Abstract
Neural stem cell self-renewal and differentiation is orchestrated by precise control of gene expression involving nuclear receptor TLX. Let-7b, a member of the let-7 microRNA family, is expressed in mammalian brains and exhibits increased expression during neural differentiation. However, the role of let-7b in neural stem cell proliferation and differentiation remains unknown. Here we show that let-7b regulates neural stem cell proliferation and differentiation by targeting the stem cell regulator TLX and the cell cycle regulator cyclin D1. Overexpression of let-7b led to reduced neural stem cell proliferation and increased neural differentiation, whereas antisense knockdown of let-7b resulted in enhanced proliferation of neural stem cells. Moreover, in utero electroporation of let-7b to embryonic mouse brains led to reduced cell cycle progression in neural stem cells. Introducing an expression vector of Tlx or cyclin D1 that lacks the let-7b recognition site rescued let-7b-induced proliferation deficiency, suggesting that both TLX and cyclin D1 are important targets for let-7b-mediated regulation of neural stem cell proliferation. Let-7b, by targeting TLX and cyclin D1, establishes an efficient strategy to control neural stem cell proliferation and differentiation.
Keywords: Let-7, cyclin D1, fate-determination
Neural stem cells are undifferentiated precursors that retain the ability to proliferate and self-renew, and they have the capacity to give rise to both neuronal and glial lineages (1). Although the functional properties of neural stem cells have been studied extensively, molecular mechanisms underlying neural stem cell self-renewal and differentiation are only beginning to be understood. One class of factors thought to be important in this process is microRNAs (miRNAs), which are short, 20–22 nucleotide RNA molecules that are expressed in a tissue-specific and developmentally regulated manner and function as negative regulators of gene expression in a variety of eukaryotes. MiRNAs are involved in numerous cellular processes including development, proliferation, and differentiation (2, 3). Studies based on expression patterns, predicted targets, and overexpression analyses suggest that miRNAs are key regulators in stem cell biology.
The lethal-7 (let-7) gene is one of the first two miRNAs discovered in Caenorhabditis elegans (C. elegans), and the first known human miRNA (4, 5). Mature let-7 is highly conserved across species in both sequence and function. It plays an important role in development and cell maturation (6–9). Let-7 is expressed in both embryonic and adult brains (10–13). Recently, increased expression and maturation of let-7 has been observed during neural cell specification (8, 9). Let-7a, one of the members of the let-7 family, has been shown to play a role in neuronal differentiation of embryonic neural progenitors (14), while let-7b has been shown to reduce the self-renewal of aging neural stem cells through targeting the high-mobility group A (HMGA) family member Hmga2 expression (15).
TLX (NR2E1) is an orphan nuclear receptor that is expressed in vertebrate forebrains (16). Mature Tlx knockout mice have significantly reduced cerebral hemispheres and exhibit increased aggressiveness and progressively violent behavior (17–20). We have shown earlier that TLX is an essential regulator of adult neural stem cell self-renewal (18). TLX maintains neural stem cells in an undifferentiated and self-renewable state by complexing with histone deacetylases to repress TLX downstream target genes, the cyclin-dependent kinase inhibitor p21, and the tumor suppressor gene Pten (21). PTEN negatively regulates cyclin D1, a positive cell cycle regulator, via its protein phosphatase activity (22). Thus, TLX induces cyclin D1 expression partly by repressing Pten (23). The TLX-positive neural stem cells play an important role in spatial learning and memory in adult brains (24). Furthermore, TLX is expressed in the periventricular neural stem cells in embryonic brains and plays an important role in neural development by regulating cell cycle progression of neural stem cells (23, 25, 26). TLX is a key regulator that acts to establish the undifferentiated and self-renewable state of both embryonic and adult neural stem cells. However, the upstream events that regulate TLX expression and function remain largely unknown.
Here we demonstrate that let-7b suppresses the expression of Tlx and cyclin D1 by binding to the 3′ untranslated region (UTR) of their mRNAs and thus serves as a key regulator of neural stem cell proliferation and differentiation. Increased expression of let-7b led to reduced neural stem cell proliferation and accelerated neural differentiation, whereas antisense-knockdown of let-7b resulted in increased neural stem cell proliferation. Moreover, in utero electroporation of let-7b to embryonic neural stem cells led to reduced cell cycle progression. Let-7b acts in neural stem cells through the control of Tlx and cyclin D1 expression. Introducing Tlx or cyclin D1 expression vectors, lacking their endogenous 3′ UTR containing the let-7b target sites, rescued the let-7b effect on proliferation deficiency in neural stem cells. These results suggest that let-7b acts as an upstream regulator of neural stem cell proliferation and differentiation by targeting Tlx and cyclin D1 expression via their 3′ UTR. Let-7b thus forms a regulatory cascade with TLX and cyclinD1 to regulate the switch of neural stem cell proliferation and differentiation.
Results
Let-7b Regulates Neural Stem Cell Proliferation and Differentiation.
Recently, miRNAs have been shown to play important roles in stem cell biology. Let-7b is expressed in mammalian brains (10–12). We tested whether let-7b is expressed in neural stem cells or their differentiated progenies. As shown in Fig. 1A, let-7b is expressed in neural stem cells isolated from adult mouse forebrains (d0). Interestingly, the expression of let-7b is upregulated significantly during neural differentiation, along with increased expression of a neuronal marker Tuj1 and an astrocyte marker Gfap (Fig. 1A). To determine whether let-7b regulates neural stem cell proliferation, we transfected neural stem cells with increasing concentrations of let-7b RNA duplexes. The transfected cells were treated with 5-bromodeoxyuridine (BrdU) and analyzed by BrdU labeling to assess cell proliferation. Transfection of let-7b led to dose-dependent inhibition of cell proliferation, as revealed by decreased percentage of BrdU labeling in let-7b-treated cells (Fig. 1B and C), whereas minimal cytotoxicity was detected in let-7b-transfected cells (Fig. 1D). These results suggest that let-7b negatively regulates neural stem cell proliferation.
Fig. 1.
Let-7b inhibits neural stem cell proliferation. (A) Expression of let-7b in a 7 day differentiation time course in mouse adult neural stem cells. Day (d) 0 represents undifferentiated neural stem cell state. U6 was included as a loading control. Relative let-7b levels were normalized by U6 levels and shown underneath the blots, with let-7b level in d0 as 1. RT-PCR analysis of Tuj1 and Gfap expression in the same differentiation time course was shown in the right panels. Gapdh was included as a loading control. (B) Neural stem cells were transfected with increasing concentrations of RNA duplexes of let-7b for 48 hr. The transfected cells were treated with BrdU for 2 hr and immunostained with a BrdU-specific antibody (Green). The merged images are BrdU staining along with phase contrast images. A let-7b mutant with mutations in the seed region was included as a control, adding to the wild type let-7b to have a total of 200 nM miRNAs in each transfection. (C) Quantification of BrdU+ cells in let-7b-treated neural stem cells. Error bars are standard deviation of the mean; assays were repeated three times. ∗p < 0.005 by one-way Anova. (D) Minimal cytotoxicity in let-7b-transfected neural stem cells. Neural stem cells were mock-transfected (-), transfected with a mutant let-7b (C, 200 nM), or wild type let-7b (200 nM), followed by LDH assays for cytotoxicity.
To determine whether let-7b regulates neural stem cell differentiation, neural stem cells were transfected with let-7b RNA duplexes and cultured in differentiation conditions. When neural stem cells were cultured in N2 media containing forskolin and fetal bovine serum to induce astrocyte differentiation, transfection of let-7b led to a 2.4-fold increase in the percentage of GFAP-positive astrocytes at day three of differentiation (Fig. 2A and B). When neural stem cells were induced for neuronal differentiation using retinoic acid and fetal bovine serum, a 2.1-fold increase in the percentage of TUJ1-positive neurons was observed in let-7b-transfected cells at day three of differentiation (Fig. 2C and D). These results together indicate that let-7b accelerates neuronal and glial differentiation of neural stem cells that are primed for differentiation.
Fig. 2.
Let-7b accelerates neural differentiation. (A) Overexpression of let-7b promotes glial differentiation. Neural stem cells were transfected with 200 nM of let-7b RNA duplexes for 48 hr. The transfected cells were induced into differentiation using forskolin and fetal bovine serum for 3 d. The treated cells were immunostained with a GFAP-specific antibody (Red). Nuclei dapi staining is shown in Blue. (B) Quantification of glial differentiation using the percentage of GFAP+ cells. (C) Overexpression of let-7b promotes neuronal differentiation. The let-7b transfected neural stem cells were induced into differentiation using retinoic acid and fetal bovine serum for 3 d. The treated cells were immunostained with a TUJ1-specific antibody (Green). Nuclei dapi staining is shown in Blue. (D) Quantification of neuronal differentiation using the percentage of TUJ1+ cells. For both B and D, error bars are standard deviation of the mean; assays were repeated three times; ∗p < 0.001 (B) and p < 0.01 (D) by student’s t-test.
Let-7b Reduces Cell Cycle Progression in Embryonic Brains.
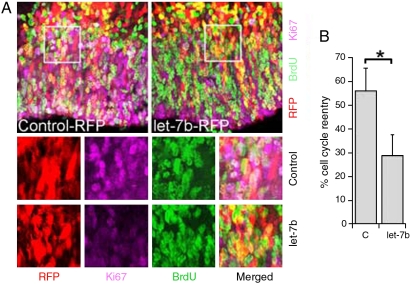
Let-7 has been shown to act as a regulator of cell cycle exit in C. elegans seam cells to control the timing of terminal differentiation (4) and to regulate cell cycles in human cancer cells (27, 28). To determine whether let-7b regulates cell cycle progression in neural stem cells of mammalian brains, we transfected let-7b into embryonic day 13.5 (E13.5) mouse brains by in utero electroporation. Cell cycle reentry of neural stem cells was determined by scoring the fraction of cells that reentered cell cycle after pulse labeling with BrdU for 24 hr before brain harvest. Cells that left the cell cycle were identified as BrdU+ and Ki67-, whereas cells that reentered the cell cycle as BrdU+ and Ki67+. At E15.5, there was a twofold decrease in the percentage of neural precursors that reentered the cell cycle in wild type let-7b-electroporated brains, when compared with the mutant let-7b control RNA-transfected brains (Fig. 3A and B). These studies suggest that let-7b functions in neural stem cells by promoting cell cycle exit.
Fig. 3.
Let-7b regulates cell cycle progression in embryonic brains. (A) Reduced fraction of cell cycle reentry in let-7b-electroportaed brains. Wild type or mutant (control) let-7b RNA duplexes were mixed with pCAGGS-RFP reporter plasmid DNA and electroparated into E13.5 mouse brains. Animals were exposed to a BrdU pulse 24 hr before brain harvest. Brain sections were stained with antibodies to BrdU (Green) and Ki67 (Purple). The transfected cells that reentered the cell cycle were labeled as RFP + BrdU + Ki67+ (Whitish Color), whereas the transfected cells that left the cell cycle were labeled as RFP + BrdU + Ki67- (Yellow). Enlarged images of the boxed region were shown in the bottom panels. (B) Quantification of cell cycle reentry. Fraction of cell cycle reentry is calculated as RFP + BrdU + Ki67+ cells out of RFP + BrdU+ cells in wild type or mutant (C) let-7b RNA dulplexes-transfected brains. Error bars are standard deviation of the mean; assays were repeated three times; ∗p < 0.01 by student’s t-test.
Let-7b Inhibits Tlx Expression by Binding to its 3′ UTR.
What is the molecular basis of let-7b-mediated cell cycle regulation? We have shown that TLX regulates cell cycle progression of neural stem cells in the developing brain (23). Using the miRanda algorithm, let-7b was identified to have a conserved target site in the 3′ UTR of Tlx gene (Fig. S1). To determine whether let-7b targets Tlx, we made a luciferase reporter construct with mouse 1.4 kb Tlx 3′ UTR containing the predicted let-7b target site and the flanking sequences inserted into the 3′ UTR of a Renilla luciferase reporter gene in a siCHECK vector. Increasing amounts of RNA duplexes of mature let-7b was transfected into human embryonic kidney HEK 293 cells along with the reporter gene. Dose-dependent repression of the reporter gene was observed in let-7b-treated cells (Fig. 4A). To test whether the predicted let-7b target site in Tlx 3′ UTR is critical for repression of Tlx expression by let-7b, point mutations disrupting the base-pairing in the predicted let-7b target site were introduced into Tlx 3′ UTR in the luciferase reporter construct. Mutation of the let-7b target site abolished the repression by let-7b (Fig. 4B). This result suggests that let-7b represses Tlx expression through the predicted target site in Tlx 3′ UTR.
Fig. 4.
Let-7b represses Tlx expression. (A) Let-7b-mediated repression of luciferase reporter gene downstream of 3′ UTR of Tlx. Tlx 3′ UTR reporter gene was cotransfected into HEK 293 cells with increasing amounts of control or let-7b RNA duplexes. (B) Mutation of let-7b target site in Tlx 3′ UTR abolished let-7b-mediated repression. Luciferase reporter gene under the control of wild type (WT) or mutant (Mut) Tlx 3′ UTR was transfected into HEK 293 cells along with control or let-7b RNA duplexes. For both A and B, data shown are means ± standard deviation of three replicates; ∗p < 0.05, ∗∗p < 0.005 by student’s t-test. (C, D) Let-7b-mediated repression of Tlx expression in neural stem cells revealed by Western blot (C) and Northern blot (D) analyses. C stands for control RNA.
Next we tested whether let-7b targets Tlx expression in neural stem cells. Mature let-7b RNA duplexes were transfected into neural stem cells. Tlx expression was examined by Western blot and Northern blot analyses. Significant reduction of both protein and mRNA levels of Tlx was detected in let-7b transfected cells (Fig. 4C and D). Repression of Tlx mRNA levels by let-7b is consistent with previous observation that regulation by let-7 results in target mRNA degradation (29). These results together indicate that let-7b down-regulates Tlx at both protein and RNA transcript levels.
Let-7b Regulates Neural Stem Cells by Targeting Tlx Expression.
To determine whether let-7b regulates neural stem cell proliferation and differentiation through targeting Tlx expression, neural stem cells were stably transduced with a TLX-expressing vector, which lacks Tlx 3′ UTR containing the let-7b recognition site (TLXΔ3′ UTR). Transfection of let-7b had no effect on the expression of the transduced TlxΔ3′ UTR, although it down-regulates endogenous Tlx expression levels (Fig. 5A). Expression of TLXΔ3′ UTR recovered the proliferative deficiency induced by let-7b considerably (Fig. 5B and C). Furthermore, while transfection of let-7b increased astroglial differentiation in control neural stem cells, no significant increase in astrocyte differentiation was detected in TLXΔ3′ UTR-transduced cells upon let-7b treatment (Fig. 5D). These results strongly suggest that let-7b regulates neural stem cell proliferation and differentiation, at least in part, by inhibiting Tlx expression through its 3′ UTR.
Fig. 5.
TLX expression vector lacking its 3′ UTR (TLXΔ3′ UTR) rescues let-7b-induced neural stem cell proliferation deficiency. (A) RT-PCR analysis of TlxΔ3′ UTR and total Tlx expression in control neural stem cells (C) and TLXΔ3′ UTR-expressing neural stem cells treated with control RNA (-let-7b) or let-7b (+let-7b). Gapdh was included as a loading control. (B) Control or TLXΔ3′ UTR-expressing neural stem cells were transfected with control or let-7b RNA duplexes. The transfected cells were treated with BrdU and immunostained with a BrdU-specific antibody (Green). The merged images are BrdU staining along with phase contrast images. (C) Quantification of BrdU-positive (BrdU+) cells in control or TLXΔ3′ UTR-expressing neural stem cells treated with control or let-7b RNA duplexes. (D) Quantitation of GFAP+ cells in control neural stem cells or TLXΔ3′ UTR-expressing neural stem cells treated with control or let-7b RNA duplexes. For both C and D, error bars are standard deviation of the mean; assays were repeated three times; ∗p < 0.005 by student’s t-test.
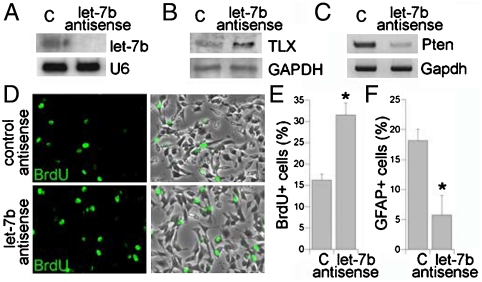
By using 2′-O-methyl antisense RNA oligonucleotides as small RNA inhibitors, the role of let-7b in neural stem cell proliferation was further investigated. 2′-O-methyl antisense oligonucleotide against let-7b was transfected into neural stem cells with 2′-O-methyl antisense oligonucleotide against GFP included as a negative control. Treatment of antisense oligonucleotide against let-7b led to significant knockdown of let-7b mature form (Fig. 6A). The expression of TLX was upregulated in let-7b antisense RNA-treated neural stem cells (Fig. 6B), along with decreased expression of Pten (Fig. 6C), a negative cell cycle regulator (30), the expression of which is repressed by TLX (21). BrdU labeling analysis revealed that knockdown of let-7b led to a significant increase in cell proliferation (Fig. 6D and E), whereas transfection of let-7b antisense RNA resulted in a substantial decrease in the percentage of GFAP-positive astrocytes when neural stem cells were induced for astrocyte differentiation (Fig. 6F). These results further support the notion that let-7b negatively regulates neural stem cell proliferation and promotes neural differentiation.
Fig. 6.
Let-7b antisense RNA promotes neural stem cell proliferation. (A) 2′-O-methyl let-7b antisense RNA knocks down let-7b mature form analyzed by Northern blot analysis. 2′-O-methyl antisense GFP RNA was included as a negative control (C). U6 was included as a loading control. (B) Expression of TLX in 2′-O-methyl let-7b antisense RNA-treated neural stem cells analyzed by Western blot analysis. GAPDH was included as a loading control. (C) Expression of Pten in 2′-O-methyl let-7b antisense RNA-treated neural stem cells analyzed by RT-PCR analysis. Gapdh was included as a loading control. (D) Neural stem cells were transfected with control RNA or 2′-O-methyl let-7b antisense RNA. The transfected cells were treated with BrdU and immunostained with a BrdU-specific antibody (Green). The merged images are BrdU staining along with phase contrast images. (E) Quantification of BrdU+ cells in control and 2′-O-methyl let-7b antisense RNA-treated neural stem cells. (F) Quantification of GFAP+ cells in control and 2′-O-methyl let-7b antisense RNA-treated neural stem cells. For both E and F, error bars are standard deviation of the mean; assays were repeated three times; ∗p < 0.05 by student’s t-test.
Let-7b Represses Cyclin D1 Expression to Regulate Neural Stem Cell Proliferation.
In addition to Tlx, we also identified a let-7b target site in cyclin D1 that is conserved in human, mouse, rat, dog, and chicken (Fig. S2) using the Targetscan algorithm. To validate whether let-7b represses cyclin D1 expression, we made a luciferase reporter construct with 1 kb mouse cyclin D1 3′ UTR containing the predicted let-7b target site and flanking sequences inserted into the 3′ UTR of Renilla luciferase gene in siCHECK vector. Repression of the reporter gene was observed in the wild type let-7b RNA duplexes-transfected cells, but not in the mutant let-7b-transfected cells. Mutation of the let-7b target site in the cyclin D1 3′ UTR abolished the repression by let-7b (Fig. 7A). These results strongly suggest that let-7b represses cyclin D1 expression through the predicted target site in cyclin D1 3′ UTR.
Fig. 7.
Let-7b represses cyclin D1 expression in neural stem cells. (A) Let-7b-mediated repression of luciferase reporter gene downstream of 3′ UTR of cyclin D1 gene. Wild type (WT) or mutant (MT) cyclin D1 3′ UTR reporter gene was cotransfected into HEK 293 cells with WT or MT let-7b RNA duplexes. Data shown are means ± standard deviation of three replicates; ∗p < 0.01 by student’s t-test. (B, C) Let-7b-mediated repression of cyclin D1 expression in neural stem cells revealed by RT-PCR (B) and Western blot (C) analyses. A mutant let-7b RNA duplex was included as the control RNA (C). (D) Cyclin D1 expression vector lacking its 3′ UTR (cyclin D1Δ3′ UTR) rescued let-7b-induced neural stem cell proliferation deficiency. A control vector (C), cyclin D1Δ3′ UTR (cycD), TLXΔ3′ UTR (TLX), or both TLXΔ3′ UTR and cyclin D1Δ3′ UTR (TLX/cycD)-transduced neural stem cells were transfected with mutant let-7b (-let-7b) or wild type let-7b (+let-7b) RNA duplexes. The proliferation rate of the transfected cells was determined by the percentage of BrdU+ cells in total living cells. (E) Quantification of GFAP+ cells in C, cycD, TLX, TLX/cycD-transduced neural stem cells (the same as in d) at day three of astrocyte differentiation. For both D and E, error bars are standard deviation of the mean; assays were repeated three times; ∗p < 0.05 by two-way Anova.
Next we tested whether let-7b targets cyclin D1 expression in neural stem cells. Wild type and mutant let-7b RNA duplexes were transfected into neural stem cells. Cyclin D1 expression was examined by RT-PCR and Western blot analyses. Cyclin D1 mRNA and protein levels were both reduced in the wild type let-7b-transfected cells (Fig. 7B and C), indicating that let-7b down-regulates cyclin D1 expression in neural stem cells.
To determine whether cyclin D1 contributes to let-7b-mediated regulation of neural stem cell proliferation, we transduced neural stem cells with a retroviral vector expressing cyclin D1 lacking its 3′ UTR (cyclin D1Δ3′ UTR). In contrast to the 4.6-fold decrease in the percentage of BrdU+ cells induced by let-7b in control vector-transduced cells, transduction of cyclin D1Δ3′ UTR rescued the proliferative deficiency induced by let-7b considerably, similar to transduction of TLXΔ3′ UTR (Fig. 7D). Transduction of both TLXΔ3′ UTR and cyclin D1Δ3′ UTR recovered the proliferative deficiency induced by let-7b to a similar extent to the transduction of cyclin D1 alone (Fig. 7D). Furthermore, while transfection of let-7b increased astrocyte differentiation significantly in control neural stem cells, no significant increase in GFAP+ cells was detected in cyclin D1Δ3′ UTR alone, TLXΔ3′ UTR alone, or double-transduced cells, upon let-7b treatment (Fig. 7E). These results together provide strong evidence that both Tlx and cyclin D1 are important target genes for let-7b function in neural stem cell proliferation and differentiation, presumably acting in the same signaling pathway.
Discussion
We show here that miRNA let-7b regulates neural stem cell fate decision. Overexpression of let-7b led to inhibition of neural stem cell proliferation and accelerated neural differentiation, whereas antisense knockdown of let-7b resulted in increased proliferation of neural stem cells. Let-7b regulates neural stem cell proliferation by reducing cell cycle progression. Furthermore, this study provides a previously undescribed link between let-7b and an essential stem cell regulator TLX. Let-7b functions in neural stem cells by repressing the expression of Tlx and its downstream effector, cyclin D1.
The brain expresses a complex miRNA signature. The role of miRNAs in brain development and function are just beginning to be uncovered. Our recent study has shown that the brain-specific miRNA miR-9 has a key role in vertebrate brain development (31). Another brain-enriched miRNA, miR-124, has also been shown to regulate neuronal differentiation in both the developing spinal cord and in the adult subventricular zones (32–34). How the brain orchestrates members of the miRNA signature to control cell differentiation through precise transitions between neural progenitors and differentiated cells remains to be explored. It is likely that the distinct temporal expression patterns of these miRNAs reflect their specific roles in coordinating gene expression profiles that characterize neural cell fate determination.
Recently, let-7b has been shown to target Hmga2 to reduce the self-renewal of neural stem cells in the aging brain (15). Our study presented here revealed that let-7b inhibits neural stem cell proliferation and promotes differentiation in embryonic brains and adult neural stem cells through targeting Tlx and cyclin D1. These studies suggest that let-7b plays a role in neural stem cell proliferation and differentiation across a spectrum of developmental stages through targeting distinct key molecules.
Using a bioinformatics approach and experimental validation, we identified Tlx as a target of let-7b in neural stem cells. The expression of Tlx is high in neural stem cells but is reduced dramatically upon differentiation (18). In contrast, the level of let-7b is increased significantly upon differentiation. In this case, Tlx is preferentially expressed at high levels when the targeting miRNA expression is low. The expression of this target is down-regulated as let-7b accumulates. These data support the concept that miRNAs that are induced during differentiation suppress stem cell regulators to ensure proper cell fate transition (35). This is consistent with the observation that the timing of let-7 expression triggers terminal cell differentiation in C. elegans and supports the hypothesis that let-7 is involved in regulating the timing of cell fate decision (2, 4, 5).
This study identified Tlx as a key target of let-7b in neural stem cells. A miRNA may have multiple target genes (36). However, one or two of these target genes may be more important targets in specific cellular context. One of the questions addressed here is whether the cell proliferation and differentiation effect mediated by let-7b in neural stem cells is directly related to repression of Tlx expression. Transfection of let-7b into neural stem cells that are stably transduced with a TLX-expressing vector lacking the let-7b target site showed that ectopically expressed TLX rescued the proliferative deficiency induced by overexpression of let-7b substantially and compromised let-7b-induced precocious differentiation. These experiments suggest that Tlx is an important target gene of let-7b in controlling neural stem cell proliferation and differentiation. However, this study does not exclude the possibility that additional target genes may also play a role in let-7b function in neural stem cells.
Indeed, we have identified cyclin D1, a key cell cycle regulator, as another let-7b target gene in neural stem cells. In a previous study, we have shown that cyclin D1 is a downstream effector of TLX signaling. Decreased cyclin D1 expression was observed in Tlx-null brains (23), presumably due to increased expression of Pten, a negative regulator of cyclin D1 (22). Targeting both Tlx and cyclin D1 by let-7b provides double insurance to inhibit neural stem cell proliferation when let-7b expression is elevated upon differentiation. Let-7b thus provides an efficient control to regulate neural stem cell fate determination by targeting two components in one pathway. Characterizing the let-7b-TLX-cyclin D1 signaling cascade provides unique insight into the role of let-7b in neural stem cell fate determination.
Materials and Methods
Neural Stem Cell Culture, BrdU Labeling, Immunostaining, Cytotoxicity Analysis.
Neural stem cells were isolated from adult mouse forebrains using percoll gradient as described (18) and cultured in DMEM F12 medium with 1 mM L-glutamine, N2 supplement (Gibco-BRL), EGF (20 ng/ml), FGF-2 (20 ng/ml), and heparin (50 ng/ml) for proliferation. For differentiation, neural stem cells were exposed to DMEM F12 media with N2 supplement (N2 media) containing 5 μM forskolin and 0.5% FBS or N2 media containing 1 μM retinoic acid and 0.5% FBS. BrdU labeling and immunostaining analysis was performed as described (18), using antibodies for BrdU (Accurate; diluted 1∶10,000), TUJ1 (Covance, 1∶2,500), and GFAP (Advance Immuno; 1∶1,000). About 4,000 cells were counted for quantification for each treatment. Cytotoxicity was determined by measuring release of the cytosolic enzyme lactate dehydrogenase (LDH) into culture medium upon cell lysis using CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega). Cytotoxicity (%) was expressed as the percent of LDH released into the medium out of the total LDH activity.
In Utero Electroporation and Cell Cycle Analysis.
All animal experiments were performed in accordance with City of Hope and National Institutes of Health guidelines. In utero electroporation was carried out as described (31). 37.5 pmol of wild type or mutant let-7b RNA duplexes were injected into the lateral ventricles of E13.5 mouse embryos along with 0.625 μg of pCAGGS-RFP reporter plasmid. Twenty-four hr after electroporation, timed-pregnant mice were injected with BrdU (Sigma) at 50 mg/kg of mouse body weight intraperitoneally and animals were allowed to survive for 24 hr. Brains of the electroporated embryos were collected for cell cycle analysis as described (23). Specifically, brain sections were immunostained with antibodies for Ki67 (Lab Vision, 1∶500) and BrdU (Accurate, 1∶10,000). Cells that left the cell cycle were identified as BrdU+ and Ki67-, and cells that reentered the cell cycle as BrdU+ and Ki67+. Percent of cell cycle reentry was expressed as BrdU + Ki67+ cells out of BrdU+ cells. About 2,000 cells were counted for quantification for each treatment. The sequences of the wild type and mutant let-7b are included in SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. John Zaia for critical comments on the manuscript. Y.S. is a Kimmel Scholar. G.S. is a Herbert Horvitz Postdoctoral Fellow. W.L. is supported by a training grant from California Institute for Regenerative Medicine. This work was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke Grant R01 NS059546 (to Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908750107/DCSupplemental.
References
- 1.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Yu F, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 8.Wulczyn FG, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 9.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 10.Kapsimali M, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miska EA, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PloS one. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 17.Monaghan AP, et al. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 19.Roy K, Thiels E, Monaghan AP. Loss of the tailless gene affects forebrain development and emotional behavior. Physiol Behav. 2002;77:595–600. doi: 10.1016/s0031-9384(02)00902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young KA, et al. Fierce: A new mouse deletion of Nr2e1; violent behavior and ocular abnormalities are background-dependent. Behav Brain Res. 2002;132:145–158. doi: 10.1016/s0166-4328(01)00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng LP, Brown JL, Eng C. PTEN coordinates G(1) arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum Mol Genet. 2001;10:599–604. doi: 10.1093/hmg/10.6.599. [DOI] [PubMed] [Google Scholar]
- 23.Li W, et al. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol Endocrinol. 2008;22:56–64. doi: 10.1210/me.2007-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behavior. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- 25.Roy K, et al. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci. 2004;24:8333–8345. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 28.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 29.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Groszer M, et al. PTEN negatively regulates neural stem cell self-renewal by modulating G0–G1 cell cycle entry. Proc Natl Acad Sci USA. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. MiR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crc Cr Rev Oncol-Hem. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.