Abstract
Metal coordination is a key structural and functional component of a large fraction of proteins. Given this dual role we considered the possibility that metal coordination may have played a templating role in the early evolution of protein folds and complexes. We describe here a rational design approach, Metal Templated Interface Redesign (MeTIR), that mimics the time course of a hypothetical evolutionary pathway for the formation of stable protein assemblies through an initial metal coordination event. Using a folded monomeric protein, cytochrome cb562, as a building block we show that its non-self-associating surface can be made self-associating through a minimal number of mutations that enable Zn coordination. The protein interfaces in the resulting Zn-directed, D2-symmetrical tetramer are subsequently redesigned, yielding unique protein architectures that self-assemble in the presence or absence of metals. Aside from its evolutionary implications, MeTIR provides a route to engineer de novo protein interfaces and metal coordination environments that can be tuned through the extensive noncovalent bonding interactions in these interfaces.
Keywords: evolution, metal coordination, protein self-assembly, protein–protein interactions, templating
Early precursors to modern single- or multidomain proteins likely emerged from intra- or intermolecular association reactions between random peptide/protein surfaces (1, 2). Only those surfaces that provided sufficient interaction enthalpy to overcome the entropic cost of association were presumably subjected to natural selection to yield stable and functional protein folds or complexes (3). Because such association surfaces comprise weak noncovalent bonds they must have been large enough to generate a sufficiently favorable binding enthalpy, which has a very low probability of occurrence by chance. In fact, it has been estimated that the likelihood of finding a foldable sequence in a random library of 80 amino-acid-long peptides that can fulfill the most basic function—binding a small molecule—is about 1 in 1011 (4).
In light of such low odds, metal coordination could have provided distinct evolutionary benefits in the emergence of folded and functional protein domains and complexes. From a structural perspective metal coordination is arguably the most efficient way to generate a folded domain while minimizing the sequence space required. Perhaps not coincidentally, Szostak and colleagues found that the best ATP-binding protein selected from a library of 6 × 1012 random peptide sequences contained a Zn coordination motif that was necessary for the structural organization of the ATP-binding site (4). Similarly, Riechmann and Winter looked for chimeras of peptide segments that could bind antilysozyme antibodies (5). They discovered that the most efficient binder incorporated an unforeseen heme group that dramatically stabilized the resulting dimer. From a functional perspective metals possess properties such as Lewis acidity and redox reactivity that enable them to carry out catalytic transformations not accessible by organic building blocks.
Given such immediate structural and functional benefits it is reasonable to ask whether metal coordination may have played a role in the early evolution of protein folds and complexes through an initial nucleation event. Motivated by this possibility we developed a rational design approach, MeTIR, which mimics the time course of a hypothetical evolutionary pathway. We show that through MeTIR, noninteracting protein surfaces can first be rendered self-associating through an initial metal-templating event followed by the generation of stability, both through a small number of mutations on the protein surface. MeTIR yields unique protein architectures that stably self-assemble in the presence—or absence—of metals, thus providing a potential route for the de novo generation of protein interfaces and tunable metal coordination centers within biological scaffolds*.
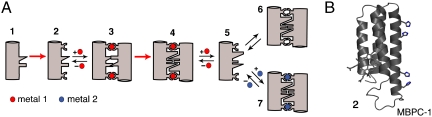
The underlying “template-and-stabilize” strategy for MeTIR is illustrated in Fig. 1. The key to MeTIR is the use of a folded, physiologically monomeric protein—instead of random polypeptide sequences or protein fragments—as a model system subject to optimization. The stability and rigidity of a folded protein ensure that its overall architecture is retained as its surface is modified and that it remains structurally tractable by crystallography. At the same time, using a monomeric protein ensures that its surface does not carry any bias towards self-association. With these in mind, we employed cytochrome cb562 (cyt cb562), a four-helix bundle heme protein, as a model system. Cyt cb562 (illustrated as Species 1 in Fig. 1) is a variant of the putative electron transfer protein cyt b562, which features an engineered covalent linkage between the heme cofactor and the polypeptide backbone (6). This linkage significantly increases the protein stability and prevents the dissociation of the heme. Importantly, cyt cb562 features a highly polar, non-self-associating surface and remains monomeric even at millimolar concentrations.
Fig. 1.
(A) Cartoon outline for MeTIR and the species involved. Red arrows represent engineering or hypothetical evolutionary steps. Red and blue spheres represent metal ions 1 and 2 with different preferential coordination geometries. 1. Protein/peptide with a non-self-associating surface; 2. 1 modified with metal-coordinating groups; 3. initial metal1-templated protein/peptide complex with noncomplementary interfaces; 4. metal1-templated protein/peptide complex with optimized, complementary interfaces; 5. protein/peptide with a self-associating surface; 6. metal-independent protein/peptide complex biased towards metal1 binding; 7. protein/peptide complex with distorted metal2 coordination. (B) The structure of MBPC-1, corresponding to species 2, showing the two i, i + 4 diHis motifs on helix3.
Results and Discussion
Metal-Driven Nucleation.
In order to nucleate protein–protein interactions through metal coordination, two sets of i, i + 4 dihistidine (diHis) motifs were incorporated near each end of cyt cb562 Helix3; the resulting variant was designated Metal Binding Protein Cytochrome-1 (MBPC-1) (Fig. 1B, Species 2 in Fig. 1A) (7, 8). Such i, i + 4 diHis motifs have been used for the assembly and stabilization of metalloproteins and peptides through coordination of divalent transition metal ions (9–12). We have previously shown that MBPC-1 self-assembles into discrete oligomeric structures upon metal coordination by the diHis motifs. Given the non-self-associating surface of cyt cb562, the oligomerization of MBPC-1 is entirely driven by metal coordination. Consequently, the supramolecular arrangement of MBPC-1 is dictated by the inner-sphere metal coordination geometry: the octahedral and the square-planar coordination preferences of Ni(II) and Cu(II) respectively lead to a C3-symmetrical trimer and a C2-symmetrical dimer (Fig. S1) (13), whereas tetrahedral Zn(II) coordination yields a D2-symmetrical tetramer (Fig. 2) (7). These three structures are collectively illustrated as Species 3 (Fig. 1A), whose interfaces lack complementary noncovalent interactions that would typically be expected to drive protein–protein association. According to our hypothetical evolutionary pathway, Species 3 therefore represents the initial peptide or protein ensemble that has assembled around a metal ion, a process that has required a minimal number of mutations in the amino acid sequence.
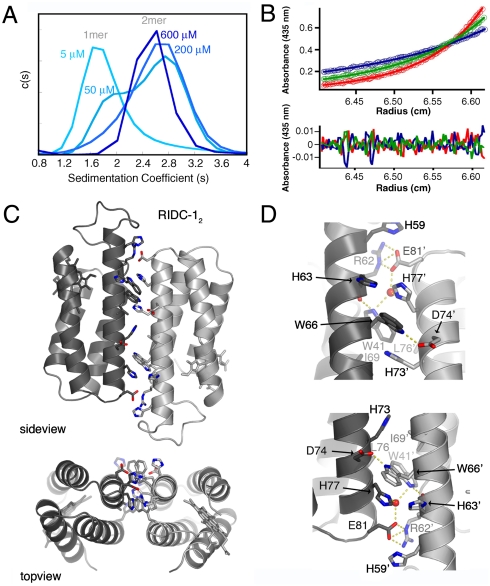
Fig. 2.
Three pairs of interfaces (i1, i2, i3) formed within the D2-symmetrical Zn4:MBPC-14 tetramer; the Zn coordination environment in each interface is listed below.
Analysis of Protein Interfaces.
The Ni- and Cu-driven MBPC-1 complexes feature small (< 1000 Å2) interfaces. The Zn-driven tetramer (Zn4:MBPC-14), in contrast, provides an extensive buried surface (∼5000 Å2) with intimate contacts among its four subunits. Hence, Zn4:MBPC-14 was chosen as the focus of surface redesign efforts to generate a stable noncovalent bonding network around the metal centers. Owing to its D2 (222) symmetry, Zn4:MBPC-14 can be dissected into three pairs of C2- symmetrical interaction surfaces (Fig. 2). As seen in most natural D2-symmetrical protein complexes (14), two of the three interfaces (i1 and i2) in Zn4:MBPC-14 are significantly more extensive than the third (i3), which is almost entirely made up by metal-coordinating residues. Each interface is held together by one or two Zn ions using alternative combinations of four coordinating residues (the engineered His73 and His77, and the native His 63 and Asp74). Importantly, the third engineered residue, His59, was found not to be involved in Zn coordination, meaning that the total number of required mutations to obtain a tetrameric complex is only two. Together the three pairs of interfaces make an interwoven assembly linked together by four Zn ions.
From a redesign perspective i1, i2, and i3 can be considered as independent targets since the amino acid side chains that make up their cores do not overlap. However, because the three interfaces constitute a cooperative assembly, their contributions to the stability of the entire Zn-mediated tetrameric architecture should be synergistic. The buried surface areas in i1, i2, and i3 in the Zn4:MBPC-14 complex are 1080, 870, and 490 Å2, respectively and comprised almost entirely of polar side chains that are forced together by Zn coordination. Given its small size we did not pursue the redesign of i3 and first sought to optimize the largest interface i1 followed by i2.
General Redesign Strategy.
To achieve a successful redesign of interfaces we developed a strategy that explicitly addressed a) preserving the fold of the cyt cb562 monomer and b) mutating the minimal number of residues that might have the maximal impact on binding affinity in the Zn4:MBPC-14 complex [see Supporting Information (SI) text for details on redesign strategy]. For the first goal, we analyzed the structures and flagged as “undesignable’’ all sequence positions in which the residues contacted the heme groups or Zn atoms (in Zn4:MBPC-14), residues that had low solvent accessible surface areas (SASA), and residues that were involved in side chain-main chain hydrogen bonds.
For the second goal, the remaining set (designable residues) were subjected to rotamer optimizations employing a variant of the RosettaDesign algorithms (15) used for optimizing multiple conformers for a single sequence (16). In this case, each monomeric subunit of the tetramer represented a conformer. The designable residues were then ranked from high to low according to the Rosetta energy values and quality of packing scores (17). It was then determined which of those residues at the top of the list (i.e., poorly packed residues and residues with high energy) were at the interface. The neighboring residues of each nonoptimal designable interface residue were then enumerated, which yielded clusters of designable residues. These clusters were subsequently used for redesign (i.e., concerted optimization of amino acid types at all cluster sequence positions). Throughout all procedures the backbones of the monomeric subunits were held fixed as intended by templating.
Redesign of i1.
The interface i1 is formed between two monomers found in a crisscrossed antiparallel arrangement that allows close contacts between four α-helices (helices 2 and 3 from each monomer), in turn yielding the most extensive and intimate interprotein surface (Fig. 2 and Fig. 3A). The average gain in solvation energy upon the formation of i1 in the native structure was calculated to be only -1.5 kcal/mol, suggesting that the noncovalent interactions in i1 are not expected to yield a stable dimer (18). Our design strategy yielded a set of six mutations for il: Arg34Ala, Leu38Ala, Gln41Trp, Lys42Ser, Asp66Trp, and Val69Ile. Visual inspection of these positions in the native structure indicates that they make up the bulk of i1, collectively burying ∼600 Å2. In the redesigned i1 interface the mutated residues were predicted to form a well-packed hydrophobic core in which Ala34/Trp66’ (where the apostrophe indicates the second monomer) and Ala38/Trp41/Ile69’ form knob-in-hole interactions (Fig. 3B). Although the potential contribution of position 42 to i1 stability is less obvious, calculations suggested a strong preference for Ser to the native Lys in this position. Consequently, all of the six prescribed mutations were incorporated into MBPC-1 to generate the construct RIDC-1 (Rosetta Interface Design Cytochrome-1).
Fig. 3.
Side chain conformations in the core regions of interfaces i1 and i2 in (A) Zn4:MBPC-14, (B) the Rosetta-calculated model, and (C) Zn4:RIDC-24. Highlighted are six positions in each interface that were subjected to redesign, as well as those involved in Zn coordination. Water molecules observed in the Zn4:RIDC-24 structure are shown as small red spheres.
Redesign of i2.
We then addressed the redesign of the interface i2. The monomers that comprise i2 are positioned side by side, whereby the majority of interprotein contacts are now made between two helices 4 aligned antiparallel to each other. As expected again from a monomeric protein, the interfacial contacts in i2 are noncomplementary and calculated to yield an average gain of solvation energy of only -3 kcal/mol. In contrast to i1, the side-by-side alignment of monomers in i2 precludes the formation of a well-packed and solvent-protected core (Fig. 3A). For this interface our design strategy converged on smaller hydrophobic residues (Ile67Leu, Gln71Ala, Ala89Lys, Gln93Leu, Thr96Ala, and Thr97Ile) (Fig. 3B) at six surface positions that contribute a large fraction (∼400 Å2) of the buried surface in i2. In the calculated model of redesigned i2, Leu93 and Ala96 side chains from each monomer interdigitate across the center of i2 to form a modest-sized hydrophobic patch. Ala71 and Ile97 side chains are predicted to form intraprotein knob-in-hole interactions, whereas Ile67 appears mostly to add hydrophobic bulk in the interface. Given the small and less-than-optimally packed core of redesigned i2, we predicted that it would not contribute significantly to the stability of the Zn-induced tetramer on its own. Hence, we constructed the second-generation variant, RIDC-2, which includes the six calculated mutations in i2 in addition to the six incorporated into RIDC-1.
Stability and Zn-Mediated Oligomerization of RIDC-1 and RIDC-2.
Both RIDC-1 and RIDC-2 involve a significant increase in surface hydrophobicity compared to the parent construct MBPC-1. We therefore asked if they would be stable and maintain the native tertiary fold. Both variants are expressed in large yields in bacterial cultures, remain soluble, and feature heme groups with wild-type absorption features, suggesting that they are correctly folded. Chemical denaturation titrations indicate that both RIDC-1 and RIDC-2 are at least as stable as MBPC-1 (Fig. S2) although RIDC-2 displays non-two-state unfolding behavior due likely to the presence of additional hydrophobic mutations on its surface.
The effect of interface redesign on the overall stability of the Zn-induced tetrameric assembly was assessed by sedimentation velocity (SV) experiments (Fig. 4A and Fig. S3). Previous SV measurements on MBPC-1 indicated that the predominant species in solution at low protein and equimolar Zn concentrations was monomeric (7). Dimeric and tetrameric species become significantly populated only at MBPC-1 and Zn concentrations over 100 μM (1∶1 protein∶Zn) with increasing concentrations favoring the population of the tetrameric form. As a benchmark, the relative populations of monomeric, dimeric, and tetrameric species at 600 μM MBPC-1 and Zn are 12%, 35%, and 50%, respectively. In the cases of RIDC-1 and RIDC-2, on the other hand, there is only a very small detectable amount (approximately 6% of total species) of the monomeric form even at 5 μM protein and equimolar Zn. By 200 μM RIDC-1 or RIDC-2 and Zn, the tetrameric form is fully (> 95% of total) populated (Fig. S3). Because protein oligomerization is intimately coupled to metal binding in the tetrameric structures, it is challenging to obtain individual stability constants for the numerous equilibria leading to their formation. Nevertheless, the SV population distributions at different protein and Zn concentrations indicate that the interfacial mutations in RIDC-1 and RIDC-2 stabilize the Zn-induced tetramer by nearly two and three orders of magnitude, respectively, over the parent MBPC-1 species.
Fig. 4.
(A) From left to right: sedimentation coefficient distributions for various concentrations of MBPC-1, RIDC-1 and RIDC-2 in the presence of equimolar Zn(II). (B) Backbone superposition of Zn4:MBPC-14 (green), Zn4:RIDC-14 (blue), and Zn4:RIDC-24. (C) Tetrahedral Zn-coordination environment in Zn4:RIDC-24, with the corresponding Fo-Fc omit electron density map (3.2σ).
Next, we set out to establish whether the supramolecular architecture of Zn4:MBPC-14 is maintained in the RIDC-1 and RIDC-2 tetramers as intended by the template-and-stabilize strategy. The crystal structures of Zn4:RIDC-14 and Zn4:RIDC-24 were determined at 2.4 Å and 2.0 Å resolution, respectively. A backbone superposition of the MBPC-1, RIDC-1, and RIDC-2 tetramers (Species 4 in Fig. 1) indicates that they are topologically identical (rmsd over 424 αC 's = 0.4 ≲ and 0.6 Å) despite a total of 24 mutations on the former and 48 on the latter complex relative to Zn4:MBPC-14 (Fig. 4B). The tetrahedral Zn-coordination geometry in Zn4:RIDC-14 and Zn4:RIDC-24 is largely unchanged from Zn4:MBPC-14 (7), although the higher resolution structures of the former two reveal that Asp74 actually coordinates Zn in a monodentate fashion (Fig. 4C). Likely due to the same reason, the plane of the His73 imidazole ring in Zn4:RIDC-14 and Zn4:RIDC-24 is found to be rotated by 90° compared to that originally modeled in the Zn4:MBPC-14 structure (7).
The redesigned i1 interface, which features the same substitutions in RIDC-1 and RIDC-2, is structurally superimposable in both Zn4:RIDC-14 and Zn4:RIDC-24 structures with small variations in side chain conformations. As predicted, the engineered Trp41 and Trp66 side chains provide the bulk of the hydrophobic core in i1, which now buries ∼1500 Å2 of surface area and is largely devoid of solvent molecules (Fig. 3C, Fig. S3a). The Zn-coordination sphere in Zn4:RIDC-14 and Zn4:RIDC-24 is intimately linked to the i1 core through interactions between the coplanar His73’ and Trp66 aromatic rings (Fig. 4C). The Trp66 sidechain is further anchored by H-bonding between the indole imine and the Ile69’ backbone carbonyl group across the interface. Interestingly, there appears to be some fluidity within i1 because the electron density corresponding to the Trp41 side chain is best accommodated with two overlapping conformations of the indole ring (Fig. S3a).
In contrast to i1, the redesigned i2 is replete with solvent molecules (Fig. 3C). As expected from the small sizes of the substituted residues, the increase in buried surface area in i2 upon redesign is minimal (20 Å2) with a modest calculated gain in solvation free energy (-1.5 kcal/mol). Taken together the structural details of i1 and i2 are consistent with the results from SV measurements that redesign of i1 leads to a significant stabilization of the Zn-induced tetramer and that of i2 has a small incremental effect.
Metal-Independent Dimerization of RIDC-1.
Since the redesign of i1 generates an extensive set of hydrophobic interactions in Zn4:RIDC-14 and Zn4:RIDC-24, we examined whether these interactions could also sustain stable monomer–monomer interactions in the absence of metal coordination. SV and sedimentation equilibrium (SE) experiments show that RIDC-1 and RIDC-2 indeed form a metal-independent dimeric species (corresponding to Species 6 in Fig. 1) (Fig. 5A, B and Fig. S4). The dimer-monomer dissociation constants (Kd) have been determined by SE measurements to be 25 μM for RIDC-12 and 55 μM for RIDC-22 under low ionic-strength conditions (20 mM Tris buffer, pH 7, 5 mM EDTA), and 43 μM and 40 μM, respectively, under high ionic-strength conditions (150 mM NaCl in addition). Similar Kd’s for RIDC-12 and RIDC-22 suggest that dimerization in both cases involves the same protein interface (i.e., i1), while the lack of an apparent ionic-strength dependence in these values implies that dimerization is mainly driven by hydrophobic interactions.
Fig. 5.
(A) Sedimentation coefficient distributions for various concentrations of RIDC-1 in the presence of 5 mM EDTA, showing the population of the dimeric species at increasing concentrations. (B) Representative sedimentation equilibrium profile for RIDC-1 (see Fig. S4 for the complete set of measurements). Shown here are the scans for 20 μM RIDC-1 in 20 mM Tris (pH 7) and 5 mM EDTA collected at 20,000, 25,000, and 30,000 rpm, which are best fit with a monomer-dimer equilibrium model. (C) Side and top views of the RIDC-12 crystal structure, whereby the interfacial residues are shown as sticks. (D) Close-up views of the two (nearly) symmetrical interaction zones in the dimer interface detailing the hydrophobic and H-bonding contacts.
Crystals of RIDC-12 were obtained at protein concentrations (> 3 mM) that should favor the formation of the metal-independent dimer. The 2.0 Å resolution structure reveals a pair of RIDC-1 molecules in the asymmetric unit that arrange in an antiparallel fashion to form a 1300-Å2 interface (Fig. 5C). This C2-symmetrical interface is formed largely along the helix 3 from each monomer, primarily utilizing the engineered hydrophobic residues and those that are originally involved in Zn coordination. As detailed in Fig. 5D, His73 and Leu76 from one monomer form a patch of hydrophobic contacts across the interface with Trp41’, His63’, Ile69’, and Trp66’, whose indole group extends out toward the solvent without being involved in obvious crystal-packing interactions. The orientations of Trp41 and Trp66 side chains are stabilized by H-bonds to the Arg62 backbone carbonyl and the Asp74’ carboxylate groups, respectively. The Trp41–Arg62 interaction is part of a larger H-bonding circuit that involves a conserved water molecule in addition to His77’ and Glu81’. The latter closes the circuit through extensive salt bridges to the Arg62 side chain.
Overall, the modest size of this interface is consistent with the measured dissociation constants in the micromolar range, which are typical of transient protein complexes (19, 20). Based on the information available, such as the ionic-strength independence of dimerization despite the observed salt-bridging interactions, we propose that RIDC-12 in solution is a collection of several structurally similar conformers afforded by the fluidity of engineered hydrophobic interactions in i1. The crystal structure of RIDC-12 likely represents one of the most stable conformations favored over others through lattice-packing interactions. These conclusions are corroborated by docking simulations that were run prior to the determination of the RIDC-12 crystal structure (see SI text for details). The simulations run in the absence of metal coordination (Fig. S5 and Table S2) reveal a shallow funnel toward the docking geometry observed in the RIDC-12 crystal structure, suggesting that the engineered hydrophobic interactions do not impose strong specificity, but that they do orient the monomers to form complexes that closely approximate the conformation induced by Zn coordination. Ultimately the observation that RIDC-1 can form a metal-independent dimer suggests that the contribution of i1 mutations to Zn4:RIDC-14 stability is not only enthalpic, but also entropic. Dimerization of RIDC-1 halves the number of protein components toward tetramerization, while preorganizing the Zn-coordinating residues (H63, H73’, and H77’) into close proximity.
Cu-Mediated Dimerization of RIDC-1.
An implied outcome of the increased stability of the Zn-induced tetramer due to interface redesign is an increased preference for the formation of tetrahedral Zn-coordination environment, which in turn should translate into greater Zn binding affinity/specificity. As mentioned previously, it is not possible to experimentally dissect metal-protein binding equilibria from protein–protein association equilibria that collectively lead to the formation of the tetramer. As an indirect means to assess the increase in Zn binding affinity/specificity due to interface redesign, we investigated the oligomerization behavior of RIDC-1 in the presence of Cu(II), which prefers nontetrahedral coordination geometries. Cu(II) was previously observed to induce the dimerization of MBPC-1 through the square-planar coordination of His59, His63, His73’, and His77’ (Fig. 6A). If the surface mutations to convert MBPC-1 to RIDC-1 indeed lead to increased preference towards Zn, this should be reflected in the distortion of the Cu(II) coordination environment in RIDC-1.
Fig. 6.
(A) The influence of Zn-templated interfacial mutations in i1 on the conformations of Cu-mediated dimeric assemblies. (B) Backbone superposition of Cu2:RIDC-12 (gray) and a dimeric half of Zn4:RIDC-14 (orange) that contains i1. Interfacial residues of Cu2:RIDC-12 are shown as sticks; see Fig. S3 for detailed comparison of Cu2:RIDC-12 and Zn4:RIDC-14 interfaces. (C) Cu coordination environment in Cu2:RIDC-12 (Site 1), highlighting the open coordination sites occupied by two water molecules. The Glu81 side chain from a crystallographic symmetry-related dimer that forms H-bonds to the coordinated water molecules is shown in light gray. The Fo-Fc omit electron density map is contoured at 3σ.
SV measurements indicate that Cu(II) binding leads to the exclusive formation of a dimer (Cu2:RIDC-12) at all RIDC-1 concentrations used (5–600 μM) (Fig. S6). The crystal structure of Cu2:RIDC-12 (Species 7 in Fig. 1) was determined at 2.2 Å resolution. The crystallographic asymmetric unit of Cu2:RIDC-12 crystals contains one dimer that displays a crisscrossed monomer–monomer alignment (Fig. 6A). This alignment is significantly distorted from the antiparallel arrangement of monomers seen in Cu2:MBPC-12 toward that observed across i1 in Zn-induced tetramers. The Cu2:RIDC-12 backbone can be superimposed with each dimeric half of Zn4:RIDC-14 that contains the i1 interface with an overall rmsd of 1.4 Å (Fig. 6B) and features the same set of hydrophobic contacts in the interface (Fig. S3c). As a consequence of the crisscrossed arrangement of monomers in Cu2:RIDC-12, His59 is now pushed out of the Cu coordination sphere, leaving only His63, His73’, and His77’ as the protein-based ligands (Fig. 6C).
The two Cu coordination sites in the dimer interface display small differences. Cu1 exhibits a slightly distorted square-pyramidal geometry with one equatorial and one axial water molecule, both of which are H-bonded to Glu81 carboxylate from a symmetry-related dimer in the crystal lattice (Fig. 6D). Cu2, in contrast, is found in an approximately trigonal bipyramidal geometry whereby the crystallographically related Glu81 side chain is now directly coordinated (Fig. S7a). Because SV studies indicate the exclusive formation of a dimer, we suggest that in solution the Cu centers in Cu2:RIDC-12 adopt a square-planar or pyramidal geometry with three equatorial His ligands and one or two solvent molecules. This coordination geometry is supported by the axial EPR spectrum of Cu2:RIDC-12 (Fig. S7b).
As previously discussed, the engineered hydrophobic interactions in i1 are sufficiently flexible that they conform to the preferred square-planar coordination geometry of Cu(II) without paying a large energetic penalty (Fig. S5a, d). Clearly the enforcement of tetrahedral or near-tetrahedral coordination geometries on Cu(II) as intended by Zn templating would require more extensive and specific noncovalent interactions. A fitting example in this case is provided by blue copper proteins that use a rigid network of noncovalent interactions (i.e., the entire protein fold) to produce the “rack-effect” or the “entatic state” for Cu (21). Nevertheless, the redesigned surface in i1 provides sufficient driving force for the formation of the crisscrossed supramolecular arrangement over the antiparallel alignment seen in Cu2:MBPC-12 forcing Cu to adopt three—instead of four—histidine ligands. Thus, the memory of Zn coordination engraved into noncovalent interactions through templating is still imposed on Cu, ultimately enforcing an unsaturated coordination environment.
Conclusions
All biologically relevant transition metal ions form stable complexes with peptides, sometimes even when they lack metal-coordinating side chain functionalities (22). This is evident from the fact that unstructured polypeptides and folded proteins alike often form aggregates in the presence of high concentrations of transition metals. In modern-day organisms cellular metal concentrations are tightly regulated and the availability of free transition metal ions is generally low (23). It is safe to assume, on the other hand, that during early evolution of folded proteins such advanced regulatory mechanisms did not exist. Therefore, the interactions between peptides/proteins and metals were likely under thermodynamic control and governed by environmental concentrations of soluble metal ions (24). Under such conditions it would have been nearly impossible for any polypeptide chain to avoid being associated with metal ions. One possible outcome of such metal-polypeptide association reactions is precipitation. At the same time it is probable that some resulted in the formation of discrete soluble structures formed around metal-coordinating nuclei. If any such metal-nucleated structures inferred some benefit to an organism (e.g., generation of stable structures that did not deleteriously associate with existing cellular components, sequestration of essential or toxic metals, reactivity), they could have been subject to natural selection and evolution into more stable architectures or advanced functions. Our findings suggest that it is feasible for a non-self-associating protein surface to assemble into higher order architectures through a small number of mutations that enable metal coordination. Once the entropic cost of association is overcome, the resulting noncovalent interfaces can be optimized through additional mutations, in turn leading to more stable architectures that can form even in the absence of metal coordination. Given the lack of detailed information regarding the cellular environment during the early emergence of proteins, any such evolutionary model is speculative. Yet, the fact that a large fraction of known proteins contain metal ions as integral components and that many metal active sites are located in interfaces between secondary substructures or domains (25), suggest that evolutionary pathways involving initial metal-mediated nucleation events are possible.
From a practical protein interface design viewpoint, MeTIR offers important advantages. In principle MeTIR can be implemented on any protein surface large enough for the incorporation of stable metal binding motifs. As demonstrated here, metal–protein interactions mediated by such motifs can be strong enough to hold together extensive—and originally repulsive—protein surfaces that are amenable to structural characterization and subsequent (and iterative) redesign to generate associative interactions. In this regard the utility of crystal structures as a starting point for designing protein structures and interfaces has been well documented (26, 27). The most successful de novo interface design efforts to date have focused on coiled-coil motifs with predetermined docking orientations and knowledge-based energy functions (28, 29). In contrast MeTIR is not restricted to any particular type of protein interface or a particular fold for the partners in the complex; the fact that the protein building blocks used in this study are helical bundles is coincidental and purely due to practical reasons (stability, solubility, crystallizability). While the dependence on crystallographic information can be regarded as a limitation, MeTIR represents a promising step toward the ultimate goal of designing arbitrary protein interfaces from scratch.
From a practical inorganic chemical viewpoint our results show that individual proteins can be utilized as large polydentate ligands that bring along the advantage of having extensive functionalizable surfaces. While the complex nature of protein surfaces renders the control of metal coordination challenging, it allows the tuning of the metal coordination environment through distant noncovalent interactions. Such interactions, after all, account for the exquisite control of metal selectivity and reactivity in natural metalloproteins and enzymes through the formation of an extensive three-dimensional bonding network.
Materials and Methods
For a detailed description of experimental (x-ray crystallography, analytical ultracentrifugation, EPR spectroscopy, and chemical denaturation) and computational methods (protein interface redesign and protein docking simulations), see the SI text.
Supplementary Material
Acknowledgments.
The authors thank Prof. Ulrich Müller for helpful discussions. This work was supported by the National Institutes of Health (Molecular Biophysics Training Grant to E.N.S, NRSA to X.I.A., and GM073960 to B.K.), the Hellman Family Foundation, National Science Foundation (CHE-0908115) and the Arnold and Mabel Beckman Foundation (F.A.T.). Portions of this research were carried out at SSRL, operated by Stanford University on behalf of the Department of Energy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes: 3HNI, 3HNJ, 3HNK, and 3HNL).
See Commentary on page 1811.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906852107/DCSupplemental.
*Previous work by several groups has shown that various preexisting folds can be employed as stable scaffolds for hosting diverse metal coordination environments. Some of such engineered metalloproteins manifest properties characteristic of highly evolved natural systems, such as metal selectivity (30), redox tunability (31, 32) as well as open coordination sites that allow for reactivity (33) and reversible small molecule binding (34).
References
- 1.Soding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. BioEssays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 2.Bogarad LD, Deem MW. A hierarchical approach to protein molecular evolution. Proc Natl Acad Sci USA. 1999;96:2591–2595. doi: 10.1073/pnas.96.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andre I, Strauss CEM, Kaplan DB, Bradley P, Baker D. Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci USA. 2008;105:16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riechmann L, Winter G. Early protein evolution: Building domains from ligand-binding polypeptide segments. J Mol Biol. 2006;363:460–468. doi: 10.1016/j.jmb.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Faraone-Mennella J, Tezcan FA, Gray HB, Winkler JR. Stability and folding kinetics of structurally characterized cytochrome cb562. Biochemistry. 2006;45:10504–10511. doi: 10.1021/bi060242x. [DOI] [PubMed] [Google Scholar]
- 7.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling protein–protein interactions through metal coordination: Assembly of a 16-helix bundle protein. J Am Chem Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgado EN, Lewis RA, Faraone-Mennella J, Tezcan FA. Metal-mediated self-assembly of protein superstructures: Influence of secondary interactions on protein oligomerization and aggregation. J Am Chem Soc. 2008;130:6082–6084. doi: 10.1021/ja8012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold FH, Haymore BL. Engineered metal-binding proteins—purification to protein folding. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 10.Ghadiri MR, Choi C. Secondary structure nucleation in peptides—transition-metal ion stabilized alpha-helices. J Am Chem Soc. 1990;112:1630–1632. [Google Scholar]
- 11.Handel TM, Williams SA, Degrado WF. Metal-ion dependent modulation of the dynamics of a designed protein. Science. 1993;261:879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- 12.Krantz BA, Sosnick TR. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nat Struct Biol. 2001;8:1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- 13.Salgado EN, Lewis RA, Mossin S, Rheingold AL, Tezcan FA. Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through CuII and NiII coordination. Inorg Chem. 2009;48:2726–2728. doi: 10.1021/ic9001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyte J. Structure in Protein Chemistry. New York: Garland Science; 2007. [Google Scholar]
- 15.Liu Y, Kuhlman B. RosettaDesign server for protein design. Nucleic Acids Res. 2006;34:W235–238. doi: 10.1093/nar/gkl163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambroggio XI, Kuhlman B. Computational design of a single amino acid sequence that can switch between two distinct protein folds. J Am Chem Soc. 2006;128:1154–1161. doi: 10.1021/ja054718w. [DOI] [PubMed] [Google Scholar]
- 17.Leaver-Fay A, Butterfoss GL, Snoeyink J, Kuhlman B. Maintaining solvent accessible surface area under rotamer substitution for protein design. J Comp Chem. 2007;28:1336–1341. doi: 10.1002/jcc.20626. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 20.Nooren IMA, Thornton JM. Structural characterisation and functional significance of transient protein–protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 21.Gray HB, Malmstrom BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 22.Martell AE, Smith RM. Critical Stability Constants. New York: Plenum Press; 1974–1989. [Google Scholar]
- 23.Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 24.Frausto da Silva JJR, Williams RJP. The Biological Chemistry of the Elements. Oxford: Oxford Univ Press; 2001. [Google Scholar]
- 25.Bertini I, Gray HB, Stiefel EI, Valentine JS. Biological Inorganic Chemistry, Structure & Reactivity. Sausalito: University Science Books; 2007. [Google Scholar]
- 26.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein–protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Bryson JW, Desjarlais JR, Handel TM, DeGrado WF. From coiled coils to small globular proteins: Design of a native-like three-helix bundle. Protein Sci. 1998;7:1404–1414. doi: 10.1002/pro.5560070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 29.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KH, et al. Control of metal coordination number in de novo designed peptides through subtle sequence modifications. J Am Chem Soc. 2004;126:9178–9179. doi: 10.1021/ja048839s. [DOI] [PubMed] [Google Scholar]
- 31.Reddi AR, Reedy CJ, Mui S, Gibney BR. Thermodynamic investigation into the mechanisms of proton-coupled electron transfer events in heme protein maquettes. Biochemistry. 2007;46:291–305. doi: 10.1021/bi061607g. [DOI] [PubMed] [Google Scholar]
- 32.Hwang HJ, Lu Y. pH-dependent transition between delocalized and trapped valence states of a Cu-A center and its possible role in proton-coupled electron transfer. Proc Natl Acad Sci USA. 2004;101:12842–12847. doi: 10.1073/pnas.0403473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koder RL, et al. Design and engineering of an O2 transport protein. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.