Abstract
Ubiquitination of histones provides an important mechanism regulating chromatin remodeling and gene expression. Recent studies have revealed ubiquitin ligases involved in histone ubiquitination, yet the responsible enzymes and the function of histone ubiquitination in spermatogenesis remain unclear. We have previously shown that mice lacking the ubiquitin ligase UBR2, one of the recognition E3 components of the N-end rule proteolytic pathway, are infertile associated with meiotic arrest at prophase I. We here show that UBR2 localizes to meiotic chromatin regions, including unsynapsed axial elements linked to chromatin inactivation, and mediates transcriptional silencing via the ubiquitination of histone H2A. UBR2 interacts with the ubiquitin conjugating enzyme HR6B and its substrate H2A and promotes the HR6B–H2A interaction and the HR6B-to-H2A transfer of ubiquitin. UBR2 and ubiquitinated H2A (uH2A) spatiotemporally mark meiotic chromatin regions subject to transcriptional silencing, and UBR2-deficient spermatocytes fail to induce the ubiquitination of H2A during meiosis. UBR2-deficient spermatocytes are profoundly impaired in chromosome-wide transcriptional silencing of genes linked to unsynapsed axes of the X and Y chromosomes. Our findings suggest that insufficiency in UBR2-dependent histone ubiquitination triggers a pachytene checkpoint system, providing a new insight into chromatin remodeling and gene expression regulation.
Keywords: chromatin inactivation, H2A, meiosis, N-end rule, ubiquitin
The N-end rule pathway is a ubiquitin (Ub)-dependent proteolytic system where E3 ligases called N-recognins recognize specific N-terminal amino acids of short-lived substrates as an essential component of degradation signals called N-degrons (1, 2). A pre-N-degron of a stable protein can be converted into an N-degron through N-terminal modifications, such as deamidation, oxidation, and arginylation (3–5). UBR1 and UBR2 are really interesting new gene (RING) finger E3s that share size (200 kDa), sequence (46% identity), and enzymatic specificities to type-1 (Arg, Lys, and His; basic) and type-2 (Phe, Leu, Trp, Tyr, and Ile; bulky hydrophobic) destabilizing N-terminal residues (3, 5, 6). Regardless of the similarity in biochemical properties, deficiency of UBR1 and UBR2 causes completely different null phenotypes in mammals. Mutations of human UBR1 cause Johanson–Blizzard syndrome (JBS), an autosomal recessive disorder with multiple developmental abnormalities including exocrine pancreatic insufficiency; knockout of mouse UBR1 also results in metabolic abnormalities such as JBS-like exocrine pancreatic defects (reviewed in ref. 2). UBR2-/- male mice are infertile associated with germ cell apoptosis and arrest of spermatocytes at meiotic prophase I, whereas UBR2-/- female mice show reduced fertility and partial lethality (5). Besides these functions, the N-end rule pathway plays a role in a variety of processes, including cardiovascular system, DNA repair, neurogenesis, olfaction, sensing oxygen and nitric oxide, G-protein signaling, chromosome segregation, peptide import, selective degradation of misfolded proteins, and apoptosis (reviewed in ref. 2; 3–8).
Spermatogenesis involves one of the most dramatic chromatin-remodeling processes, including synapsis and transcriptional silencing. During the pachytene stage of meiosis, the X and Y chromosomes are located into the nuclear periphery and form the XY body, where they achieve partial synapsis in the pseudoautosomal region (PAR). In the XY body, the transcription of genes linked to unsynapsed XY axes are silenced through a process called meiotic sex chromosome inactivation (MSCI) (9). Unpaired chromosomal regions are silenced during meiosis not only in the sex chromosomes but also in autosomal chromosomes through a pachytene checkpoint system called meiotic silencing of unsynapsed chromatin (MSUC) (9). Meiotic chromatin silencing involves differential histone modifications, including ubiquitination, phosphorylation, methylation and acetylation (10). Ubiquitinated histone H2A (uH2A) represents the most abundant ubiquitination substrate in mammals, comprising 5–15% of total H2A. uH2A is also the major Ub conjugate in male meiosis, which accumulates in the XY body and unsynapsed chromosomal regions undergoing MSUC (11). However, E3 ligases for H2A ubiquitination and the function of this modification in meiosis remain unknown. In somatic cells, Ring1B, a Polycomb group protein, mediates monoubiquitination of H2A, for chromatin inactivation processes, including X inactivation of female somatic cells and transcriptional silencing of Hox genes (12–15). Recently, 2A-HUB/hRUL138 has been shown to mediate H2A monoubiquitination for transcriptional repression of specific chemokine genes (16). In yeast, the E2 enzyme Rad6 and the E3 ligase Bre1 promote H2B monoubiquitination (17). Rad6 and its mammalian homologs, HR6A and HR6B, can function as E2 for the N-end rule proteolytic pathway (3; reviewed in ref. 2). In mammals, the human homolog of yeast Bre1, RNF20/RNF40, mediates H2B monoubiquitination (18, 19).
Results
UBR2 Localizes to Meiotic Chromatin Regions in Spermatocytes, Including Unsynapsed Axes Linked to Transcriptional Silencing.
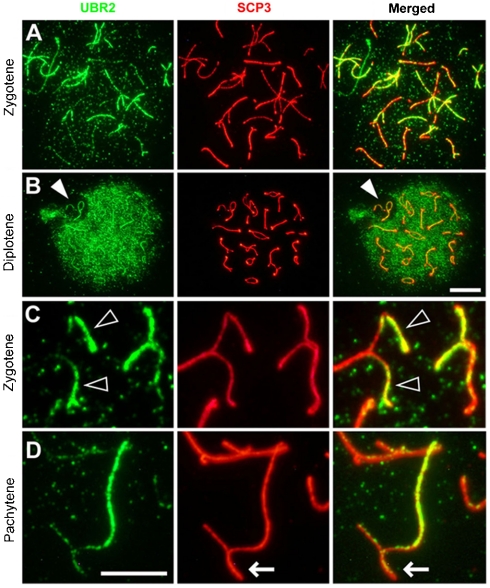
We tested whether UBR2 localizes to meiotic chromatin by using immunohistochemistry on spread chromosomes. UBR2 staining was detected in specific regions of meiotic chromosomes with dynamic changes in spatiotemporal localizations throughout meiotic prophase (Fig. 1 and Fig. S1). The specificity of anti-UBR2 antibody was verified by using a peptide competition assay (Fig. S2). At the leptotene stage when spermatocytes enter meiosis, UBR2 staining appeared as foci in the chromatin and as segment-like staining along the newly emerging axial elements (Fig. S1A). By the zygotene stage when homologous chromosomes are in the process of synapsing, UBR2 staining was enriched on unsynapsed axial regions (Fig. 1A and C). As synapsis of autosomal chromosomes expands throughout the axes, UBR2 staining gradually disappears from the axes that have achieved synapsis (Fig. S1C and F) and, instead, became enriched on the unpaired axes of the X and Y chromosomes that have already established synapsis along the PAR at late zygotene. In the XY body, UBR2 accumulated on unpaired XY axes but was not detected in the XY chromatin domain, a pattern that persisted through diplotene (Fig. 1B, arrowhead). These results reveal UBR2 as a chromatin-associated E3 ligase that marks MSCI.
Fig. 1.
Localization of UBR2 to meiotic chromosomes. (A) At zygotene, UBR2 staining is enriched on unsynapsed axial regions. (B) At diplotene, the number of UBR2 foci surge throughout the entire chromatin except for the XY chromatin domain (arrowhead). (C and D) Enlarged images for UBR2 localization to unsynapsed axial regions of autosomes at zygotene (C) and the sex chromosomes at early pachytene (D) Arrow, PAR. [Scale bar (A and B), 10 μm; (C and D), 5 μm.]
By the late pachytene stage when homologous chromosomes have completed synapsis and recombination, the number of UBR2 foci surged throughout the entire chromatin except for the XY chromatin domain (Fig. 1B). The intense UBR2 staining in postsynaptic chromatin correlated, spatially and temporally, to that of ubiquitinated histone H2A.
UBR2, in Combination with HR6B, Mediates Ubiquitination of Histone H2A.
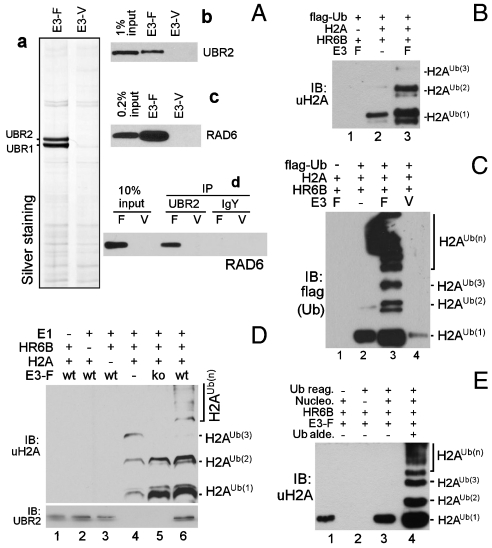
Given that UBR2 and uH2A mark MSCI (Fig. 1 and Fig. S1), we speculated that UBR2 may promote H2A ubiquitination as part of MSCI and MSUC. Loss-of-function experiments to determine the role of UBR2 and its homolog UBR1 in H2A ubiquitination in normally growing or DNA-damaged somatic cells were inconclusive, perhaps due to the presence of other E3 ligases (12–15, 20). To determine directly whether UBR2 and UBR1 can mediate H2A ubiquitination, we developed a synthetic-degron-based affinity procedure that allows rapid purification of N-recognins from mammalian tissues at low (75–150 mM) salt concentrations (see Methods), in which protein–protein interactions are preserved. By using Phe-peptide bearing a type-2 destabilizing N-terminal residue as an affinity ligand, we partially purified endogenous UBR2 (as a mixture with UBR1) by 1,000–3,000 folds from rat testes (Fig. 2Aa). An analogous procedure with Val-peptide bearing a stabilizing N-terminal residue typically yielded no detectible N-recognins (Fig. 2Ab).
Fig. 2.
UBR2, in combination with HR6B, mediates ubiquitination of H2A. (A) Partial purification of UBR2 from rat testes by using degron-bearing peptides (a). UBR2 interacts with RAD6 in the testes (B–D). Protein samples prepared by Phe-peptide (E3-F) or Val-peptide (E3-V) were subjected to immunoblotting for UBR2 (B), RAD6 (C), and anti-UBR2 immunoprecipitation and anti-RAD6 immunoblotting (D). F, Phe-peptide; V, Val-peptide. (B) In vitro ubiquitination assay (20 μL) with 100 ng E3-F prepared from rat testes, 30 ng HR6B, and 1 μg H2A in the presence of Ub activating reagents, including 1 μg FLAG-Ub and 100 ng E1, followed by immunoblotting for uH2A. (C) Same as (B) except that H2A ubiquitination was detected by using anti-FLAG immunoblotting. (D) In vitro ubiquitination assay with 100 ng E3-F prepared from +/+ (wt) and UBR2-/- mouse tissues (ko) and 2 μg free H2A. (E) Same as (B) except that 50 ng HR6B and 5 μg nucleosome (nucleo.) were used in the presence or absence of 0.05 mM Ub aldehyde (alde.).
Ub ligase requires E2 to promote protein ubiquitination. Amongst approximately 50 mammalian E2s, HR6B and its close homolog, HR6A, are the only E2s known to localize to meiotic chromatin and involved in MSCI (10, 21). (The mammalian genome encodes two HR6 isoforms, HR6A and HR6B, with 95% identity to each other, which we refer to as RAD6 when their biochemical properties cannot be distinguished.) Anti-RAD6/HR6B immunoblotting of precipitates indicated that RAD6 was copurified by Phe-peptide but not by Val-peptide (Fig. 2Ac), and immunoprecipitation of endogenous UBR2 from captured proteins brought down RAD6 (Fig. 2Ad). Thus, endogenous UBR2 and HR6B/RAD6 form an E2–E3 complex in these extracts.
To determine whether UBR2 and RAD6 cooperatively mediate the ubiquitination of H2A, we performed in vitro ubiquitinationassays by using free H2A. Human HR6B alone was able to mediate ubiquitination of H2A (Fig. 2B, lane 2), consistent with a previous observation that E2-14K/RAD6 can mediate E3-independent ubiquitination of histones in vitro (22). HR6B-dependent H2A ubiquitination was mainly restricted to monoubiquitination, except for weak bands that appear to be H2A conjugated with two Ub molecules (Fig. 2B, lane 2; Fig. 2C, lane 2). Importantly, E3 samples prepared with Phe-peptide (E3-F) promoted significantly not only monoubiquitination and diubiquitination (Fig. 2B, lanes 3 vs. 2) but also polyubiquitination (Fig. 2C, lanes 3 vs. 2). In contrast, protein samples prepared in parallel with Val-peptide (E3-V) did not promote and, moreover, often inhibited H2A ubiquitination (Fig. 2C, lane 4); under the same conditions, UbcH8 and UbcH10 did not show detectible H2A ubiquitination activity either in the presence or absence of E3-F. To determine whether UBR1 and UBR2 are responsible for H2A ubiquitination activity of E3-F, in vitro H2A ubiquitination assays were performed with E3-F, in which UBR1 and/or UBR2 were immunodepleted. Fig. S3A shows that immunodepletion of UBR1 and UBR2 from E3-F reduces significantly H2A ubiquitination. E3-F showed a moderate activity for H2A ubiquitination of chicken oligonucleosome (Fig. 2E, lanes 3 vs. 1) but was markedly increased in the presence of Ub aldehyde, an inhibitor of deubiquitinating enzymes (DUBs) (Fig. 2E, lanes 4 vs. 3), suggesting that E3-F-dependent H2A polyubiquitination may be controlled by DUBs. To determine whether UBR2-/- testes contain reduced E3 activity for H2A, E3-F was prepared after combining various tissues from wild-type and UBR2-/- mice. Compared with wild-type mice (Fig. 2D, lanes 6 vs. 4), E3-F from UBR2-/- mice showed markedly reduced E3 activity for ubiquitination of free H2A (Fig. 2D, lanes 5 vs. 6). Likewise, E3-F purified from UBR2-/- tissues produced reduced polyubiquitinated histone species from nucleosome as determined by anti-FLAG immunoblotting (Fig. S3B, lanes 3 vs. 2). Thus, UBR1 and UBR2, in combination with HR6B, can mediate monoubiquitination and polyubiquitination of H2A in vitro.
In situ hybridization and immunostaining on testicular cross sections revealed that UBR1 and UBR2 are prominently expressed in spermatogonia and spermatocytes, respectively (5). While mutually exclusive tissue distribution of UBR1 and UBR2 explains why UBR2-/- testes exhibit null phenotypes in meiosis, we do not exclude the possibility that UBR1 also participates in H2A ubiquitination during meiosis, as the localization of UBR1 to meiotic chromosomes has not been characterized extensively.
UBR2 Interacts with HR6B and H2A and Promotes the HR6B–H2A Interaction and the Discharge of Ubiquitin from HR6B ∼ Ub.
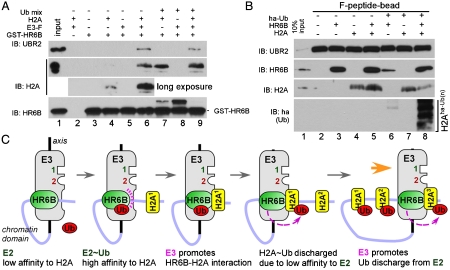
We determined the interaction of HR6B with UBR2 and H2A by using GST-pulldown assays with purified GST-HR6B, H2A, and UBR2 (as a mixture with UBR1). Fig. 3A shows that HR6B interacts with both UBR2 (Fig. 3A, lane 6) and its substrate H2A (Fig. 3A, lane 4, long exposure). The HR6B–H2A interaction appears to be intrinsically weak (Fig. 3A, lane 4) but can be drastically enhanced by E3-F (Fig. 3A, lanes 6 vs. 4, third from top), suggesting that the activity of E3-F in H2A ubiquitination involves the enhanced interaction and/or affinity between E3-bound HR6B and H2A. Likewise, the amount of UBR2 immobilized by GST-HR6B was increased in the presence of H2A (Fig. 3A, lanes 5 vs. 6 and lanes 8 vs. 9), indicative of a three-way cooperative interaction between E3, E2, and substrate. These results suggest that UBR2 binds both HR6B and H2A and enhances the interaction of HR6B with H2A, promoting the transfer of Ub from HR6B ∼ Ub to H2A.
Fig. 3.
Cooperative interactions between UBR2, HR6B, and H2A. (A) GST-pulldown assay (300 μl) with 200 ng GST-HR6B, 250 ng E3-F from rat testes, and 1 μg H2A, followed by immunoblotting for UBR2, H2A, and HR6B. Input: UBR2, 20%; H2A, 10%; HR6B, 10%. (B) Ubiquitination-coupled E3 binding assay. UBR2 (as a mixture with UBR1) immobilized on F-peptide-beads were mixed with HR6B and/or H2A in the presence of E1 and Ub activating reagents, followed by immunoblotting of proteins (HR6B, H2A, and uH2A) retained by X-peptide. (C) A model for H2A ubiquitination in the XY body by the UBR2-HR6B complex.
We determined the interaction of UBR2 with HR6B and H2A by using a ubiquitination-coupled binding assay, in which UBR2 (as a mixture with UBR1) immobilized on F-peptide-beads were mixed with HR6B or H2A in the presence or absence of E1 and Ub activating reagents (Fig. 3B). This assay can measure the levels of HR6B and H2A in complex with E3s while histone is being ubiquitinated. HR6B showed a robust retention (up to approximately 50%) to the F-peptide (Fig. 3B, lane 3) but not to the V-peptide (Fig. S3C). In the presence of E1 and Ub activating reagents, H2A promoted the binding of HR6B/HR6B ∼ Ub to E3 (Fig. 3B, lanes 8 vs. 6), which was also observed in an independent GST-pulldown assay (Fig. 3A, lanes 9 vs. 8). We also observed that a large portion (up to approximately 30%) of H2A binds to immobilized E3 (Fig. 3B, lane 4), and the UBR2–H2A interaction is further enhanced when E3 is engaged with E2 (Fig. 3B, lanes 5 vs. 4). In the presence of E1 and Ub activating reagents, H2A molecules were massively converted to polyubiquitin-conjugated species while they remain bound to the E3–E2 complex (Fig. 3B, lane 8, bottom). Concurrently, the steady-state level of E3-bound, unmodified H2A was markedly depleted, as determined by immunoblotting with anti-H2A antibody that cannot detect ubiquitinated H2A, yet another evidence for the E2-H2A transfer of Ub. Thus, the cooperative interaction of UBR2, HR6B/HR6B ∼ Ub, and H2A enhances the overall affinity between each other, promoting the HR6B-H2A transfer of Ub.
One notable property of HR6B is that the interaction of HR6B to H2A is drastically accelerated when HR6B is charged with Ub (Fig. 3A, lanes 7 vs. 4), suggesting that the HR6B-substrate affinity is reduced once HR6B ∼ Ub transfers its cargo to H2A. This Ub-dependent oscillation of the E2-substrate affinity is consistent with a model (Fig. 3C), in which HR6B ∼ Ub (with high affinity to H2A) binds to H2A, and Ub-discharged HR6B (with low affinity to H2A) dissociates from H2A ∼ Ub, in part explaining how an E2–E3 pair mediates histone ubiquitination cycles while scanning chromatin (see Discussion).
Chromosome-Wide Ubiquitination of H2A is Disrupted in UBR2-Deficient Spermatocytes.
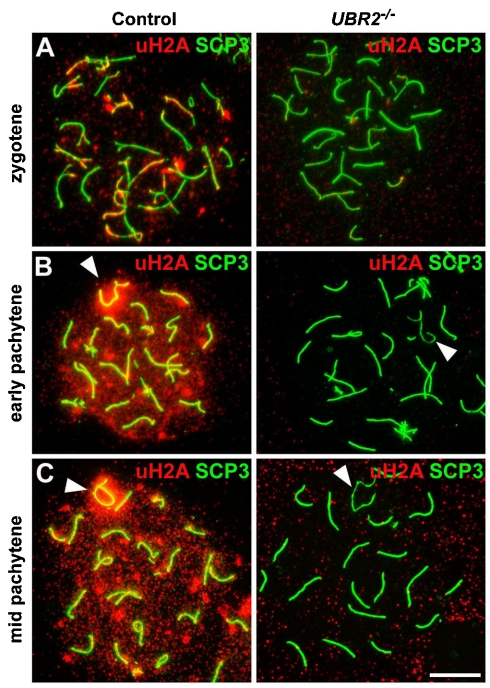
To determine whether the inactivation of UBR2 impairs the ubiquitination of H2A during meiosis, an antibody to uH2A, which can detect both monoubiquitinated and polyubiquitinated H2A (see Fig. 2E), was applied on spread chromosomes from control and UBR2-/- spermatocytes. In control cells, uH2A staining was first detected at late-zygotene mainly on unsynapsed axes and as foci-like signals on the chromatin domain (Fig. 4A). As spermatocytes enter pachytene, uH2A signals surged throughout the entire chromatin, with the highest level in the XY body (Fig. 4B and 4C, arrowheads). Overall, uH2A showed remarkable similarity to UBR2 in temporal and spatial localization on meiotic chromosomes at zygotene and pachytene, except that the UBR2 staining is almost completely excluded from the XY chromatin domain (Fig. 4C vs. Fig. S1D, arrowhead), consistent with the model presented in Fig. 3C (see Discussion). Importantly, the uH2A staining both in axes and the chromatin domain was drastically diminished in UBR2-/- spermatocytes (Fig. 4, right panels) throughout the entire meiotic stages, which was reproducibly observed in 154 nuclei from P16 testes. Downregulation of uH2A in mutant spermatocytes prior to the onset of meiotic defects eliminates the possibility that H2A ubiquitination is nonspecifically affected due to developmental arrest. These results suggest that chromosome-wide ubiquitination of H2A is disrupted in UBR2-/- spermatocytes.
Fig. 4.
UBR2-deficient spermatocytes are impaired in chromosome-wide ubiquitination of H2A. Immunostaining of uH2A (red) was performed for meiotic chromosomes from control and UBR2-/- spermatocytes at zygotene (A), early pachytene (B), and midpachytene (C). The X–Y pair is indicated by arrowheads. The majority of UBR2-/- spermatocytes are arrested at or before early pachytene (5). The pachytene nuclei from UBR2-/- mice shown in this study are typical for those surviving spermatocytes. (Scale bar, 10 μm.)
UBR2 Is Required for Transcriptional Silencing of Genes Linked to Unsynapsed Axes of the Sex Chromosomes in Meiosis.
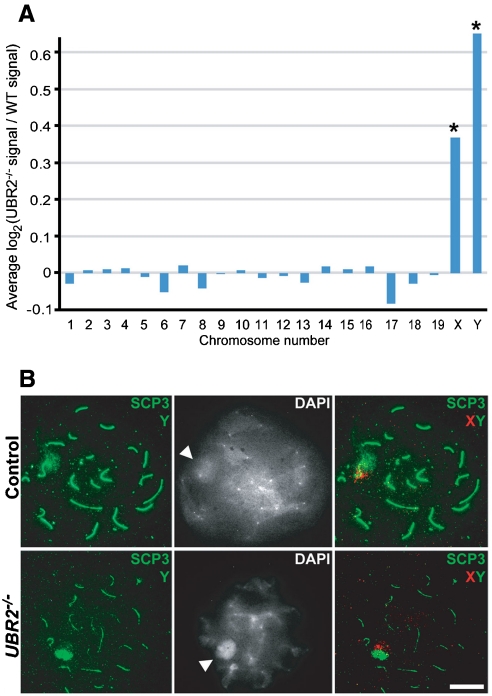
As both UBR2 and uH2A mark meiotic chromatin inactivation (Figs. 1 and 4), we determined whether UBR2-knockout would impair MSCI. When spermatocytes enter the pachytene stage, the X and Y chromosomes condense to form the sex body, where the chromatin linked to unpaired XY axes is transcriptionally silenced. We employed quantitative real-time PCR to examine the expression of XY-linked genes in 4-week-old +/+ and UBR2-/- testes. Fig. S4A shows that UBR2 inactivation results in marked upregulation for the genes (MeCP2, Hprt, Hdac6, and Rbmy) known to be silenced at pachytene, but not for those (Ube2a, Ube1x, and Pctk1; indicated by asterisks) not subject to MSCI (23), indicating a strong correlation between UBR2 function and MSCI. To characterize the role of UBR2 in silencing at the chromosome level, we performed microarray-based analysis of approximately 22,000 probe sets by using +/+ and UBR2-/- testes at P17 when meiosis has advanced to late pachytene (the accession no. GSE18824 at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/)). When the data were sorted for individual chromosomes, the average mRNA levels of genes encoded by autosomal chromosomes were very similar in two groups (Fig. 5A). Strikingly, UBR2-/- testes showed overall upregulation of 542 X-specific and 15 Y-specific probes when compared to +/+ testes (Fig. 5A; Table S1), indicative of chromosome-wide failure in MSCI. Consistent with these results is previous Northern blot analysis, in which five (Fthl17, Nxf2, Taf2q, Tktl1, and Rbmy) out of seven randomly selected XY-linked genes (Fsc1, Fthl17, Nxf2, Pramel3, Taf2q, Tktl1, and Rbmy) are upregulated in 6-week-old UBR2-/- testes compared to +/+ controls (5). These results suggest that insufficiency in UBR2-dependent histone ubiquitination causes defects in transcriptional silencing of genes linked to the sex chromosomes.
Fig. 5.
UBR2 is required for transcriptional silencing of the X and Y chromosomes in male meiosis. (A) Microarray analysis of control and UBR2-/- testes at P16. (B) Chromosome painting for X (red, arrowhead) and Y (green, arrowhead) coupled with SCP3 immunostaining (green) in control and UBR2-/- spermatocytes. Dense peripheral DAPI staining indicates the sex body (arrowheads). (Scale bar, 10 μm.)
Transcriptional silencing of XY-linked genes is associated with physical exclusion of proteins involved in transcription, such as RNA polymerase II, from the chromatin. We determined whether the exclusion of RNA polymerase II from XY is impaired in UBR2-/- mice using immunostaining on spread chromosomes. In contrast to +/+ spermatocytes at pachytene wherein RNA polymerase II was found to be excluded from the XY chromosomes, the majority of UBR2-/- spermatocytes showed a uniform distribution of RNA polymerase II throughout the entire nucleus (Fig. S4C). We next determined whether UBR2 is required for XY pairing and XY body formation. Fluorescent in situ hybridization (FISH) analysis using probes for the X and Y chromosomes showed that in the majority of UBR2-/- spermatocytes at the pachytene-like stage, X–Y pairing normally occurred within the DAPI-rich sex body (Fig. 5B). Moreover, H- and E-stained sections of seminiferous tubules from 4-week-old +/+ and UBR2-/- mice revealed that UBR2-/- spermatocytes contained distinct XY bodies at the nuclear periphery (Fig. S4B). Although we do not exclude the possibility of subtle defects in the mutants, these results suggest that XY pairing and XY body formation do not strictly require UBR2. Collectively, our results suggest that UBR2 is required for MSCI but is not critical for XY pairing and XY body formation.
The Localization Pattern of UBR2 to Unsynapsed Axes is Distinct and Independent from the BRCA1-Dependent MSCI Pathway.
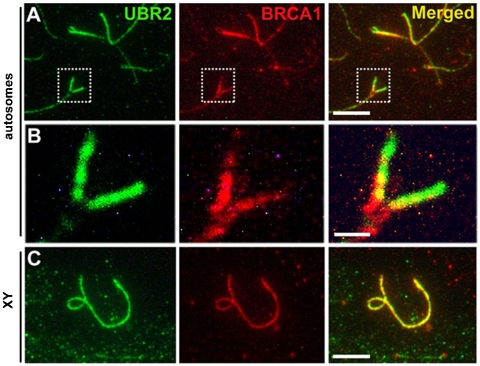
Recent studies revealed that meiotic chromatin inactivation involves a histone phosphorylation pathway, in which BRCA1 is accumulated on unsynapsed XY axes where it recruits ATR, which in turn phosphorylates the histone H2AX, an essential histone modification for MSCI and possibly MSUC as well (9). To determine whether UBR2 functionally interacts with this histone phosphorylation, we compared the spatiotemporal localizations of UBR2, BRCA1, and ATR on meiotic chromosomes. These MSCI proteins accumulated on unpaired axes of autosomes and sex chromosomes in a temporally synchronized manner (Figs. 6 and S5A–C). Intriguingly, they showed distinct spatial localization patterns along the unsynapsed axes, in particular in autosomes, in that the UBR2 staining on unsynapsed axes was virtually excluded from the synaptic folk, whereas the axial staining of BRCA1 and ATR was relatively weaker in the region proximal to the telomere (Figs. 6B and S5B). Thus, histone ubiquitination and phosphorylation may be differentially regulated along the chromosomal axes as illustrated in Fig. S5D.
Fig. 6.
The localization pattern of UBR2 to unsynapsed axial regions of autosomes is distinct from that of BRCA1. (A and C) Localization of UBR2 (green) and BRCA1 (red) on unsynapsed axes of the autosomes at zygotene (A) and the sex chromosomes at pachytene (C). (B) Enlarged images for chromosomal regions indicated by the insets in (A). [Scale bar (A and C), 5 μm; (B) 1 μm.]
We determined whether the recruitment of UBR2 and BRCA1 to meiotic chromosomes requires each other’s function. The staining patterns of BRCA1 and ATR were not altered significantly in UBR2-/- spermatocytes at zygotene through pachytene (Fig. S6A–C). Moreover, phosphorylated H2AX (γH2AX) normally localized to the XY body of pachytene-like UBR2-/- spermatocytes (Fig. S6D). Next, we examined the impact of BRCA1 deficiency on UBR2 localization by using spermatocytes from BRCA1Δ11/Δ11p53+/- mice (24), which produce a truncated BRCA1 polypeptide lacking the exon 11-encoded amino acids in the background of p53 haploinsufficiency. Fig. S7 shows that the UBR2 staining on meiotic chromosomes is not significantly altered in BRCA1-deficient spermatocytes. These results suggest that the recruitments of UBR2 and BRCA1/ATR to unsynapsed axes are mutually independent and that UBR2-dependent histone ubiquitination is not essential for the function of BRCA1-dependent histone phosphorylation pathway during MSCI.
Discussion.
In this study we demonstrate that UBR2, the major component of the N-end rule proteolytic pathway (reviewed in ref. 2; 3, 5), plays a critical role in chromatin inactivation and chromosome-wide transcriptional silencing during meiosis via ubiquitination of histone H2A. Our results also provide molecular basis for several seemingly unrelated observations that UBR2-/- mice are infertile associated with meiotic arrest and germ cell apoptosis (5) and that HR6B-/- mice are infertile associated with impaired transcriptional silencing of some of XY-linked genes (10; see also references therein). Notably, the S. cerevisiae homolog of HR6B, Rad6, in complex with Ubr1 can promote the proteolysis of N-end rule substrates, whereas the same E2 paired with Bre1 can mediate gene silencing via monoubiquitination of H2B, the most abundant histone modification in yeast, as opposed to H2A ubiquitination which is not detected in yeast (15, 25). Thus, HR6B has an evolutionarily conserved function in N-end rule-dependent proteolysis and histone ubiquitination in the context of chromatin inactivation.
Numerous studies revealed fairly detailed mechanisms of how an E3–E2 complex mediates the ubiquitination of short-lived proteins in the cytoplasm via Lys48 residue of Ub, which usually leads to degradation through the 26S proteasome complex. In contrast, analogous mechanisms for substrates associated to chromatin (or in a nucleosome) remain poorly understood. We here demonstrate that a three-way cooperative interaction of UBR2, HR6B/HR6B ∼ Ub, and H2A promotes H2A ubiquitination. It is generally accepted that a RING E3 ligase is a bisubstrate enzyme that binds both substrate and E2 and accelerates the E2-substrate transfer of Ub. Under this paradigm, the E3–substrate and E2–E3 interactions are the primary determinant in substrate specificity and processivity, whereas the E2–substrate interaction is usually not detectible or transiently induced by E3. We note that HR6B shows significant affinity to H2A in the absence of E3, although the interaction of H2A with a free HR6B molecule is drastically enhanced by E3, suggesting that HR6B has an intrinsic property to interact with H2A and mediate monoubiquitination of H2A in vitro. What is then the role of UBR2 in H2A ubiquitination? We propose a model (Fig. 3C), in which H2A ubiquitination in the XY chromatin domain is initiated by long-distance chromatin interaction between UBR2 on chromosomal axes (see Fig. 1B, arrowhead) and an E2 molecule (for example, HR6B) on the chromatin domain (10, 21), the E2 being associated with a processivity factor that allows rapid moving through chromatin. Once charged with Ub, HR6B ∼ Ub gains high affinity to H2A and, thus, binds and transfers Ub to H2A1. In these sequential events, UBR2 promotes the HR6B–H2A interaction and the discharge of Ub from HR6B. Once Ub is discharged, HR6B (now with low affinity to H2A) is dissociated from H2A1, charged again with Ub by E1, and pushed though chromatin to the position H2A2 by the action of the processivity factor, where HR6B ∼ Ub (with high affinity to H2A) binds H2A2. This HR6B ∼ Ub–H2A2 interaction will temporarily halt the movement, providing enough time for ubiquitination of H2A2. After each HR6B-H2A transfer of ubiquitin, HR6B may be repositioned on the surface of E3 or dissociated from E3 until a new E2–E3 interaction is established.
Materials and Methods
For descriptions of materials and methods, including antibodies and PCR primers, see SI Text and Table S2. Staining on spread chromosomes, quantitative RT-PCR, microarray analysis, and FISH were performed by using standard techniques, which are described in SI Text. Total RNA from UBR2-/- testes at P16 and littermate controls were used for microarray analysis with 22,000 probes. Quantitative PCR was performed in duplicates by using Platinum SYBR Green (Invitrogen) with ABI 7300/7500 real-time PCR system and primer sequences listed in Table S2. FITC-labeled Y chromosome probe and Cy3-labeled X chromosome probes from Cambio (Cambridge, UK) were used for FISH according to the manufacturer’s instruction. Endogenous UBR2 was partially purified from extracts prepared from rat or mouse testes by using synthetic degrons essentially as described (6), with major modifications. All procedures for in vitro ubiquitination assays, ubiquitination-coupled E3 binding assays, and GST-E2 binding assays are described in SI Text.
Supplementary Material
Acknowledgments.
We are grateful to James Turner at Medical Research Council of National Institute for Medical Research (UK) for slides containing spread meiotic chromosomes from Brca1Δ11/Δ11p53+/- mice, to Jai Wha Seo for genotyping and maintaining mouse strains, to the Genomics and Proteomics Core Laboratories of the University of Pittsburgh for microarray analysis, and to Takafumi Tasaki and other Kwon lab members for critical reading of the manuscript. This work was in part supported by National Institutes of Health grants (to Y.T.K. and Y.J.) and the World Class University Program Grant R31-2008-000-10103-0 (to I.M.J. and Y.T.K.) from the Korean Ministry of Education, Science and Technology and the National Research Foundation of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910267107/DCSupplemental.
References
- 1.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Tasaki T, Kwon YT. The mammalian N-end rule pathway: New insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 5.Kwon YT, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An JY, et al. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 10.Baarends WM, et al. Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J Cell Sci. 2007;120:1841–1851. doi: 10.1242/jcs.03451. [DOI] [PubMed] [Google Scholar]
- 11.Baarends WM, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 14.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, et al. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 21.van der Laan R, et al. Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J Cell Sci. 2004;117:5023–5033. doi: 10.1242/jcs.01368. [DOI] [PubMed] [Google Scholar]
- 22.Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988;263:13268–13275. [PubMed] [Google Scholar]
- 23.Namekawa SH, et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 24.Turner JM, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 25.Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.