control region (19). Attempts to identify these mtDNA interaction sites in more detail revealed a region in the NCR around the major transcription promoters, as well as two other elements outside the NCR (15). Our findings that imported DNA containing part of the NCR shows higher affinity for the membrane fraction are in good agreement with these observations. It is also intriguing to note that human mtDNA has been reported to be indirectly associated with cytoskeletal elements, which are revealed after extraction of mitochondrial membranes (36). Such interactions mediated across the inner and outer membrane have also been shown in S. cerevisiae and Trypanosoma (37,38).
As the four adjacent probes that we have derived from the NCR showed similar membrane avidities, we believe it is unlikely that we are measuring binding to a sequence-specific membrane receptor. Previous reports showing preferential interactions of imported DNA with the endogenous mtDNA when carrying NCR sequence are consistent with these data and suggest that the unusual D-loop structure of much of this region may act as the binding partner. However, we were unable to demonstrate any strand specificity that would have been in agreement with a strand displacement mode of interaction. Alternatively, it cannot be ruled out that a membrane-associated DNA anchoring protein with modest sequence selectivity could act as the receptor. ATAD3 has been postulated to interact with mtDNA in vivo and has been shown to bind to triplex sequence (39) but more recent experiments have argued against such a role (40). Thus, the exact nature of these DNA binding sites remains enigmatic but they may be essential for facilitating mtDNA transmission and replication in addition to directing mtDNA repair.
In these studies, BER occurred solely in the membrane compartment. Consistent with these findings, Stuart et al. (16) showed that most of the spBER machinery is associated with the mitochondrial inner membrane-containing particulate fraction. More specifically, the glycosylase, polymerase and ligase activities were found to be associated with the particulate fraction, with only marginal activity found freely soluble in a matrix extract. In the first instance, this looks consistent with the mtDNA itself being anchored to the membrane. However, the situation looks more complicated. First, membrane interaction of spBER enzymes seems to be independent of the mtDNA (16). Second, the AP endonuclease activity was mostly found in the matrix fraction, which is quite unexpected since AP endonuclease is important for releasing the glycosylase from the damage site and recruiting the downstream enzymes in the patch repair (41). It is even more striking when considering uracil repair, as uracil–DNA glycosylase is believed to be a monofunctional enzyme deprived of lyase activity and AP endonuclease is required to incise the AP site generated upon uracil excision. Third, localization to the membrane does not seem to mediate the formation of stable BER complexes (17). Finally, our in organello assays show that in intact mitochondria, activities relevant for base excision and single strand incision are also present in the soluble compartment. Altogether, it appears that the physiological localization of spBER reactions is at the membrane compartment but with a relaxed organization based on sequential, and in some cases transient, recruitment of the successive enzymes.
Recent work has revealed even more complexity to the overall process of mtDNA repair, as in addition to spBER, lpBER has been clearly demonstrated to occur in mammalian mitochondria (11,14). This adds a new layer of organization to the mtDNA repair process, as more enzymes need to be recruited to complete the lpBER, such as the DNA helicase/nuclease DNA2 and the endonuclease FEN1 (Flap EndoNuclease 1). The former acts in cooperation with polymerase γ to unwind the DNA and generate a single strand displaced structure or flap. This flap structure is non-ligatable. It is processed by the conjugating activity of DNA2, which cleaves in the middle of the structure, allowing FEN1 to cut at the flap junction and generate a ligatable end for DNA ligase III (13). Information on the sub-mitochondrial localization of these two proteins is scarce. However, we have performed some preliminary western blot experiments and found the majority of the mitochondrial FEN1 protein in the organelle particulate fractions (data not shown).
In summary, our DNA import and repair experiments are the first to address both the localization of the DNA and the repair reactions simultaneously with the advantage of using intact mitochondria rather than protein extracts. This has enabled us to demonstrate that canonical BER occurs on DNA that is anchored to the inner membrane, whereas nicked DNA in the matrix soluble compartment acts as a template for generic nick-directed DNA synthesis. The latter activity can cover substantially more than the lpBER range of 6–8 nt, suggesting that a very long stretch of DNA can be displaced. We believe that in our in organello experiments, DNA polymerase γ is able to perform strand displacement in both the membrane-anchored and the matrix-localized nicked DNA but in the latter case the interactions which result in recognition of the flap structure are lost.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Wellcome Trust (Ref 078900 to R.N.L.); French Centre National de la Recherche Scientifique (CNRS, UPR2357); University of Strasbourg (UdS); Agence Nationale de la Recherche (ANR, Rare Diseases programme). PhD fellowships from the Lebanese International University and the Lebanese Conseil National de la Recherche Scientifique (to N.I.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Bernard Connolly (ICAMB, Newcastle University) for assistance with RP-HPLC.
REFERENCES
- 1.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl Acad. Sci. USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic. Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 3.Yang JL, Weissman L, Bohr VA, Mattson MP. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair. 2008;7:1110–1120. doi: 10.1016/j.dnarep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 5.Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech. Ageing Dev. 2008;129:383–390. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Souza-Pinto NC, Wilson DM, III, Stevnsner TV, Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair. 2008;7:1098–1109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–1058. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 10.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999;27:3712–3719. doi: 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akbari M, Visnes T, Krokan H, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Jackson DA, Bartlett J, Cook PR. Sequences attaching loops of nuclear and mitochondrial DNA to underlying structures in human cells: the role of transcription units. Nucleic Acids Res. 1996;24:1212–1219. doi: 10.1093/nar/24.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akbari M, Otterlei M, Peña-Diaz J, Krokan H. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145:1201–1212. doi: 10.1016/j.neuroscience.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Koulintchenko M, Temperley R, Mason P, Dietrich A, Lightowlers R. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 19.Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl Acad. Sci. USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domena JD, Mosbaugh DW. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985;24:7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- 21.Slupphaug G, Markussen FH, Olsen LC, Aasland R, Aarsaether N, Bakke O, Krokan HE, Helland DE. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tell G, Crivellato E, Pines A, Paron I, Pucillo C, Manzini G, Bandiera A, Kelley MR, Di Loreto C, Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat. Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 23.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boesch P, Ibrahim N, Paulus F, Cosset A, Tarasenko V, Dietrich A. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009;37:5690–5700. doi: 10.1093/nar/gkp606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber-Lotfi F, Ibrahim N, Boesch P, Cosset A, Konstantinov Y, Lightowlers RN, Dietrich A. Developing a genetic approach to investigate the mechanism of mitochondrial competence for DNA import. Biochim. Biophys. Acta. 2009;1787:320–327. doi: 10.1016/j.bbabio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butow RA, Henke RM, Moran JV, Belcher SM, Perlman PS. Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun. Methods Enzymol. 1996;264:265–278. doi: 10.1016/s0076-6879(96)64026-9. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki T, Kurokawa S, Watanabe KI, Ikuta K, Ohama T. Shared molecular characteristics of successfully transformed mitochondrial genomes in Chlamydomonas reinhardtii. Plant Mol. Biol. 2005;58:515–527. doi: 10.1007/s11103-005-7081-3. [DOI] [PubMed] [Google Scholar]
- 29.Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE. The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc. Natl Acad. Sci. USA. 2001;98:2284–2288. doi: 10.1073/pnas.031430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 31.Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CM, Palmieri L, Scarcia P, et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim. Biophys. Acta. 2006;1757:1249–1262. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Floyd S, Favre C, Lasorsa FM, Leahy M, Trigiante G, Stroebel P, Marx A, Loughran G, O'C;allaghan K, Marobbio CM, et al. The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell. 2007;18:3545–3555. doi: 10.1091/mbc.E06-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferraro P, Nicolosi L, Bernardi P, Reichard P, Bianchi V. Mitochondrial deoxynucleotide pool sizes in mouse liver and evidence for a transport mechanism for thymidine monophosphate. Proc. Natl Acad. Sci. USA. 2006;103:18586–18591. doi: 10.1073/pnas.0609020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YL, Lin DW, Chang ZF. Identification of a putative human mitochondrial thymidine monophosphate kinase associated with monocytic/macrophage terminal differentiation. Genes Cells. 2008;13:679–689. doi: 10.1111/j.1365-2443.2008.01197.x. [DOI] [PubMed] [Google Scholar]
- 35.Milon L, Meyer P, Chiadmi M, Munier A, Johansson M, Karlsson A, Lascu I, Capeau J, Janin J, Lacombe ML. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 36.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley M, I, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 41.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.