Abstract
The DNA polymerases involved in DNA replication achieve high processivity of nucleotide incorporation by forming a complex with processivity factors. A model system for replicative DNA polymerases, the bacteriophage T7 DNA polymerase (gp5), encoded by gene 5, forms a tight, 1∶1 complex with Escherichia coli thioredoxin. By a mechanism that is not fully understood, thioredoxin acts as a processivity factor and converts gp5 from a distributive polymerase into a highly processive one. We use a single-molecule imaging approach to visualize the interaction of fluorescently labeled T7 DNA polymerase with double-stranded DNA. We have observed T7 gp5, both with and without thioredoxin, binding nonspecifically to double-stranded DNA and diffusing along the duplex. The gp5/thioredoxin complex remains tightly bound to the DNA while diffusing, whereas gp5 without thioredoxin undergoes frequent dissociation from and rebinding to the DNA. These observations suggest that thioredoxin increases the processivity of T7 DNA polymerase by suppressing microscopic hopping on and off the DNA and keeping the complex tightly bound to the duplex.
Keywords: DNA replication, processivity, facilitated diffusion, sliding, single-molecule imaging
The bacteriophage T7 replisome is an elegantly simple, well-studied model system of DNA replication (1). Replicative DNA synthesis is supported by T7 DNA polymerase (gp5), in a tight, 1∶1 complex with Escherichia coli thioredoxin, which will henceforth be referred to as gp5/trx. By itself, the 80-kDa gp5 synthesizes DNA in a distributive fashion and is capable of adding only a few nucleotides to the 3′ end of a primer before dissociating from the primer template (2). Association of gp5 with the 12-kDa E. coli host protein thioredoxin results in a stable 1∶1 complex (KD = 5 nM) and drastically increases the processivity of DNA synthesis. This interaction is critical for replication of T7 DNA—the phage cannot reproduce in thioredoxin null strains of E. coli (3, 4). The affinity of gp5/trx for primer-template DNA is increased at least 20-fold compared to that of gp5 alone (5), resulting in a complex that is capable of polymerizing hundreds of nucleotides before dissociation (2).
Thioredoxin binds to gp5 via a 71-amino acid loop that resides between two alpha helices in the thumb domain and is called the thioredoxin-binding domain (TBD) (6). This loop is absent from other members of the Pol I family of DNA polymerases, but when inserted into E. coli DNA polymerase I at the homologous site, the resulting chimera displays a thioredoxin-dependent increase in processivity (7). The TBD also functions as a scaffold for the assembly of the replisome. The binding of thioredoxin structures the domain and creates two small solvent exposed loops containing binding sites for the T7 single-stranded DNA-binding protein and the T7 helicase (8, 9). Thioredoxin itself does not bind to DNA, and only one residue on thioredoxin has been implicated in binding to any of the other replisome components other than gp5 (10).
Crystal structures of gp5/trx bound to primer-template DNA reveal that the TBD is extended over the duplex region of the primer-template DNA (6, 11). However, they do not reveal an obvious structural mechanism by which the binding of thioredoxin increases the processivity of gp5. Thioredoxin is rotated away from the DNA, and does not appear to directly interact with it. The protein complex does not completely encircle the duplex region of the DNA in the crystal structure, but the TBD has a high degree of conformational mobility compared to the rest of the protein and a shift of this domain could potentially result in the formation of such a clamp-like structure (6). Remodeling of the TBD may also orient several basic residues toward the DNA, providing additional contacts to the DNA backbone, and leading to a higher affinity for the primer-template through increased electrostatic interactions (5). In the absence of a more direct way to observe the dynamic interplay between the T7 DNA polymerase and DNA, it has been difficult to study the molecular nature of the polymerase–DNA interactions and to understand the role of thioredoxin in processivity.
In recent years, single-molecule tools have greatly contributed to our understanding of DNA–protein interactions and nucleic-acid enzymes. The ability to observe, in real time, interactions of a single protein with a single DNA molecule allows for the detection of transient intermediates and kinetic details that are difficult to obtain with ensemble-averaging biochemical assays. Single-molecule techniques have been used to study diffusion of a variety of proteins along DNA, such as transcription factors (12, 13), DNA repair factors (14, 15), processivity factors (16, 17), RNA polymerases (18–20), and restriction enzymes (21). In this report, we present single-molecule imaging experiments that visualize the interaction of individual T7 DNA polymerases with DNA, demonstrate that the protein complex diffuses along duplex DNA, and show that a change in the diffusional mode of T7 DNA polymerase is mediated by the binding of its processivity factor, thioredoxin. To our knowledge, our results represent a unique reported observation of a DNA polymerase diffusing along DNA and provide a unique example of a protein or protein complex that is capable of converting between a sliding and hopping mode in response to a specific trigger. In addition, they permit us to glean insight into the mechanisms underlying thioredoxin-mediated processivity.
Results
Single-Molecule Observation of T7 DNA Polymerase Diffusing Along Duplex DNA.
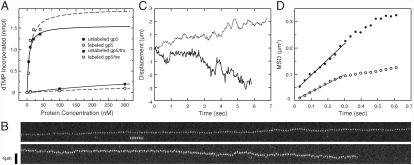
To understand the role of thioredoxin in stabilizing polymerase–DNA interactions, we use single-molecule fluorescence imaging to visualize individual, fluorescently labeled T7 DNA polymerase molecules interacting with double-stranded DNA. Copurified gp5/trx was labeled with an amine-reactive derivative of Alexa Fluor 555 as described in Materials and Methods. Gp5 purified free of thioredoxin was similarly labeled. We used a solution-phase DNA polymerization assay to demonstrate that all labeled proteins retained the ability to add nucleotides to the 3′ end of a primer, similar to that of unlabeled proteins (Fig. 1A).
Fig. 1.
Labeled T7 DNA polymerase retains primer extension activity and diffuses along the length of λ-phage DNA. (A) Amount of dTMP incorporated as T7 DNA polymerase extends a primer annealed to circular, single-stranded M13 DNA is dependent on protein concentration. Labeled and unlabeled gp5/trx show nearly identical activity. Labeled and unlabeled gp5 also have similar activity, but are substantially less active than gp5/trx due to the absence of thioredoxin. (B) Kymographs of gp5/trx complexes diffusing along DNA. Each kymograph represents 222 exposures of 30 msec each. The direction of flow is down. (C) Trajectories of the gp5/trx complexes obtained from the kymographs shown in (B). (D) MSD vs. time calculated from the trajectories in (B) and (C). Solid circles correspond to the solid line trajectory and open circles correspond to the dotted trajectory. The first ten data points are fit with a linear dependence and the slope was used to determine the linear diffusion coefficient.
To observe the interactions of the fluorescently labeled T7 DNA polymerase with double-stranded DNA, we tethered biotinylated, duplex, linearized λ-phage DNA to a streptavidin-coated coverslip (SI Text). Aqueous buffer is flowed through a flow channel assembled onto the coverslip, stretching the DNA close and parallel to the glass surface by hydrodynamic force. Derivatization of the glass surface with high molecular-weight poly (ethylene glycol) minimizes nonspecific interactions between glass and DNA or protein. We use total internal reflection (TIR) illumination to reduce the background contributed by fluorescently labeled proteins in solution while exciting the fluorescently labeled proteins that are bound to the DNA.
When introduced into the flow cell at subnanomolar concentrations, fluorescently labeled gp5/trx binds nonspecifically along the length of the double-stranded λ DNA. This binding does not require the presence of free nucleotides or magnesium, nor does it take place exclusively at 3′ termini. At protein concentrations of 4–8 picomolar, so few fluorescently labeled molecules bind to the DNA that, on average, we observe no more than one fluorescently labeled gp5/trx complex bound to any one DNA molecule at a time. Likewise, we can observe single molecules of labeled gp5, without thioredoxin, binding to the DNA at protein concentrations of 40–80 picomolar. In both cases, these molecules were observed to move randomly and bidirectionally along the DNA. We recorded movies of proteins undergoing one-dimensional Brownian diffusion while bound to the DNA (Fig. 1B). We determined protein positions with high precision by fitting their observed point-spread functions to a 2-dimensional Gaussian profile and subsequently traced the position trajectories of individual proteins (Fig. 1C). The diffusion trajectories were corrected for flow-induced drift by determining the drift velocity of a large number of particles under the same experimental conditions and subtracting the mean value from each individual trajectory (see SI Text).
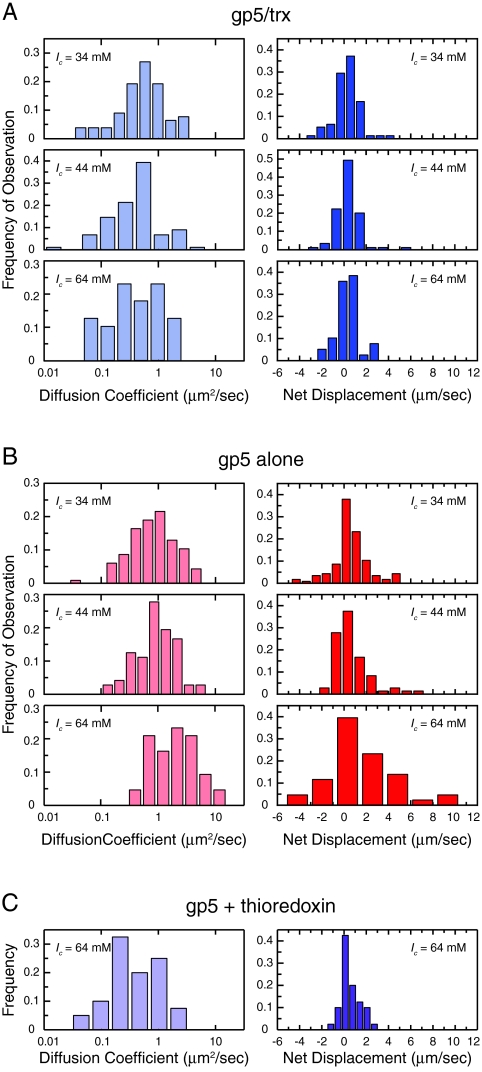
For most protein trajectories the mean-squared displacement (MSD) versus time was approximately linear, indicating diffusive movement over the timescale included in the analysis (Fig. 1D). At shorter timescales, longitudinal DNA fluctuations dominate the observed protein movement. Because the linear diffusion coefficient, D1, can be obtained from the proportionality constant, MSD = 2D1t, the slope of the linear fit to the MSD versus time for the first ten time points was used to determine D1 for each individual molecule. This fit was not constrained to pass through the origin to exclude the effect of the short-lived DNA fluctuations on the measurement (Fig. 1D). Histograms of the observed diffusion coefficients of many individual molecules reveal broad distributions (Fig. 2), caused by an increased variance of the MSDs for shorter trajectories. However, because a large number of individual measurements have been obtained, the average diffusion coefficient can be determined with a high degree of confidence.
Fig. 2.
Observed diffusion coefficients and net displacements. Histograms of observed diffusion coefficients, plotted on a log scale (Left), and net displacements, plotted on a linear scale (Right). The total displacement from the start of each trajectory to its end is normalized for the length of the trajectory. Positive displacement is in the direction of the buffer flow. Each histogram consists of data collected under identical conditions. (A) For gp5/trx, the diffusion coefficient distribution remains essentially unchanged over all experimental conditions. Net displacement distributions are symmetric, centered near zero, and unaffected by the ionic strength of the experimental buffer. (B) For gp5, in the absence of thioredoxin the diffusion coefficient distribution shifts toward higher values as the ionic strength of the experimental buffer increases. The normalized net displacement distributions are skewed in the direction of buffer flow, with the mean value increasing with the ionic strength of the experimental buffer. (C) The distributions of the observed diffusion coefficients and net displacements of gp5 + thioredoxin at a buffer ionic strength of 64 mM are most similar to those of gp5/trx.
Salt Dependence of Diffusion Reports on Translocation Mode.
A protein that diffuses along DNA can use one of two mechanisms—sliding or hopping (22). A sliding protein remains nonspecifically bound to the DNA while transferring from one binding site to another along the DNA. Because the movement does not require dissociation from the DNA, the diffusion coefficient of a protein that uses the sliding mechanism is not expected to be affected by the ionic strength of the experimental buffer. On the other hand, a protein that diffuses by hopping microscopically dissociates from the DNA many times during each macroscopic binding event. The protein moves just far enough away from the DNA for cations to recondense on the phosphate backbone of the DNA, but remains close enough that it is highly likely to rapidly rebind to the DNA at a nearby site. These microscopic dissociation events are much too small to be observed in our experiments, but increasing ionic strength is expected to increase the fraction of the time the protein is not electrostatically interacting with DNA, leading to an effective increase of the measured diffusion constant.
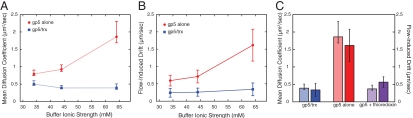
We studied the diffusive behavior of T7 DNA polymerase in experimental buffers with ionic strengths (Ic) ranging from 34–64 mM (Materials and Methods). For gp5/trx, the mean diffusion coefficient remains constant at approximately 0.4 μm2/ sec (3.5 × 106 bp2/ sec), even as the ionic strength of the experimental buffer is increased (Figs. 2A and 3A and SI Text). This suggests that gp5/trx uses the sliding mechanism to diffuse along DNA, and does not dissociate from the DNA during the diffusion events. However, the mean diffusion coefficient of gp5 in the absence of thioredoxin increases with the ionic strength of the experimental buffer (Figs. 2B and 3A and SI Text), from 0.79(+0.11,-0.044) μm2/ sec (6.9 × 106 bp2/ sec) at Ic = 34 mM to 1.86(+0.44,-0.17) μm2/ sec (16 × 106 bp2/ sec) at Ic = 64 mM. Error is the standard error of the geometric mean, which is asymmetric (see SI Text for calculation method and table of values). This observation indicates that gp5 uses the hopping mechanism and transiently dissociates from the DNA during diffusion events.
Fig. 3.
Salt dependence of diffusion coefficient and flow-induced drift. (A) In the absence of thioredoxin, the diffusion coefficient of gp5 increases with the ionic strength of the experimental buffer; gp5/trx diffusion is insensitive to changes in experimental buffer composition. Data points and error bars represent the geometric mean and standard error of the geometric mean, respectively, and are calculated as described in SI Text.
(B) The mean normalized net displacement indicates the amount of flow-induced drift in the diffusion of molecules along the DNA. As the ionic strength of the buffer increases, gp5 trajectories increasingly drift in the direction of the buffer flow. Trajectories of gp5/trx show minimal drift that remains constant with increasing buffer ionic strength. Data points and error bars represent the mean and the SEM, respectively.
(C) Summary of data at a buffer ionic strength of 64 mM for gp5/trx, gp5 alone, and gp5 + thioredoxin. Light colored bars represent the mean diffusion coefficient observed, and darker colored bars represent the flow-induced drift for each protein. Error bars correspond to the SEM. In the absence of thioredoxin, gp5 has a significantly higher mean diffusion coefficient, and greater degree of flow-induced drift. When an excess of unlabeled thioredoxin is added to the experiment, the diffusion coefficient and flow-induced drift of gp5 both revert to values indistinguishable from those observed for labeled gp5/trx.
Theoretical Estimates of Diffusion Kinetics Suggest Helical Sliding of gp5/trx.
The upper limit for the diffusion coefficient of a protein diffusing along the DNA is described by the Einstein relation for Brownian motion
 |
where ξ is the frictional coefficient representing the frictional coupling between the protein and solution, kB the Boltzmann constant, and T the absolute temperature (298 K). For simple, one-dimensional movement, the frictional coefficient is dependent only on the Stokes drag force experienced by the protein as it moves: ξ = 6πηa, where η is the viscosity of the surrounding solution ( for water at 25 °C) and a is the radius of the protein [4.6 nm for gp5/trx (23)]. A protein that slides on DNA while maintaining electrostatic contact with the backbone would be expected to rotate as it tracks along the DNA helix. Any linear movement will therefore also include a rotational component, increasing the rate of energy dissipation. Schurr (24) derived the effective friction factor for a protein that is forced to rotate as it diffuses along DNA
for water at 25 °C) and a is the radius of the protein [4.6 nm for gp5/trx (23)]. A protein that slides on DNA while maintaining electrostatic contact with the backbone would be expected to rotate as it tracks along the DNA helix. Any linear movement will therefore also include a rotational component, increasing the rate of energy dissipation. Schurr (24) derived the effective friction factor for a protein that is forced to rotate as it diffuses along DNA
 |
For gp5/trx, rotation around the DNA represents a 97-fold increase in the effective friction factor, with a predicted upper limit for the diffusion coefficient of 0.55 μm2/ sec (4.8 × 106 bp2/ sec). The mean diffusion coefficient of gp5/trx does not exceed this limit for any experimental condition. However, the mean diffusion coefficient for gp5 in the absence of thioredoxin exceeds this limit even at the lowest buffer ionic strength. This suggests that while gp5/trx is forced to rotate along the helical pitch of the DNA as it diffuses, gp5 without thioredoxin is free to move along the DNA in a manner unconstrained by the helical nature of the DNA. This is consistent with the notion that gp5/trx slides along the DNA, while gp5 alone hops.
Flow-Induced Drift Reports on Diffusion Mechanism.
For a protein diffusing linearly along a DNA molecule, the distribution of the net displacement for each trajectory, normalized for its length, should be centered at zero. Deviation from net zero movement suggests a bias in the direction the protein moves at each step in its random walk along the DNA. In our experiments, proteins will be subjected to a small amount of Stokes drag force due to the fact that buffer solution is flowing along the DNA. The Stokes drag force for an object close to the surface is  , where ν is the flow velocity and d is the distance from the surface. Based on our experimental geometry we estimate an average protein distance from the surface of 0.2 µm (SI Text). Assuming a parabolic flow velocity profile within the 100 µm high flow channel, and an average flow velocity of 0.83 cm/ sec, the flow velocity at 0.2 µm from the bottom surface of the flow cell will be 100 μm/ sec (13). Given the radius of the protein, the estimated Stokes drag force due to buffer flow at this velocity is around 8 fN. A protein that is dissociated from the DNA will be free to move along with the buffer, while one that is bound to the DNA will simply be biased in its direction of movement. Proteins that slide along the DNA should, therefore, exhibit relatively small drift in the direction of the buffer flow, and the degree of drift should be insensitive to the ionic strength of the experimental buffer. Proteins that hop, however, would be expected to exhibit a greater degree of drift in the direction of the buffer flow, and that drift should increase as the ionic strength of the experimental buffer increases. As a measure of this flow-induced drift, we use the mean of the net, normalized displacements for the set of diffusion trajectories obtained at each ionic strength. For gp5/trx, there is a slight bias in the normalized net displacement in the direction of buffer flow, but the mean remains constant at approximately 0.3 μm/ sec (900 bp/ sec) as the ionic strength of the experimental buffer is increased (Figs. 2A and 3B and SI Text). In the case of gp5 alone, the flow-induced drift is more pronounced, and the mean increases with buffer ionic strength (Figs. 2B and 3B and SI Text), from 0.60( ± 0.16) μm/ sec (1700 bp/ sec) at Ic = 34 mM to 1.62( ± 0.46) μm/ sec (4800 bp/ sec) at Ic = 64 mM. Error is the standard error of the arithmetic mean. These observations further support the hypothesis that gp5 slides in close contact with the DNA when complexed with thioredoxin, but that in the absence of thioredoxin, gp5 microscopically hops as it diffuses along the DNA.
, where ν is the flow velocity and d is the distance from the surface. Based on our experimental geometry we estimate an average protein distance from the surface of 0.2 µm (SI Text). Assuming a parabolic flow velocity profile within the 100 µm high flow channel, and an average flow velocity of 0.83 cm/ sec, the flow velocity at 0.2 µm from the bottom surface of the flow cell will be 100 μm/ sec (13). Given the radius of the protein, the estimated Stokes drag force due to buffer flow at this velocity is around 8 fN. A protein that is dissociated from the DNA will be free to move along with the buffer, while one that is bound to the DNA will simply be biased in its direction of movement. Proteins that slide along the DNA should, therefore, exhibit relatively small drift in the direction of the buffer flow, and the degree of drift should be insensitive to the ionic strength of the experimental buffer. Proteins that hop, however, would be expected to exhibit a greater degree of drift in the direction of the buffer flow, and that drift should increase as the ionic strength of the experimental buffer increases. As a measure of this flow-induced drift, we use the mean of the net, normalized displacements for the set of diffusion trajectories obtained at each ionic strength. For gp5/trx, there is a slight bias in the normalized net displacement in the direction of buffer flow, but the mean remains constant at approximately 0.3 μm/ sec (900 bp/ sec) as the ionic strength of the experimental buffer is increased (Figs. 2A and 3B and SI Text). In the case of gp5 alone, the flow-induced drift is more pronounced, and the mean increases with buffer ionic strength (Figs. 2B and 3B and SI Text), from 0.60( ± 0.16) μm/ sec (1700 bp/ sec) at Ic = 34 mM to 1.62( ± 0.46) μm/ sec (4800 bp/ sec) at Ic = 64 mM. Error is the standard error of the arithmetic mean. These observations further support the hypothesis that gp5 slides in close contact with the DNA when complexed with thioredoxin, but that in the absence of thioredoxin, gp5 microscopically hops as it diffuses along the DNA.
Reconstituted gp5/trx Diffuses Along DNA by Sliding.
We also observed the diffusion of labeled gp5 in the presence of 50 nM unlabeled thioredoxin. Given the estimated KD for the binding of thioredoxin of 5 nM (2), it is likely that all gp5 molecules observed have an unlabeled thioredoxin bound to them. At an ionic strength of 64 mM, the observed diffusion coefficients and flow-induced drag were reduced to values indistinguishable from those of labeled gp5/trx (Figs. 2C and 3C), confirming that the difference in diffusional properties between gp5 and gp5/trx arise from the absence or presence of thioredoxin.
Quantification of Electrostatic Interactions Between T7 DNA Polymerase and Duplex DNA.
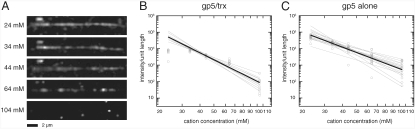
At protein concentrations higher than those used for the single-molecule diffusion experiments, we observe many fluorescently labeled T7 DNA polymerase molecules binding at random sites along the length of the stretched DNA molecules. At constant protein concentration, the amount of protein that is bound to the DNA decreases as the ionic strength of the experimental buffer is increased, manifested as a decrease in the fluorescence intensity per unit length along the tethered DNA. We use the integrated fluorescence intensity along the entire DNA length as a proxy for the affinity of protein for DNA (SI Text).
For both gp5/trx and gp5, we found that, as would be expected for nonspecific DNA binding, which is mainly mediated by electrostatic interactions, the binding affinity decreases with increasing buffer ionic strength (Fig. 4A). For a binding reaction mediated by electrostatic interactions, it has been shown that KD is related to the number of charge–charge interactions involved
where [M+] is the concentration of monovalent cations (equivalent to the ionic strength of the buffer for our experiments), m′ is the number of interactions between basic residues on the protein and the negatively charged phosphate backbone of the DNA, and ψ is the ion condensation parameter (0.88 for double-stranded DNA) (25). Log–log plots of the average intensity per unit length for each DNA molecule as a function of the concentration of monovalent cations were linear over most of the range of experimental conditions (Fig. 4B and C). The average slope of the linear fits to the data for 10 DNA molecules over this range was -4.34 (± 0.26 SEM) for gp5/trx, which corresponds to 4.93 (± 0.29) charge–charge interactions. For gp5 alone, the average slope of the fits to the data for 16 DNA molecules was -3.29 (± 0.26 SEM), which corresponds to 3.74 (± 0.29) charge–charge interactions. Therefore, for the nonspecific binding mode observed here, the binding of thioredoxin to gp5 leads to approximately one additional electrostatic interaction between gp5/trx and the DNA backbone.
Fig. 4.
T7 DNA polymerase binds along the length of λ-phage DNA. (A) At a constant protein concentration the amount of fluorescently labeled protein bound to a single, tethered DNA molecule decreases as the concentration of monovalent cations in the experimental buffer increases. Each panel contains a view of gp5/trx binding to the same DNA molecule at a different monovalent cation concentration (Left). Experiments performed with gp5 in the absence of thioredoxin yielded similar results. (B) Log–log scaled plots of the observed intensity per unit length of DNA vs. the total monovalent cation concentration for gp5/trx. Each data point (open circles) represents the average intensity per unit length for one DNA molecule in one experimental condition; linear fits through all the data obtained for a single DNA molecule are shown in gray. The thick black line is defined by the means of the fit parameters for all the DNA molecules, and has a slope of -4.34 (± 0.26 SEM), indicating five charge–charge interactions between the protein and the DNA backbone. (C) Log–log scaled plots of the observed intensity per unit length of DNA vs. the total monovalent cation concentration for the gp5 free of thioredoxin, colored as in (B). In this case, the mean fit has a slope of -3.29 (± 0.26 SEM), indicating four charge–charge interactions between the protein and the DNA backbone.
Discussion
Diffusive Behavior of T7 DNA Polymerase.
The binding of T7 DNA polymerase to long, duplex DNA that does not contain a recessed 3′ end has not been previously directly observed, likely due to its transient nature—the binding event trajectories we recorded lasted only a few seconds each. In characterizing this previously unobserved binding mode for T7 DNA polymerase, we have shown that gp5/trx slides along the DNA. During each binding event, the complex maintains very close contact with the DNA, does not dissociate from the DNA during transfer from one nonspecific binding site to another, and likely tracks along the DNA helix as it moves. However, the observed diffusion coefficients are very close to the theoretical limits, suggesting a low activation barrier for sliding. On the other hand, in the absence of thioredoxin, gp5 diffuses along the DNA using a hopping mechanism, taking microscopic excursions away from the DNA during each binding event. Hopping along the DNA does not require helical motion around the DNA, consistent with the observation that the mean diffusion coefficients were higher than the theoretical limit for such movement.
Our results indicate that the differences between the diffusional properties of gp5/trx and gp5 are due to the binding of thioredoxin. At an ionic strength at which the behavior of gp5 in the absence of thioredoxin differs significantly from that of gp5/trx, the mean diffusion coefficients and degree of flow-induced drift for labeled gp5 in the presence of excess unlabeled thioredoxin were nearly identical to the values obtained for gp5/trx. This finding indicates that the binding of thioredoxin to gp5 is sufficient to restore the behavior of gp5 to that of gp5/trx purified as a complex. Therefore, it is the binding of thioredoxin that suppresses microscopic hopping during diffusive binding events.
The Role of Sliding in DNA Synthesis.
It is possible that the sliding activity described in this report plays a role in the association of the T7 DNA polymerase with the 3′ end of a primer. The idea that reduction of dimensionality, in the form of one- or two-dimensional diffusion, could play a role in molecular interactions was suggested long before such phenomena could be directly observed (26). The observation of LacI repressor association with its cognate site at a rate faster than 3-dimensional diffusion (27) stimulated the development of theoretical work describing the potential mechanisms by which proteins might be able to diffuse along DNA to speed up searching processes (22). However, we feel that the change of the mode of diffusion along duplex DNA that is triggered by the binding of thioredoxin to the T7 DNA polymerase may play another important role. After the addition of a nucleotide to the primer, the polymerase must move forward to the new 3′ end of the nascent DNA strand, before another nucleotide can be added. The diffusive binding mode we have reported here may come into play during this realignment of the protein on the DNA. A polymerase that is capable of taking steps along the DNA backbone with a low probability of dissociating would arrive at the correct position to engage in another round of nucleotide addition more efficiently than one that is not held in close contact with the DNA. In the absence of thioredoxin, the T7 DNA polymerase would be less likely to complete the maneuver correctly, as it does not track the DNA helix. This event would have a strong negative effect on the processivity of synthesis, especially if a failure to correctly reposition the active site at the primer terminus leads to an increased likelihood of complete dissociation from the primer template.
Electrostatic Interactions of T7 DNA Polymerase with Duplex DNA.
Our measurements indicate that nonspecific binding to duplex DNA involves five charge–charge interactions between solvent exposed basic residues on the protein and the negatively charged DNA backbone for the gp5/thioredoxin complex, and four such interactions for gp5 in the absence of thioredoxin. Under conditions permitting catalysis, gp5/trx has been previously shown to form 7 charge–charge interactions between the protein and the DNA backbone, while gp5 alone forms only 2 charge–charge interactions (5).
We cannot at this time determine precisely which residues mediate the nonspecific binding of T7 DNA polymerase to double-stranded DNA, but there are several good candidates. Several contacts between active site residues and the DNA backbone were visible in the structure of gp5/trx bound to a primer template (6). Likewise, several basic residues within the TBD could form charge–charge interactions with the DNA backbone. Conversion of three of these residues (Lys300, Lys302, and Lys304) to glutamic acid reduces the affinity of the polymerase for thioredoxin as well as for DNA (28). The portion of the TBD containing these residues is disordered in the original T7 DNA polymerase crystal structure (6), but the entire loop is visible in a later structure of the gp5/trx opposite a lesion (11). The side chains of five lysine residues in the TBD are pointing toward the DNA. Charge neutralizing mutations of three of these significantly reduced gp5/trx processivity (8). These residues may be involved in the nonspecific, diffusive binding mode we observe here.
Observation of single molecules of gp5 binding to duplex DNA requires a tenfold increase in protein concentration over that adequate to permit observation of the binding of single molecules of gp5/trx to duplex DNA suggesting that thioredoxin binding results in at least a tenfold decrease in the KD for T7 DNA polymerase binding to duplex DNA. The contribution to the free energy of binding resulting from the release of cations condensed along the phosphate backbone of the DNA can be estimated as
where R is the gas constant (25). The change in this quantity resulting from a single additional charge–charge interaction ranges from 2.0 kcal/mol at an ionic strength of 24 mM to 1.2 kcal/mol at an ionic strength of 104 mM. Noting that a change in binding energy of 0.59 kcal/mol (equivalent to 1 RT) corresponds to a 2.7-fold (equivalent to e1) change in KD, a single additional charge–charge interaction could account for a tenfold difference in the resulting KD under these experimental conditions. As a result, the increased affinity of the gp5/thioredoxin complex for duplex DNA may be, in large part, the result of increased electrostatic interactions. Further structural, biochemical, and biophysical studies are needed to further elucidate the exact relation between polymerase structure and DNA-binding properties.
Materials and Methods
Reagents.
Biotinylated lambda DNA constructs were made by linearizing phage-λ DNA (New England Biolabs) and annealing oligonucleotides to the resulting 12-nucleotide-long, single-stranded complimentary ends. Annealed oligonucleotides were ligated to the λ DNA using T4 DNA ligase (New England Biolabs). For more details, see SI Text.
T7 DNA polymerase was expressed in E. coli, and purified as a 1∶1 complex of gp5 and bacterial thioredoxin, as described previously (2). T7 gp5 was expressed in E. coli strain A307(DE3) lacking endogenous thioredoxin and purified as described (9). Proteins were labeled with Alexa Fluor 555 carboxylic acid, succinimidyl ester (Invitrogen). The conjugation reactions were biased to the amino terminals by adjusting the pH (SI Text) The degree of labeling was determined by measuring the absorbance spectrum of the sample and was found to be approximately 4 fluorophores/complex for gp5/trx, and 2 fluorophores/molecule for gp5 alone.
Protein activity was assessed by measuring the amount of 3H-dTMP added by the polymerase to the 3′ end of a primer annealed to circular, single-stranded viral DNA isolated from phage M13mp18 (New England Biolabs) (2). Details included in SI Text.
Single-Molecule Assay.
Glass coverslips were functionalized with 3-aminopropyltriethoxysilane and coated with a 100∶1 mixture of methoxy-terminated and biotin-terminated NHS ester derivatized PEG molecules. This functionalized surface was then coated with streptavidin (29).
Flow cells were constructed with a 2 mm wide by 100 μm tall channel as previously described (29). The biotinylated DNA constructs were flowed through the channel to permit binding to the surface and then excess, unbound DNA was washed away. Experimental buffer is flowed continuously through the channel at a rate of 0.1 ml/ min during experiments, creating shear force that stretches the DNA parallel to the glass surface. Fluorescently labeled proteins were illuminated with a 532 nm laser by through-objective TIR, and image streams captured by an EM-CCD camera (Hamamatsu C9100-13) using Meta Vue imaging software (Molecular Devices).
Experimental buffers contained 20 mM Tris (pH 7.5), 2 mM EDTA, 1 mg/ml BSA (New England Biolabs), 2 mM potassium phosphate, 5 mM sodium chloride, 0.5% glycerol, varying amounts of additional sodium chloride or potassium glutamate, and the fluorescently labeled protein of interest. See SI Text for ionic strength calculations.
Data Analysis.
To determine the average fluorescence intensity of protein bound per unit length, a region of interest containing the DNA molecule was selected. Background was subtracted from the measured intensity within the selected region including the DNA for each frame in each movie (SI Text). The intensity per pixel length along each DNA molecule was calculated for each movie frame, and the final value for each DNA molecule is the average over all 500 frames of the movie.
Particle positions were determined by fitting a two-dimensional Gaussian to each single-molecule fluorescence image. Single-molecule trajectories were traced using a particle tracking code built upon code developed by Daniel Blair and Eric Dufresne, obtained from http://physics.georgetown.edu/matlab/. To determine the linear diffusion coefficient (D1) of the tracked particles, the MSD longitudinal to the stretched DNA (MSDx) for each particle was calculated for several time windows (nΔt). The slope of a linear fit of the resulting data equals 2D1.
Supplementary Material
Acknowledgments.
We thank Sharmistha Ghosh for graciously providing purified thioredoxin. This work was supported by National Institutes of Health Grants R01 GM54397 (C.C.R.) and R01 GM077248 (A.M.v.O.) and the National Science Foundation Grant CAREER MCB-0543784 (A.M.v.O.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912664107/DCSupplemental.
References
- 1.Richardson CC. Bacteriophage T7: Minimal requirements for the replication of a duplex DNA molecule. Cell. 1983;33(2):315–317. doi: 10.1016/0092-8674(83)90411-7. [DOI] [PubMed] [Google Scholar]
- 2.Tabor S, Huber H, Richardson C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262(33):16212–16223. [PubMed] [Google Scholar]
- 3.Mark DF, Richardson CC. Escherichia coli thioredoxin: A subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1976;73(3):780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modrich P, Richardson CC. Bacteriophage T7 deoxyribonucleic acid replication invitro. Bacteriophage T7 DNA polymerase: An enzyme composed of phage- and host-specific subunits. J Biol Chem. 1975;250(14):5515–5522. [PubMed] [Google Scholar]
- 5.Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987;262(33):16224–16232. [PubMed] [Google Scholar]
- 6.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391(6664):251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 7.Bedford E, Tabor S, Richardson CC. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA. 1997;94(2):479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27(4):539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Hamdan SM, et al. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci USA. 2005;102(14):5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, Hamdan SM, Cook TE, Richardson CC. Interactions of Escherichia coli Thioredoxin, the Processivity Factor, with Bacteriophage T7 DNA Polymerase and Helicase. J Biol Chem. 2008;283(46):32077–32084. doi: 10.1074/jbc.M805062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brieba LG, et al. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. Embo J. 2004;23(17):3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys Rev Lett. 2006;97(4):048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 13.Tafvizi A, et al. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J. 2008;95(1):L01–03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graneli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc Natl Acad Sci USA. 2006;103(5):1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103(15):5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komazin-Meredith G, Mirchev R, Golan DE, van Oijen AM, Coen DM. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc Natl Acad Sci USA. 2008;105(31):10721–10726. doi: 10.1073/pnas.0802676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochaniak AB, et al. Proliferating Cell Nuclear Antigen Uses Two Distinct Modes to Move along DNA. J Biol Chem. 2009;284(26):17700–17710. doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabata H, et al. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262(5139):1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 19.Harada Y, et al. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys J. 1999;76(2):709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JH, Larson RG. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35(11):3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnet I, et al. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36(12):4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 23.Randahl H, Slaby I, Holmgren A. An improved purification method and a physical characterization of phage T7 DNA polymerase. Eur J Biochem. 1982;128(2-3):445–449. doi: 10.1111/j.1432-1033.1982.tb06984.x. [DOI] [PubMed] [Google Scholar]
- 24.Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys Chem. 1979;9(4):413–414. [PubMed] [Google Scholar]
- 25.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 26.Adam G, Delbruck M. Reduction of dimensionality in biological diffusion processes. In: Rich A, Davidson N, editors. Structural Chemistry and Molecular Biology. San Francisco: W. H. Freeman and Company; 1968. pp. 198–215. [Google Scholar]
- 27.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 28.Yang XM, Richardson CC. Amino acid changes in a unique sequence of bacteriophage T7 DNA polymerase alter the processivity of nucleotide polymerization. J Biol Chem. 1997;272(10):6599–6606. doi: 10.1074/jbc.272.10.6599. [DOI] [PubMed] [Google Scholar]
- 29.Lee J-B, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.