Abstract
We investigated co-transcriptional recruitment of pre-mRNA processing factors to human genes. Capping factors associate with paused RNA pol II at the 5′ ends of quiescent genes. They also track throughout actively transcribed genes, and accumulate with paused polymerase in the 3′ flanking region. 3′ processing factors CstF and CPSF are maximally recruited 0.5-1.5 kb downstream of poly(A) sites where they coincide with capping factors, Spt5, and Ser2 hyperphosphorylated, paused pol II. 3′ end processing factors also localize at transcription start sites, and this early recruitment is enhanced after polymerase arrest with DRB. These results suggest that promoters may help specify recruitment of 3′ end processing factors. We propose a dual pausing model where elongation arrests near the transcription start site and in the 3′ flank to allow co-transcriptional processing by factors recruited to the pol II ternary complex.
Keywords: capping enzyme, CstF, CPSF, Spt5, pol II pausing, DRB, cleavage/polyadenylation, histone 3′ processing
INTRODUCTION
An intricate web of interactions between the RNA polymerase II (pol II) elongation complex and pre-mRNA processing factors ensures efficient and accurate mRNA biogenesis by coordinating maturation with synthesis of the transcript 1-4. Co-transcriptional engagement of processing factors involves recognition of consensus sites in the nascent pre-mRNA and protein-protein interactions with components of the pol II ternary complex including the C-terminal domain (CTD) 2,5 6. The pol II CTD comprises heptad repeats (YSPTSPS) that are reversibly phosphorylated at the S2 and S5 positions as polymerase traverses a gene 5,7, and these modifications have been suggested to regulate recruitment of pre-mRNA processing factors. The CTD is essential for efficient pre-mRNA processing in mammalian systems 2-4 but has only a minor role in budding yeast 8, therefore recruitment mechanisms for processing factors almost certainly differ between these species. In contrast to yeast, relatively little is known about the timing and order of recruitment of different processing factors during the pol II transcription cycle in metazoan cells though splicing factors and 3′ end processing factors have been localized at active transcription sites 9-12.
There are two human enzymes for maturation of 5′ ends: human capping enzyme (HCE), a bifunctional triphosphatase-guanylyltransferase, and RNA 7-methyltransferase (MT) 13. Capped pol II transcripts as short as 25-40 bases are made in transcription extracts and isolated nuclei suggesting that capping is a very early co-transcriptional event 14-17. Both HCE and MT can bind directly to pol II that is phosphorylated on the CTD 18 but in vitro studies suggest that these interactions with actively transcribing pol II are weak 16,17, and their in vivo significance remains unknown.
Co-transcriptional recruitment of capping enzymes may be coordinated with promoter-proximal pausing 15-17,19, a common feature of pol II transcription of mammalian, but not yeast, genes 20,21. In support of this idea, pol II pauses at the 5′ end of the DHFR gene, and HCE localizes in the same region 22. Promoter-proximal pausing is mediated by negative elongation factor (NELF) and DRB sensitivity inducing factor (DSIF) comprising Spt4 and Spt5. DRB arrests pol II elongation by inhibiting Cdk9 that phosphorylates the pol II CTD, DSIF, and NELF 20,21. Spt5 binds directly to HCE and stimulates capping 23. These observations led to the hypotheses that promoter-proximal pausing facilitates capping 17,19 or alternatively, that recruitment of capping enzyme facilitates release from the pause 24. These possibilities have yet to be investigated in detail in vivo by localizing capping enzymes relative to pol II and Spt5.
Three different processing machineries carry out 3′ end maturation of polyadenylated mRNAs, non-adenylated histone mRNAs and U snRNAs. Cleavage/polyadenylation requires cleavage stimulation factor (CstF) and cleavage polyadenylation specificity factor (CPSF) 25 that both share common subunits with the histone 3′ end processing complex, heat labile factor, (HLF) 26,27. U snRNA 3′ ends are processed by a distinct CTD binding complex 28. CstF binds to the pol II CTD through its 50kd subunit and to G/U rich sequences adjacent to the poly (A) site through its 64kD subunit. CstF77 bridges the other two subunits and contacts CPSF160, which binds the AAUAAA sequence 25. CPSF73 is an endonuclease that cleaves poly (A) sites 29 and histone 3′ ends 26. CPSF can bind to TFIID and to the body of pol II suggesting the hypothesis that CPSF binds first to the promoter via TFIID and is then transferred to pol II 30, 31. During elongation, CPSF is proposed to associate with pol II in a manner that excludes CstF and later, CstF is recruited, at a pause site following transcription of the AAUAAA element 30,32. Pausing has been detected downstream of the β-actin gene where it has been linked to transcription termination 33. It is not clear whether the phenomenon of 3′ pausing is general among mammalian genes, or how it is related to recruitment of 3′ end processing factors in vivo. While cleavage/polyadenylation factors have been detected previously on metazoan genes 10-12, the 5′-3′ distribution and order of recruitment of CPSF and CstF on genes in vivo have yet to be determined.
The basis for specific recruitment of different mRNA 3′ end processing complexes to different genes is a major unresolved question. Two possible solutions are: 1) that signals in the nascent RNA recruit the appropriate processing factors and 2) that elements in the promoter dictate recruitment of appropriate factors 31,34,35 perhaps by specifying different modifications of the pol II CTD. It is not clear whether 3′ end processing factors localize to transcription start sites in vivo, but if they do, it would lend support to the notion that promoters help to specify their recruitment. In this report, we localized pre-mRNA 5′ and 3′ end formation factors relative to pol II on human genes to determine where on the gene, and when in the transcription cycle, that they are recruited.
RESULTS
Capping enzymes co-localize with pol II paused at promoters
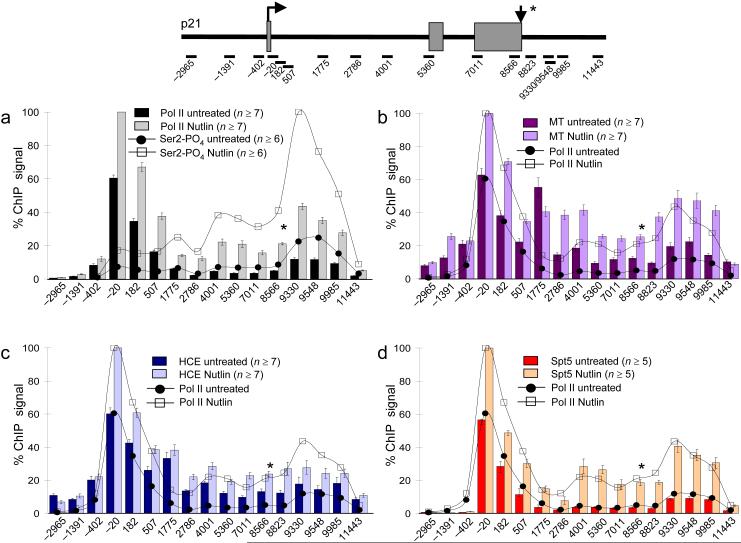
To investigate co-transcriptional recruitment of capping factors, we mapped HCE, MT, and pol II by high resolution ChIP on the human p21 gene before and after transcriptional activation. Protein occupancy was assayed at 17 positions within the p21 gene and its flanking sequences, in HCT116 cells where the gene can be activated by p53 36 (Fig. 1, map). Each position corresponds to a 50-125 bp amplicon that was quantified by real time PCR using SybrGreen fluorescence (see Methods). Prior to transcriptional activation, under conditions where p21 mRNA is very low by real-time RT-PCR, a high density of pol II is loaded at the p21 transcription start site 36 (amplicon −20, Fig. 1A) where it is precisely co-localized with MT and HCE (Fig. 1B, C). Spt5 also co-localized with HCE, MT, and pol II near the transcription start site of uninduced p21 (Fig. 1D) and c-fos genes (Supplementary Fig. 1). In sum, these results indicate that prior to activation, the p21 and c-fos genes are primed at their 5′ ends with paused pol II transcription complexes that are associated with Spt5 and capping factors.
Figure 1. Capping Enzymes localize with paused pol II at 5′ and 3′ ends and throughout the p21 gene.
Diagram of the p21 gene with the center of real-time PCR products marked relative to the transcription start site as in ref. 36. Relative ChIP signals in HCT116 cells plotted for A) total pol II and CTD Ser2-PO4, B) cap methyltransferase (MT) C) human capping enzyme (HCE) and D) Spt5 before (untreated, dark bars) and after activation by Nutlin-3a (pale bars). Down arrow and * indicates poly(A) signal at +8570. Flanking primers −2965 and +11443 serve as intergenic background controls. Pol II traces from A are included in B-D.
Values are normalized to the maximal value in the Nutlin-3a data set. For reasons we do not understand, the density of capping enzymes relative to pol II and Spt5 was reproducibly higher at amplicon +1775 in intron 1 than at other positions. Mean values from n PCR reactions (n) with standard errors of mean (SEM) are shown.
Capping enzymes and paused pol II at the 5′ and 3′ ends of activated p21
When p21 transcription was induced by activating p53 with the HDM2 inhibitor, Nutlin-3a, pol II and Spt5 occupancy increased at the transcription start site and throughout the length of the gene but not in the intergenic control regions (Fig. 1). Unexpectedly, we consistently observed coincident peaks of pol II and Spt5 density at amplicons +9330, +9548, and +9985 located 1-1.5 kb downstream of the poly (A) site at +8570 (see Fig. 1A, D). Pol II and Spt5 occupancy fell off between +9985 and +11443 as transcription terminated. Spt5 (DSIF) therefore appears to be a constitutive component of pol II transcription complexes at initiation, elongation and termination phases of the transcription cycle.
In summary, ChIP revealed a distribution of pol II and Spt5 with twin peaks at the p21 transcription start site and well downstream of the poly (A) site. This distinctive distribution was specific to pol II and Spt5 and was not observed for TFIIB or trimethylated histone H3K4 that display peaks near the 5′ end, but not in the 3′ flanking region (Figs. 2A, Supplementary Fig. 2A). The peak of pol II downstream of p21 coincided with a valley in histone H3 occupancy, consistent with histone displacement associated with high pol II density 37 (Fig. 2A). We interpret this 3′ peak of pol II that precedes termination as the result of pausing. Remarkably, the highest level of CTD S2 phosphorylated pol II on p21 occurred at this 3′ pause site located about 1 kb downstream of the poly (A) site (Fig. 1A).
Figure 2.
A) Pol II pausing and histone localization at 5′ and 3′ ends of p21. Pol II, Spt5, histone H3 and TFIIB localization by ChIP on the Nutlin-3a activated p21 gene. Displacement of H3 occurs at sites of pol II accumulation at the 5′ and 3′ ends. Note that unlike pol II and Spt5, TFIIB peaks near the 5′ end but not at the 3′ end. B) DRB inhibits pausing and accumulation of MT downstream of the p21 poly (A) site. ChIP of pol II and MT after induction by Nutlin-3a or DRB normalized to the maximum value in each data set to emphasize differences in 5′-3′ distribution. Mean values from n PCR reactions with standard errors of mean (SEM) are shown.
When p21 was activated, HCE and MT occupancy increased with pol II not only at the 5′ end but also at positions within the gene (Fig 1B, C). Consistent with early capping of nascent transcripts, the cap binding complex, CBC, was also present at the 5′ end of activated p21 (Supplementary Fig. 2B) similar to previous results at c-fos 9. HCE and MT levels normalized to pol II, remain relatively high throughout the gene and 3′ flank (Supplementary Table 1a) demonstrating that their association with pol II complexes persists during elongation and termination. Although methylation is delayed relative to guanylation in coupled transcription and capping reactions in vitro 16,17, we did not observe any delay in recruitment of MT relative to HCE. Remarkably, MT, and to a lesser extent HCE, also accumulated with paused pol II and Spt5 downstream of the poly (A) site (Fig. 1 B, C).
Unlike most genes, p21 is activated by the elongation inhibitor DRB (Fig. 2B) resulting in transcription by pol II that is hypophosphorylated on CTD S2 (Supplementary Table 1a) 36. We asked whether DRB activation altered the association of capping factors with the p21 gene. Relative to Nutlin-3a, DRB suppressed the 3′ pause and this loss of pol II from the 3′ flanking region correlated with reduced MT (Fig. 2B) and Spt5 (Supplementary Fig. 2C) occupancy. Despite this inhibition of pausing, DRB did not prevent termination as evidenced by the loss of pol II between positions +9330 and +11443 (Fig. 2B). In summary, the experiments in Figures 1 and 2 show that capping factors track with pol II and Spt5 in transcription complexes that are paused at 5′ and 3′ ends, as well as those that are elongating through the gene. They therefore appear to remain in association with pol II long after addition of the cap which is expected to occur within the first 50 bases 15.
5′ and 3′ pol II pausing with capping factors on multiple genes
To determine whether the twin peaks of pol II and capping factors at the 5′ and 3′ ends of p21 are common to other genes, we also analyzed c-Myc and GAPDH (Fig. 3). The maximum occupancy of capping factors on c-Myc and GAPDH coincided with the peaks of pol II and Spt5 at the transcription start sites, as previously reported for DHFR 22, but they were also detected at low levels within the genes (Fig. 3B, D). Downstream of the poly (A) sites, MT density increased coincident with paused pol II that is highly phosphorylated on CTD S2 residues (Figs. 3A, C). Similar results were obtained in HCT116 and Hela cells (Supplementary Fig. 3A). Likewise, increased HCE density was detected downstream of c-Myc, but not GAPDH (Fig. 3B, D). The absence of HCE from the 3′ end of GAPDH may be due to a gene-specific effect of the sequence of the poly (A) site or 3′ flanking region. Together these results suggest that a quite general feature of human genes is that elongation is punctuated by dual pauses at the transcription start site and in the 3′ flanking region where pol II accumulates in association with Spt5 and one or both capping factors.
Figure 3. Capping Enzymes track with pol II on c-Myc and GAPDH genes.
A) Positions of real-time PCR products are indicated relative to the c-Myc P2 start site. Relative ChIP signals are shown for total pol II, CTD Ser2-PO4, and Spt5 B) MT and HCE on c-Myc in HCT116 cells. Traces of pol II and Spt5 from A serve as reference. C) total pol II, CTD Ser2-PO4, and Spt5 on GAPDH. D) MT and HCE on GAPDH. Traces of pol II and Spt5 from C serve as reference. * and down arrow indicate poly(A) signals at +5166 relative to the c-Myc P2 transcription start site, and +3830 for GAPDH. Mean values from n PCR reactions with standard errors of mean (SEM) are shown..
The co-localization of HCE and MT with paused pol II at 5′ ends of uninduced p21 and c-fos (Figs. 1, Supplementary Fig. 1) suggests that they may associate with polymerases that are not engaged in active elongation. We investigated this idea further by asking whether capping factors localize with pol II that has been arrested by DRB. As expected, inhibition of Cdk9 with DRB increased pol II occupancy at the transcription start site and decreased the CTD phospho-S2 signal at the 3′ end of GAPDH (data not shown) but it did not entirely eliminate pol II from positions near the 3′ end of the gene (Fig. 4A) consistent with previous results 38. Importantly, both capping factors accumulated with DRB-arrested pol II at the transcription start site, suggesting that pol II complexes which are not actively transcribing, are still associated with capping factors (Figs. 4A, B).
Figure 4.
A, B. Association of capping factors with DRB-arrested pol II complexes on GAPDH. Positions of real-time PCR products are indicated. Relative ChIP signals are shown for pol II, MT and HCE on the GAPDH gene in untreated and DRB treated HCT116 cells.
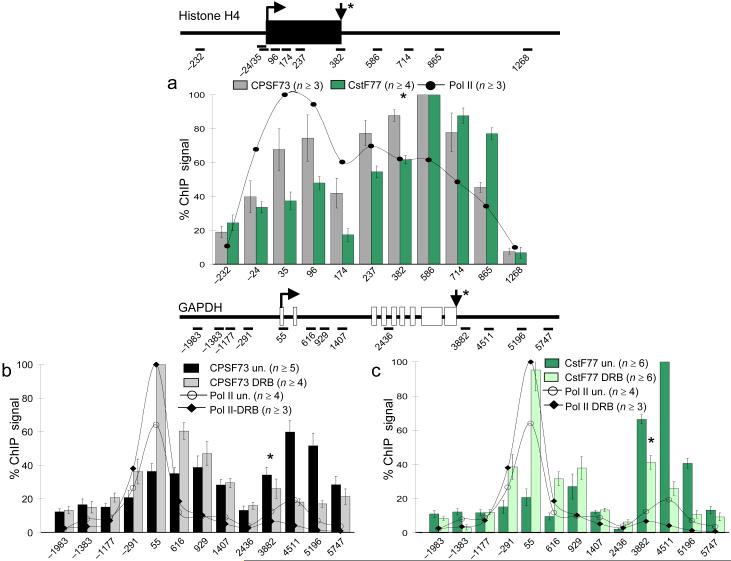
C, D. Localization of capping factors, pol II and Spt5 on histone H4 and U2 snRNA genes. C) pol II, Spt5 MT and HCE on histone H4/d (Accession no. X60483). D) pol II, Spt5, MT, HCE and CBC on U2 snRNA (Accession no. U57614). CBC data is the sum of 4 determinations each with anti-CBC20 and 80. Mature RNA 3′ ends (down arrow and *) map to +387 for H4/d and +190 for U2. Mean values from n PCR reactions with standard errors of mean (SEM) are shown.
Capping factors and Spt5 at the Histone H4 and U2 snRNA genes
Histone mRNAs and U snRNAs are among the shortest pol II transcripts, and it is not known whether they are capped co-transcriptionally or post-transcriptionally. We found that capping factors and Spt5 were co-transcriptionally recruited to the histone H4/d and U2 snRNA genes at levels comparable to poly (A)+ mRNA coding genes after normalizing to pol II (Supplementary Table 1b). On H4/d, HCE, MT, and Spt5 peaked near the 5′ end and persisted with pol II through the ORF and into the 3′ flanking region (Fig. 4C) similar to genes for poly (A)+ mRNAs.
On U2, in contrast to other genes, the 5′-3′ profiles of capping factors and Spt5 did not overlap precisely with pol II. Pol II density remained approximately constant across the U2 gene with no evidence of promoter-proximal pausing (Fig. 4D +1, +87, +219). In contrast, HCE, MT, Spt5 and CBC levels rose between the beginning and the end of the gene (Fig. 4D) peaking at +219, close to the 3′ box. In summary, recruitment of Spt5, capping factors and CBC to the U2 gene is delayed relative to pol II. These results are consistent with co-transcriptional capping of U2 and furthermore suggest that capping factors and Spt5 are coordinately recruited 23 without detectable promoter-proximal pausing. The apparent lack of pausing is consistent with our finding that recruitment of NELF-A, relative to pol II, is approximately 5 times less at U2 than at histone H4/d (data not shown).
Co-localization of 3′ processing factors and pol II at 5′ and 3′ ends
To investigate the timing and order of recruitment of cleavage/polyadenylation factors, we localized CstF and CPSF by ChIP with anti-CstF77 and -CPSF73 antibodies. Peaks of CstF and CPSF recruitment exactly coincided with one another and with paused pol II that accumulated 0.5-1.5 kb downstream of the c-Myc and GAPDH poly (A) sites (Fig. 5A, B). These factors were not detected on the U2 snRNA gene (Supplementary Fig. 3B). High levels of CPSF were also recruited independently of CstF near the transcription initiation sites, and within the c-Myc and GAPDH genes (Fig. 5A, B). These results show that CPSF recruitment starts early in the transcription cycle, probably close to the time of initiation. Most CstF recruitment, on the other hand, occurs late, after synthesis of the poly (A) site .
Figure 5. Localization of 3′ end processing factors at 5′ and 3′ ends of p21, c-Myc and GAPDH.
Positions of real-time PCR products on each gene are indicated. Relative ChIP signals in HCT116 cells are shown for A) CPSF73 and CstF77 on c-Myc and B) GAPDH. C) CstF77 and D) CPSF73 on untreated and Nutlin-3a activated p21 normalized to the maximum value for the Nutlin-3a data set. * and down arrow indicate poly(A) sites. Mean values from n PCR reactions with standard errors of mean (SEM) are shown.
We also examined recruitment of CstF and CPSF to the p21 gene before and after transcriptional activation by Nutlin3a (Fig. 5c, d). After activation, CstF77 was elevated 3-4 fold in the 3′ flank where recruitment was maximal. Low levels were also detectable within the gene and at the transcription start site (Fig. 5c). Unlike CstF77, CPSF 73 and CstF64 accumulated near the start site of the uninduced p21 gene (Fig. 5D, Supplementary Fig. 4a). Transcription activation had little effect on CPSF at the start site, but enhanced its recruitment downstream of the poly (A) site, coincident with paused pol II and CstF (Fig. 5D). Notablyly, the binding of CPSF73 and CstF64 near the 5′ end of p21 resembles that of TBP, which is also mostly recruited before activation 36. In summary, the results in Figure 5 and Supplementary Fig. 4a show that CPSF73 and CstF64 can be recruited to a 5′ end independently of CstF77 (see Discussion). Furthermore CPSF can associate with transcription complexes both at the 3′ pause downstream of poly (A) sites and close to the transcription start site. Similarly, the CPSF73 and CstF77 subunits of the histone 3′ end processing complex, HLF, were recruited near the 5′ end of the H4/d gene and persisted throughout its length with maximal accumulation about 200 bases downstream of the 3′ cleavage site (Fig. 6A). Sm proteins, presumably associated with snRNP’s, such as U7, were also localized throughout the H4/d gene Supplementary Fig. 4).
Figure 6.
A) Co-transcriptional recruitment of 3′ end processing factors to a histone gene. ChIP of CPSF73 and CstF77 subunits of HLF and pol II on histone H4/d in HCT116 cells. Down arrow and * indicates mRNA 3′ end. B, C) DRB enhances recruitment of 3′ end processing factors at the 5′ end of GAPDH. Relative ChIP signals are shown for B) CPSF73 and C) CstF77 on GAPDH untreated (un) and after DRB treatment. Pol II and CstF77 profiles for untreated cells are the same as Fig. 5B. The untreated CPSF73 data is a subset of that in Fig. 5B. Data in B are normalized to the maximum value for the DRB sample at +55, and in C they are normalized to the maximum value for the untreated sample at +4511. Mean values from n PCR reactions with standard errors of mean (SEM) are shown.
Inhibitors of Cdk9 cause elongational arrest and can disrupt cleavage/polyadenylation 38,39, but it is not clear how they affect recruitment of 3′ end processing factors. We investigated how DRB affects pol II, CstF and CPSF localization. DRB enhanced the 5′ pause and suppressed the 3′ pause on GAPDH (Fig. 6B), as we observed on p21 (Fig. 2B). DRB reduced CstF and CPSF levels downstream of the GAPDH, p21, and c-Myc poly (A) sites (Fig. 6B, C, Supplementary Fig. 5A, B, Supplementary Table 2). It also reduced HLF levels downstream of the histone H4/d gene (Supplementary Fig. 5C, D). Most of this suppression can be attributed to reduced pol II occupancy, but we did detect an additional reduction in CstF77 and CPSF73 relative to pol II, downstream of p21, c-Myc and H4/d (Supplementary Table 2). The most remarkable effect of DRB is that it elevated CPSF and CstF binding to the 5′ ends of GAPDH, p21 and c-Myc relative to pol II (Fig. 6B, C, Supplementary Table 2). This result therefore implicates Cdk9 phosphorylation in controlling the recruitment of 3′ end processing factors at 5′ ends.
DISCUSSION
Co-localization of capping factors with pol II at 5′ and 3′ ends
Pre-mRNA processing factors can, in principle, be recruited to transcribed genes in two ways that are not mutually exclusive: a) by binding to processing signals in nascent pre-mRNA and b) by binding to a protein “landing pad” in the pol II elongation complex. Our ChIP analysis demonstrates extensive co-localization of 5′ and 3′ end processing factors with pol II even at positions quite distant from where capping and cleavage/polyadenylation take place. Capping enzymes were situated not only at the 5′ end where capping is thought to occur 15, but also throughout the gene and in 3′ flanking regions more than a kilobase downstream of poly (A) sites (Figs. 1, 3). In other words capping factors appear to linger on the pol II “landing pad” long after addition of the cap. This association could be due to stable binding initiated at the 5′ end, or dynamic exchange throughout the length of the gene. The function of these factors within genes and in their 3′ flanking regions is unclear, but they are appropriately situated to influence elongation, termination, and 3′ end maturation. There are several precedents for viral and cellular capping enzymes influencing transcription initiation, elongation and termination 24,40,41. It is also of interest that HCE promoted poly (A) site cleavage in a transcription-coupled in vitro system 42. The recruitment patterns we observed for human capping enzymes differ from their yeast counterparts. Yeast Ceg1 and Abd1 localize at 5′ ends, and Abd1 is also found within genes, but neither has been detected in 3′ flanking regions 7,43.
Human capping factors co-localized with pol II poised at promoters, before transcriptional activation and after inhibiting elongation with DRB (Figs. 1B, C, 4A, B). Association of capping factors with pol II is therefore not sufficient to trigger release from a 5′ pause. Conversely, our results suggest the possibility that capping factors may collaborate with negative elongation factors, Spt4/5 and NELF, in mediating promoter-proximal pausing. Pol II pausing may not be a universal pre-requisite for co-transcriptional capping however, because capping factors and CBC were efficiently recruited to the U2 snRNA gene (Fig. 4D, Supplementary Table 1b) where we did not detect pausing. Maximal capping factor and CBC density on U2 mapped close to the 3′ box coincident with a peak of Spt5 (Fig. 4D). This result is therefore consistent with the idea that Spt5 facilitates recruitment of capping factors 23,24. Such a mechanism could also aid capping factor recruitment before the CTD has been extensively phosphorylated such as at the 5′ ends of uninduced genes (Figs. 1, Supplementary Fig. 1).
Recruitment of 3′ processing factors at 3′ and 5′ ends
Major peaks of accumulation of CstF and CPSF coincide with pol II paused 0.5-1.5kb downstream of poly (A) sites (Fig. 5). Similarly, HLF occupancy peaked downstream of the histone H4/d gene (Fig. 6A). This 3′ recruitment probably involves recognition of the processing sites in the nascent transcript. 3′ end processing factors also localized far upstream of their cognate processing sites at positions within genes and at 5′ ends. This 5′ recruitment in anticipation of transcription of processing sites, suggests an additional recruitment mechanism distinct from RNA recognition; namely protein:protein binding to pol II transcription complexes. 3′ End processing factors therefore appear to differ from splicing factors whose recruitment seems to require transcription of consensus splice sites 44.
CPSF73 and CstF64 localize close to transcription start site suggesting that their initial recruitment is linked to initiation (Fig. 5A, Supplementary Fig. 4A, ref. 10). In contrast, high levels of CstF77 were only detected downstream of the c-myc, GAPDH and p21 poly (A) sites (Figs. 5A, B) suggesting that it is recruited later. On p21, most CPSF73 and CstF64 recruitment preceded transcriptional activation and CstF77 appeared to be recruited only after the gene was activated (Figs. 5C, D, Supplementary Fig. 4A). . The low level of CstF77 at the p21 transcription start site is reproducibly elevated in response to activation by Nutlin-3a or DRB (Fig. 5C, Supplementary Fig. 5B). These results suggest the possibility that the trimeric CstF complex may assemble during transcription with recruitment of CstF64 before CstF77.
When elongation was inhibited by DRB, the normal order of recruitment of cleavage/polyadenylation factors was disrupted. In DRB, CstF was recruited early, along with CPSF, near the 5′ end (Figs. 6B, C). DRB-arrested pol II complexes at 5′ ends may therefore adopt a conformation that stabilizes interactions with both these 3′ end processing factors. DRB could also enhance the accumulation of 3′ processing factors at 5′ ends by influencing the proposed handoff of CPSF from TFIID to pol II (Fig. 7)31.
Figure 7. Dual pausing model for co-transcriptional 5′ and 3′ end maturation of polyadenylated mRNAs.
This model is based on data reported here and in references 30,31.
Two lines of evidence suggest that promoter elements may specify the recruitment of 3′end processing factors to 5′ ends independent of the RNA signals that they recognize. First, a U snRNA promoter, which is recognized by a distinct TBP-containing complex, is important for U snRNA 3′ end processing 23,24 by the CTD-bound integrator complex 36. Second, Dantonel et al 31 suggested that CPSF binds initially with TFIID to the promoter of mRNA genes followed by hand off to pol II with which it travels into the gene (Fig. 7). Our results strongly support this model by showing that CPSF73 is recruited close to transcription start sites and persists throughout the length of genes (Figs. 5, 6). 3′ end processing factors also localized near the 5′ end of the histone H4/d gene (Figs. 6A, Supplementary Table 2). Together our results imply that promoter elements could play a general role in specifying the recruitment of 3′ end processing factors.
Pausing, termination, and recruitment of 3′ end processing factors
Transcriptional pausing has been implicated both in enhancing cleavage/polyadenylation and as a prelude to termination 33,45 46 but the relationship between pausing, 3′ end formation, and termination, is still unclear. Interestingly Spt5, which has negative elongation properties, co-localized with pol II at 3′ pause sites and during subsequent termination (Figs. 1, 3, Supplementary Fig. 2C). A major conclusion of this study is that cleavage/polyadenylation factors are maximally recruited at the 3′ pause 0.5-1.5 kb downstream of poly (A) sites. This downstream pause detected by ChIP may correspond to the pause in live cells detected by FRAP in the 3′ portion of a reporter gene 47. The 3′ pause was normally followed by termination (Figs. 1, 3, Supplementary Fig. 2C) but termination could be uncoupled from pausing by DRB (Figs. 2B, 6B).
The 3′ pause coincides with maximal CTD S2 phosphorylation (Figs. 1A, 3A, 3C) and is suppressed by DRB, an inhibitor of S2 phophorylation (Figs. 2B, 6B, Supplementary Table 1a). In contrast, the 5′ pause occurs when pol II is hypophosphorylated on S2 and is enhanced by DRB. CTD S2 phosphorylation is usually regarded as a signature of elongating pol II within a gene 5, 7 however our results suggest a role for this modification well downstream of the gene where 3′ processing factors are recruited. How DRB inhibits cleavage/polyadenylation 38,39 is not completely resolved, but it correlates with both reduced recruitment of CstF and CPSF in 3′ flanking regions and inhibition of 3′ pausing (Figs. 6B, C, Supplementary Table 2).
The torpedo model for coupling termination with 3′ end processing stipulates that RNA cleavage at the poly(A) site must precede termination to permit entry of a 5′-3′ RNA exonuclease that degrades the transcript and facilitates polymerase release 48. Whether poly(A) site cleavage normally occurs co-transcriptionally or post-transcriptionally is unclear 1,49. Our demonstration that the endonuclease, CPSF73, localizes on pol II complexes paused downstream of the poly(A) site indicates that the necessary factors are in place to carry out co-transcriptional cleavage prior to termination, as required by the torpedo model.
A dual pausing model of co-transcriptional pre-mRNA processing
We propose a dual pausing model for coordination of pol II transcription with pre-mRNA processing in metazoans (Fig. 7). In this model, 5′ and 3′ transcriptional pause sites mark where recruitment of capping and cleavage/polyadenylation factors occurs, and probably also where 5′ and 3′ end maturation of the nascent transcript takes place. HCE and MT were both recruited at 5′ pause sites (Figs. 1, 3). This observation therefore supports in vitro data suggesting a link between co-transcriptional capping and promoter-proximal pausing 17,24. We also suggest that co-transcriptional poly(A) site cleavage takes place in the context of a pol II complex at the 3′ pause site where maximal recruitment of CstF and CPSF occurs (Fig. 5), approximately 1kb downstream of the poly(A) site. Localization of the active pol II-3′ end processing complex well downstream of the poly(A) consensus sequence, agrees well with the cleavage of Chironomus BR1 transcripts after 600 bases of downstream sequence has been transcribed 1. This model is also supported by the fact that efficient poly(A) site cleavage requires an intact RNA tether linking the poly(A) site with the downstream polymerase 50,51.
The co-localization of capping and cleavage/polyadenylation factors with paused pol II at both ends of human genes is quite distinct from the recruitment patterns of homologous processing factors in budding yeast. We speculate that during evolution, co-transcriptional recruitment of processing factors has adapted to altered patterns of pol II transcription including the punctuation of the transcription cycle in metazoans by prominent pause sites.
METHODS
Antibodies
Rabbit anti-pan CTD against pol II, anti-CstF77, anti-histone H3 and anti - H3K4me3 have been described (see Supplemental Methods). Rabbit antibodies were raised against His-tagged mouse capping enzyme MCE (211-597), mouse MT (141-465), human Spt5 (703-1087) and full-length human GST-TFIIB. Rabbit anti-human CPSF73 was raised against the N-terminal peptide NH2-MSAIPAEESDQLLIRPLGAGQES-COOH and affinity purified. Rabbit anti-CTD S2-PO4 was raised against a phosphorylated peptide with two heptad repeats. Anti-Sm and -CBC antibodies were gifts from T. Blumenthal and I. Mattaj.
ChIP
HCT116 human colon cancer cells (p53 positive) were treated with DRB (50μM, Sigma) or Nutlin-3a (10μM, Calbiochem) for 8hrs before ChIP as described previously 36 (Supplemental Methods). In all cases data from side-by-side cultures + and − DRB are shown. High resolution was achieved by shearing chromatin into approximately 250 bp fragments.
Real-time PCR
PCR reactions were performed with SybrGreen using the Roche LC-480 (Roche Applied Science) as described in Supplemental Methods. Primer sequences for p21 have been described 36 and additional primers are in Supplementary Table S3. To generate 5′-3′ profiles of occupancy, ChIP values were normalized relative to the amplicon with the highest fluorescence value for each gene. Where untreated and Nutlin-3a or DRB treated samples were compared, the highest DNA value between the two sets of data was used as the normalization point. Note where two peaks of approximately equal ChIP signals were detected, neither is exactly 100% after normalization and taking the mean of multiple experiments. Points were then averaged and the standard error of mean (SEM) was calculated for each primer set. Each PCR determination was made on an independent plate relative to a standard curve on the same plate. n values refer to the number of PCR determinations from at least 3 independent IP’s.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH grants GM063873 and GM58613 to D.B. and CA117907 to J.E. K. G-C was supported by NIH fellowship 5F31 GM072099 and S. K by F32 GM076951. We thank I. Mattaj (EMBL, Heidelberg) and T. Blumenthal (U. Colorado, Boulder) for antibodies. We also thank T. Blumenthal, J. Jaehning, R. Davis, T. Evans, N. Gomes, G. Bjerke. G. Bilousova, and members of the Bentley and Espinosa labs for helpful suggestions.
Contributor Information
Kira Glover-Cutter, Dept. Biochemistry and Molecular Genetics, University of Colorado School of Medicine, UCHSC, MS8101, P.O. Box 6511, Aurora CO. 80045.
Soojin Kim, Dept. Biochemistry and Molecular Genetics, University of Colorado School of Medicine, UCHSC, MS8101, P.O. Box 6511, Aurora CO. 80045.
David L. Bentley, Dept. Biochemistry and Molecular Genetics, University of Colorado School of Medicine, UCHSC, MS8101, P.O. Box 6511, Aurora CO. 80045.
REFERENCES
- 1.Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–69. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 4.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 5.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 6.Greenleaf AL. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–9. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- 7.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 9.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 10.Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16:912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci U S A. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen EB, Lis JT. In-vivo transcriptional pausing and cap formation on 3 drosophila heat-shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Mol Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 17.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell. 2002;10:585–97. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 18.Shuman S. Origins of mRNA Identity: Capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 20.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 22.Cheng C, Sharp PA. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol Cell Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominski Z, Yang XC, Marzluff WF. The Polyadenylation Factor CPSF-73 Is Involved in Histone-Pre-mRNA Processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nag A, Narsinh K, Martinson HG. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14:662–669. doi: 10.1038/nsmb1253. [DOI] [PubMed] [Google Scholar]
- 31.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 32.Nag A, Narsinh K, Kazerouninia A, Martinson HG. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA. 2006;12:1534–1544. doi: 10.1261/rna.103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3896. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuman de Vegvar H, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- 36.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 38.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 39.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vos JC, Sasker M, Stunnenberg HG. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagler J, Luo Y, Shuman S. Factor-dependent transcription termination by vaccinia RNA polymerase. Kinetic coupling and requirement for ATP hydrolysis. J Biol Chem. 1994;269:10050–10060. [PubMed] [Google Scholar]
- 42.Adamson TE, Shutt DC, Price DH. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J Biol Chem. 2005;280:32262–32271. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- 43.Schroeder S, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 46.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 49.Osheim YN, Proudfoot NJ, Beyer AL. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- 50.Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 51.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme Cleavage Reveals Connections between mRNA Release from the Site of Transcription and Pre-mRNA Processing. Mol Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.