Abstract
Structural analyses of the extracellular region of stem cell factor (SCF) receptor (also designated KIT) in complex with SCF revealed a sequence motif in a loop in the fourth Ig-like domain (D4) that is responsible for forming homotypic receptor contacts and for ligand-induced KIT activation and cell signaling. An identical motif was identified in the most membrane-proximal seventh Ig-like domain (D7) of vascular endothelial growth factor receptor 1 (VEGFR1), VEGFR2, and VEGFR3. In this report we demonstrate that ligand-induced tyrosine autophosphorylation and cell signaling via VEGFR1 or VEGFR2 harboring mutations in critical residues (Arg726 or Asp731) in D7 are strongly impaired. We also describe the crystal structure of D7 of VEGFR2 to a resolution of 2.7 Å. The structure shows that homotypic D7 contacts are mediated by salt bridges and van der Waals contacts formed between Arg726 of one protomer and Asp731 of the other protomer. The structure of D7 dimer is very similar to the structure of D4 dimers seen in the crystal structure of KIT extracellular region in complex with SCF. The high similarity between VEGFR D7 and KIT D4 in both structure and function provides further evidence for common ancestral origins of type III and type V RTKs. It also reveals a conserved mechanism for RTK activation and a novel target for pharmacological intervention of pathologically activated RTKs.
Keywords: angiogenesis, cancer, phosphorylation, protein kinases, surface receptors
Vascular endothelial growth factors (VEGFs) regulate blood and lymphatic vessel development and homeostasis by binding to and activating the three members of the VEGF-receptor (VEGFR) family of receptor tyrosine kinases (RTKs) (1). VEGFR1 (Flt1), VEGFR2 (KDR/Flk1) and VEGFR3 (Flt4) are members of type-V RTK; a family containing a large extracellular region composed of seven Ig-like domains (D1–D7), a single transmembrane (TM) helix and cytoplasmic region with a tyrosine kinase activity, and additional regulatory sequences. The second and third Ig-like domains, D2 and D3, of VEGFR ectodomains function as binding sites for the VEGF family of cytokines (i.e., VEGF-A, -B, -C, -D, and placenta growth factor) (2, 3). These growth factors are covalently linked homodimers. Each protomer is composed of four-stranded β-sheets arranged in an antiparallel fashion in a structure designated cysteine-knot growth factors (4). It was shown that VEGF-A stimulation of VEGFR2 induces endothelial cell proliferation, survival, and migration resulting in blood vessel formation and sprouting (5). On the other hand, VEGF-C activation of VEGFR3 plays an important role in the formation of the lymphatic vessel system (6, 7). Aberrant activation or expression of VEGF receptors and their ligands has been implicated in tumor angiogenesis, coronary artery disease, diabetic blindness, and other diseases (5).
Other members of the cysteine-knot family of cytokines include nerve growth factor and platelet-derived growth factors (PDGFs). However, the ectodomains of the PDGF-receptor (PDGFR) family of RTKs (type III) are composed of five Ig-like repeats of which D1, D2, and D3 function as ligand-binding regions of PDGFR and other members of the family (i.e., KIT, CSF1R, and Flt3). Structural and biochemical experiments have shown that SCF binding to the extracellular region induces KIT dimerization, a step followed by homotypic contacts between the two membrane-proximal Ig-like domains D4 and D5 of neighboring KIT molecules (8). Biochemical studies of wild-type (WT) and oncogenic KIT mutants have shown that the homotypic D4 and D5 contacts play a critical role in positioning the cytoplasmic regions of KIT dimers at a distance and orientation that facilitate transautophosphorylation, kinase activation, and cell signaling. A variety of activating mutations in KIT were implicated in different cancers including acute myeloid leukemia, gastrointestinal stromal tumors, and melanoma (9).
In this report we present structural and biochemical evidence demonstrating that homotypic contacts between the most membrane-proximal Ig-like domain of the ectodomain (D7) of VEGF-receptors play a critical role in VEGF-induced activation and cell signaling via VEGF receptors.
Results and Discussion
Sequence Analysis of VEGFR2 D4 and D7.
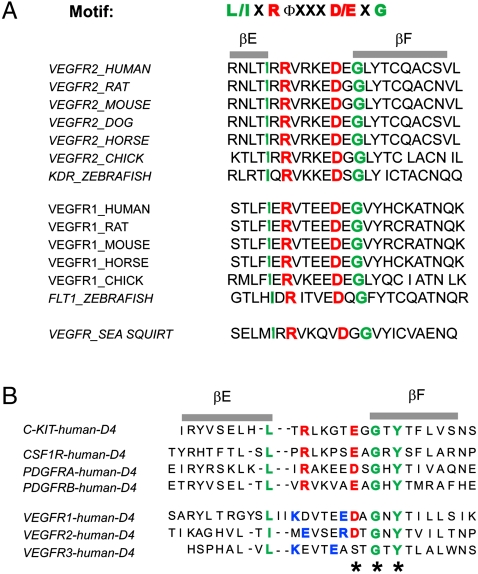
An evolutionarily conserved sequence motif (L/IxRΦxxxD/ExG) responsible for mediating homotypic contacts between Ig-like domains was identified by structure-based sequence alignment of D4 of KIT, PDGFRα, PDGFRβ, and colony stimulating factor 1 receptor (CSF1R) (8, 10). To determine which domain of VEGFR2 may be functionally related to D4 of KIT, we searched the sequences of D4 and D7 of VEGFRs for this conserved motif. The sequence alignments presented in Fig. 1 shows that a similar motif exists in the most membrane-proximal Ig-like domain (D7) of VEGFR1, VEGFR2, and VEGFR3. The conserved sequence motif is located at the loop region linking βE and βF strands of D7, a region shown to be responsible for mediating salt bridges required for homotypic D4 KIT contacts. The Arg and Asp are evolutionarily conserved from sea squirt to human in both VEGFR1 and VEGFR2 (Fig. 1), indicating functional importance of these residues in VEGFR activity. However, in contrast to D4 of KIT and PDGFR, D7 of VEGFR1 and D7 of VEGFR2 contain two conserved cysteine residues at positions B5 and F5 that form a disulfide bond between the β strands, an interaction contributing to the hydrophobic core of I-set Ig-like domains (11).
Fig. 1.
Amino Acid sequence analysis of D7 and D4 of VEGFR2 extracellular region. (A) Structure-based multiple sequence alignment of predicted EF-loop region of D7 of VEGFR1 and VEGFR2 from different species. Key amino acids in the I-set Ig frame are highlighted in green, and the conserved Arg/Asp pair in the EF loop is highlighted in red. (B) Comparison of predicted EF-loop region of D4 from VEGFR and D4 of KIT, CSF1R, and PDGFRs (type-III RTK). Key amino acids in the I-set Ig frame are highlighted in green, and the conserved Arg/Asp or Glu pair in the EF loop is highlighted in red. Nonconserved amino acids with opposite charge in the EF loop are highlighted in blue. The conserved Y-corner motif is marked with an asterisk (*).
Similar to D4 of PDGFR and KIT, D4 of VEGFR2 lacks the conserved cysteines responsible for disulfide bond formation between β strands at positions B5 and F5. In D4 of VEGFR2, the region connecting βC with βE is shorter compared to other typical I-set Ig domains, possibly because this region lacks one of the β-strands. Amino acid sequence analysis showed that VEGFR2 D4 is homologous to myomesin domain D13 (2R15) and telokin (1TLK) with sequence identity of 30 and 33%, respectively. Manual sequence alignment revealed 20% sequence identity between D4 of VEGFR2 and D4 of PDGFRα. Both D4 of KIT and D4 of PDGFR contain conserved “D/E-x-G” amino acids around the “Y-corner motif” in βF consisting of D/E-x-G/A/D-x-Y-x-C motif (12) (in D4 the Cys residue is replaced by hydrophobic amino acids) (Fig. 1B). In D4 a salt bridge is formed between a Glu residue in one molecule with an Arg residue at the -5 position of a second KIT molecule (Fig. 1B). An Asp residue is found in D4 of VEGFR2, but instead of an Arg at the -5 position this Ig-like domain contains a pair of amino acids with opposite charges at the -2 and -6 positions relative to the Asp residue (Fig. 1B). Although direct contacts between D4 have been observed in electron microscopy (EM) images of VEGF-A–induced dimers of the ectodomain of VEGFR2 (13), the function of VEGFR2 D4 remains unclear. D7 is thus more similar than D4 of VEGFR2 in the EF-loop region to corresponding sequences in D4 of KIT and PDGFR.
Homotypic D7 Contacts Are Essential for Ligand-Induced VEGFR2 Activation.
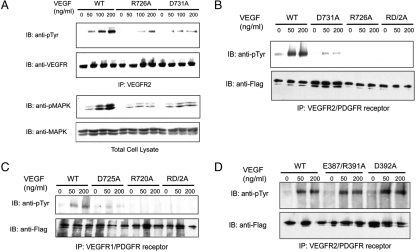
This sequence analysis and comparison with KIT suggests that residues R726 and D731 of VEGFR2 can mediate interreceptor salt bridge formation following ligand-induced receptor dimerization. To investigate the role of the conserved residues in D7 region in ligand-induced VEGFR2 activation and signal transduction, we generated VEGFR2 mutants in which Arg726, Asp731, or both amino acids were replaced by Ala residues (R726A, D731A, and RD2A). HEK293 cells were transiently transfected with pCDNA3 expression vectors that direct the expression of WT VEGFR2 or VEGFR2 harboring D7 mutations. After incubation for 24 h, the transfected cells were starved overnight prior to VEGF-A stimulation. Tyrosine autophosphorylation of VEGFR2 and MAPK of VEGF-A stimulated or unstimulated cells were analyzed using antiphosphotyrosine antibodies (anti-pTyr) or anti-phospho-MAPK antibodies, respectively. The experiment presented in Fig. 2A shows that mutations of the Arg or Asp residues predicted to be involved in mediating interreceptor salt bridge formation markedly reduced VEGF-A–induced VEGFR2 autophosphorylation and MAPK stimulation.
Fig. 2.
Ligand-induced activation of VEGFR2 is compromised by mutations in EF-loop region of D7 but not affected by mutation in EF-loop region of D4. (A) HEK293 cells transiently expressing WT VEGFR2, the R726A, or the D731A VEGFR2 mutants were stimulated with indicated amount of VEGF-A for 5 min at 37 °C. Lysates from unstimulated or VEGF-A–stimulated cells were subjected to immunoprecipitation with anti-VEGFR2 antibodies followed by immunoblotting (IB) with anti-pTyr, or with anti-VEGFR2 antibodies. Total cell lysate from the same experiment was analyzed by SDS-PAGE followed by immunoblotting with anti-phosphoMAPK (pMAPK) or anti-MAPK antibodies. (B) Serum-starved 3T3 cells stably expressing a VEGFR2-PDGFRβ chimeric receptor or chimeric receptors harboring mutations in D7 region (R726A, D731A, or an R726/D731 double mutation designated RD/2A) were stimulated with VEGF-A for 5 min at 37 °C. Lysates from unstimulated or VEGF-A–stimulated cells were subjected to immunoprecipitation with antibodies against the cytoplasmic region of the chimeric receptor followed by immunoblotting with either anti-pTyr or anti-tag (FLAG) antibodies, respectively. (C) Serum-starved 3T3 cells stably expressing a VEGFR1-PDGFR chimeric receptor or chimeric receptors harboring mutation in the D7 region (R720A, D725A, or R720D725/2A double mutations) were analyzed as described in (B). (D) 3T3 cells expressing a VEGFR2-PDGFR chimeric receptor or chimeric receptors harboring mutations in D4 region (D392A or E387/R391A double mutations) were analyzed as described in (B).
To overcome the relatively weak kinase activity of VEGFR2, we have employed a chimeric receptor approach that has been successfully applied in previous studies (14, 15). A chimeric receptor composed of the ectodomain of VEGFR2 (amino acid 1-761) connected to the TM and the cytoplasmic region of PDGFR (amino acid 528-1106) was prepared and used to further explore the role played by D7 in VEGF-A–induced VEGFR2 activation. Lysates from VEGF-A stimulated or unstimulated NIH-3T3 cells stably expressing a chimeric VEGFR2/PDGFR or chimeric VEGFR2/PDGFR harboring D7 mutations were subjected to immunoprecipitation with anti-PDGFR antibodies followed by immunoblotting with anti-pTyr antibodies. The experiment presented in Fig. 2B shows robust tyrosine autophosphorylation of the chimeric VEGFR2/PDGFR in response to VEGF-A stimulation (Fig. 2B, WT). In contrast, VEGF-A–induced tyrosine autophosphorylation of chimeric receptor harboring the D7 mutations (R726A, D731A, RD2A) was strongly compromised (Fig. 2B). We also generated a chimeric receptor composed of the extracellular region of VEGFR1 fused to the TM and intracellular region of PDGFR. 3T3 cells stably expressing WT chimeric receptors showed autophosphorylation in response to ligand stimulation (Fig. 2C, WT). By contrast, ligand-induced stimulation of kinase activity was strongly compromised in 3T3 cells expressing chimeric receptor (harboring mutations in Arg720, Asp725, or in both amino acids) R720A, D725A and RD2A, respectively (Fig. 2C). We conclude that homotypic contacts between the membrane-proximal Ig-like domains of type-III and type-V RTKs are essential for ligand-induced receptor activation and cell signaling.
As previously described (16), we have used a covalent cross-linking agent to explore the effect of D7 mutations on ligand-induced receptor dimerization by cross-linking ligand-stimulated cells followed by SDS-PAGE analysis of lysates from ligand-stimulated or unstimulated cells. This experiment demonstrated that VEGF-A–induced dimerization of the chimeric receptors was not affected by the D7 mutations (Fig. S1). Similar to what we previously reported for PDGFR and KIT (8, 10), D7-mediated homotypic contacts are necessary for receptor activation but dispensable for receptor dimerization. Moreover, ligand-induced receptor dimerization is necessary but not sufficient for tyrosine autophosphorylation and receptor activation. By contrast, VEGF-A–induced tyrosine autophosphorylation of chimeric receptor harboring mutations in D4 of VEGFR2, including a D392A mutation or mutations in which both Glu387 and Arg391 were substituted by Ala residues (ER2A), remained unchanged (Fig. 2D), suggesting that a different interface might be involved in mediating D4 interactions seen in EM images of VEGF-A–induced VEGFR2 ectodomain dimers (13).
We next used analytical centrifugation to determine the dissociation constant for dimerization of isolated D7 region. Analytical centrifugation experiments performed using 4 × 10-5, 8 × 10-5, and 1.6 × 10-4 M protein concentrations showed that isolated D7 remained monomeric in solution at a concentration as high as 10-4 M, indicating that the dissociation constant of D7 dimerization exceeds 10-4 M. A similar high dissociation constant was found for dimerization of isolated D4 or D5 of KIT or PDGFR. We have previously shown that following SCF- or PDGF-induced dimerization the local concentration of two neighboring KIT or PDGFR protomers in the cell membrane is in the range of 4–6 × 10-4 M. This together with the reduced dimensionality enables efficient lateral interactions and formation of stable homotypic contacts between pairs of Ig-like domains, which bind to each other with low affinity in the cell membrane. Moreover, the homotypic contacts between membrane-proximal Ig-like domain in type-III and type-V RTKs are supported by additional lateral interactions that take place between the TM and cytoplasmic regions of neighboring receptors in a cooperative manner.
Structure of VEGFR Extracellular Domain D7.
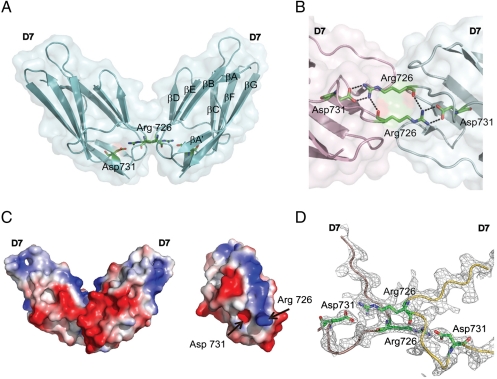
In order to determine the molecular basis underlying the role played by D7 in ligand–induced VEGFR2 activations, we next determined the crystal structure of this Ig-like domain. Crystals were obtained in space group I Centered Orthorhombic I212121, with a single D7 molecule per asymmetric unit together with 35 water molecules. D7 structure consists of amino acids 667 to 756 of VEGFR2 and diffracts x-rays to 2.7-Å resolution. The structure was determined by molecular replacement with model based on the structure of telokin [Protein Data Bank (PDB) code: 1TLK] (17). D7 assumes a typical IgSF fold that consists of a β-sandwich formed by two four-stranded sheets, one comprised of strands A, B, D, and E, and the second comprised of strands A′, G, F, and C (Fig. 3A). The first half of the A strand forms a hydrogen bond with the B strand and the A′ strand forms hydrogen bonds with the G strand, similar to the structure of Ig-like domain Ig1 and Ig2 from the extracellular region at RTK MuSK (18). The crossover connection between strand βE and βF includes a single helical turn at residues 729–731. D7 of VEGFR2 displays several characteristics of the IgSF fold including a conserved disulfide bond between Cys688 of βB and Cys737 of βF and a signature tryptophan residue that packs against disulfide bond to form the hydrophobic core. Structural comparison using DALI (19) shows that among the Ig-like domains of VEGFR2, D7 is most similar to telokin (PDB code: 1TLK) (17), with a Z score of 13.4 and an rmsd of 1.5 Å for the 89 aligned Cβ residues. D7 contains 16 of the 20 key positions in the V-frame profile that defines the I set (11). An additional exposed cross-strand disulfide bond is formed by a pair of Cys located in the βF (Cys740) and βG (Cys745). This feature is highly conserved in VEGFR2 and VEGFR3 but not in VEGFR1.
Fig. 3.
Structure of VEGFR2 D7 homodimer. (A) A ribbon diagram and a transparent molecular surface of D7 homodimer structure (side view). Asp731 and Arg726 are shown as a stick model. (B) A close view of the homotypic D7 interface of the two neighboring molecules (pink and green). Salt bridges formed between Asp731 and Arg726 are shown as dashed lines. (C) Charge distribution of D7 homodimer (side view) is shown as a surface potential model (Left). View of D7 surface that mediates homotypic contacts (Right). (D) 2Fo-Fc electron density map contoured at 1.1σ level showing a view of the D7–D7 interface. The backbones of VEGFR D7 protomers are represented as pink and yellow tubes, respectively.
The crystal structure demonstrates that homotypic D7 contacts are mediated by two β-sheets formed by the ABED strands of D7 of each protomer in which Arg726 of one protomer points toward Asp731 of the other, resulting in a buried surface area of approximately 360 A2. Fig. 3B shows that Arg726 and Asp731 form salt bridges and van der Waals contacts. The electron density map clearly demonstrates homotypic contacts between symmetry mates through the EF-loop regions (Fig. 3D). The structure of D7 dimer is very similar to the homotypic D4 contacts seen in KIT extracellular dimer structure (PDB code: 2E9W) (8). In addition D7 of VEGFR2 exhibits strong polarization of electrostatic field with an overall negatively charged surface with the exception of a positively charged center strip right along the D7–D7 interface (Fig. 3C). The strongly charged interface may prevent aberrant association of monomeric receptor molecules prior to ligand stimulation.
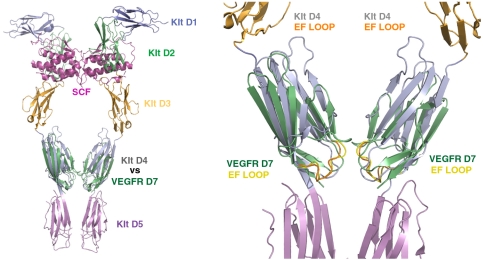
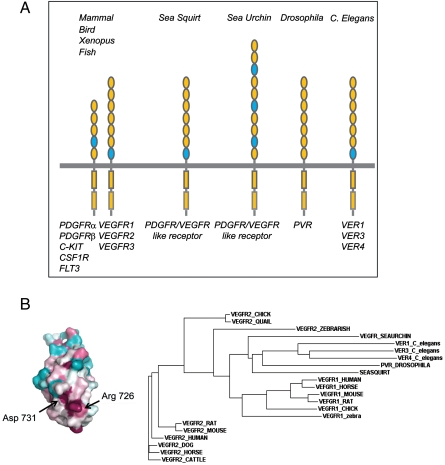
Comparison of KIT D4 and VEGFR2 D7 structures using DALI (19) showed a remarkable similarity with a Z score of 10.4 and an rmsd of 1.8 Å for the 83 aligned Cα residues. The position of the EF loop in the two structures is nearly identical, and the distance between the C-termini is approximately 15 Å for both D4 and D7 dimeric structures (Fig. 4). The high similarity between VEGFR D7 and KIT D4 in both structure and function suggests a well-conserved mechanism for RTK activation and provides further evidence for common ancestral origins of type-III and type-V RTKs. Interestingly, the Drosophila (20), Caenorhabditis elegans (21), and sea squirt (22) genomes contain a single family of VEGFR/PDGFR-like RTK composed of seven Ig-like domains in its extracellular region (Fig. 5A). In humans, the genes for type-III and type-V RTKs are found in three clusters on chromosomes 4q12 (KIT, PDGFRα and VEGFR2), 5q33 (FMS, PDGFRβ and VEGFR3) and 13q12 (Flt3 and VEGFR1). Type-III and type-V RTK genes were functionally segregated in vertebrates but are located adjacent to each other on the chromosomes. Phylogeny of type-III and type-V RTKs suggests that the five type-III and three type-V RTKs of vertebrates were generated by two rounds of cis duplication and two rounds of trans duplication (23, 24). It is of note that the conserved motif in EF-loop region exists in D7 VEGFR/PDGFR-like receptor genes (VER3 and VER4) of C. elegans, in D7 of a VEGFR/PDGFR-like receptor of sea squirt, but not in the VEGFR/PDGFR-like receptor of Drosophila. Interestingly, the extracellular region of the VEGFR/PDGFR-like receptor of sea urchin (25) is composed of 10 Ig-like domains of which D3, D6, and D4 contain typical EF-loop sequence motifs. It is possible that multiple homotypic contacts between D3, D6, and D9 of the sea urchin receptor are required for ligand-induced activation of an RTK containing 10 Ig-like domains in its extracellular region (Fig. 5A).
Fig. 4.
Superposition of the structure of D7 of VEGFR2 with the structure of D4 of dimeric KIT-SCF complex. Overlay of VEGFR D7 structure (PDB ID code: 3KVQ) and the structure of KIT dimer in complex with SCF (PDB ID code: 2E9W) (Left). A closer view of superimposed D7 and D4 regions reveals high similarity in domain arrangement and homotypic contacts (Right). VEGFR2 D7 is illustrated in green and the EF loop is in yellow. D4 of KIT is illustrated in gray and its EF loop is in orange.
Fig. 5.
Phylogenetic analysis of VEGFR1 and VEGFR2. (A) Location of the conserved EF loop in type-III and type-V RTKs from various species. Ig-like domains containing a conserved EF-loop motif are marked in blue. (B) Color-coded conservation pattern of VEGFR2 D7 region. Amino acid sequences of human VEGFR2 were used as query to search nonredundant database for homologous sequences, using PSI-BLAST. Sequence alignment of D7 was performed using ClustalW, manually adjusted based on the IgSF fold restrains for 20 key residues. The alignment of amino acid sequences was submitted to the Consurf 3.0 server to generate maximum-likelihood normalized evolutionary rates for each position. Cyan through maroon is used for labeling from variable to conserved amino acids. Phylogenetic tree of VEGFR1 and VEGFR2 are generated by the neighboring-joining method based using Clustal W2.
Conclusions
The experiments presented in this and our previous studies show that type-III and type-V RTK are activated by a common mechanism in which homotypic contacts mediated by membrane-proximal Ig-like domains ensure that the TM and cytoplasmic regions of two receptor monomers are brought to a close proximity and correct orientation to enable efficient transautophosphorylation, kinase activation, and cell signaling. The combination of ligand-induced receptor dimerization together with multiple low-affinity homotypic associations between membrane-proximal Ig-like domains provide a simple but efficient mechanism for ligand-induced transmembrane signaling. Moreover, the low binding affinity of individual Ig-like domains toward each other prevents accidental receptor activation of receptor monomers prior to ligand engagement.
It is now well established that KIT, VEGFR2, and the other members of type-III and type-V RTKs represent key drivers of a variety of cancers and other severe pathologies. The experiments presented in this report show that homotypic contact regions in type-III, type-V, and other RTKs provide ideal targets for pharmacological intervention of pathological RTK activation in cancer and other diseases.
Materials and Methods
Protein Expression and Purification.
D7 of VEGFR2 (amino acid 657-765) containing an N-terminal 6xHis-tag was expressed in Escherichia Coli using PET28a vector. Inclusion bodies were collected and solubilized in 6 M guanidine hydrochloride (pH8.0). D7 was refolded by dropwise dilution of the protein into refolding buffer containing 10 mM Tris (pH8.0), 2 mM reduced glutathione, and 0.2 mM oxidized glutathione with final protein concentration at 80–100 μg/mL. The refolding was carried out at 4 °C for 48 h with stirring. The refolding solution was cleared by filtration using a 0.45-μm filter unit and purified by FastQ sepharose column followed by size exclusion (S200, GE Healthcare) and anion exchange chromatography (Mono Q, GE Healthcare). N-terminal 6xHis tag was removed by thrombin digestion. D7 protein was concentrated to 15 mg/mL in buffer containing 25 mM Tris (pH 8.0) and 200 mM NaCl and was subjected to extensive screening for crystallization and optimization (Hampton research, crystal screening).
Crystallization and Structure Determination.
Crystals of ectodomain D7 were grown in 0.2 M succinic acid and 16% PEG3350 using the sitting drop vapor diffusion method at 4 °C. Crystals were harvested at approximate dimensions of 300 × 75 × 20 μm and cryofrozen in nitrogen stream using crystallization solution supplemented with 5–18% glycerol as cryoprotectant. Diffraction data was collected to a resolution of 2.7 Å with a quantum-210 CCD detector (Area Detector System) at the X29A beamline of National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. All datasets were processed and scaled using the HKL2000 program package (26). The crystals belonged to the I212121 space group with unit cell dimersions of a = 39.476 Å, b = 76.991 Å, and c = 102.034 Å. The asymmetric unit contains one molecule with solvent content of 64.1%. The structure was solved by molecular replacement with MolRep using models based on the structures of Ig domains from MUSK (2IEP) (18), telokin (1TLK) (17), and D4 of KIT (2E9W) (8) as search models. The structure refinement and model-building cycles were performed using RefMac (27) and COOT (28). The refined structure has a crystallographic R-value of 0.22 and free R-value of 0.29. Crystallographic statistics are summarized in Table 1. The atomic coordinates of VEGFR2 D7 were deposited in PDB with accession code 3KVQ. Molecular images were produced using Pymol (http://pymol.sourceforge.net/) and CCP4MG software (29).
Table 1.
Data collection and refinement statistics for VEGFR-D7
| Data collection | |
| Space group | I212121 |
| Unit cell dimensions | |
| a (Å) | 39.476 |
| b (Å) | 76.991 |
| c (Å) | 102.034 |
| Resolution (Å) | 50-2.7 |
| Unique reflections | 4561 |
| Completeness (%)* | 99.8 (97.8) |
| Rsym (%)*† | 4.3 (7.5) |
| Redundancy | 13.4 (12.9) |
| Refinement | |
| Rwork (%)‡ | 22 |
| Rfree (%)§ | 29 |
| Protein residues | 89 |
| Water molecules | 27 |
| Average B factors (Å2) | 37.39 |
| RMS deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (degree) | 1.2 |
| Ramachandran plot statistics | |
| Core (%) | 91.2 |
| Allowed (%) | 8.8 |
| Generous (%) | 0 |
*Values in parentheses are statistics of the highest resolution shell for SEG (2.8–2.7 Å).
†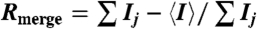 , where Ij is the intensity of an individual reflection and is the average intensity of the reflection.
, where Ij is the intensity of an individual reflection and is the average intensity of the reflection.
‡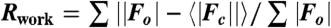 , where Fc is the calculated structure factor.
, where Fc is the calculated structure factor.
§Rfree is as Rwork but calculated for a randomly selected 10% of the reflection not included in the refinement.
VEGFR Expression Vectors and Generation of Chimeric Receptors.
cDNA of human VEGFR1 and VEGFR2 were kindly provided by Masabumi Shibuya (30). VEGFR2 cDNA was subcloned into pcDAN3 expression vector by PCR and inserted into XhoI/XbaI sites. Chimeric receptors composed of the extracellular regions of either VEGFR1 or VEGFR2 were fused to the TM and cytoplasmic region of PDGFR-β. A flag-tag was added to the C-terminus and the chimeric receptor was cloned into EcoRI/XhoI sites of pLXSHD retroviral expression vector.
Supplementary Material
Acknowledgments.
This article is dedicated to the memory of Dr. Peng Xie. This work was supported by National Institutes of Health (Grants R01-AR051448, R01-AR051886, and P50-AR054086 to J. S). The authors thank the staff of X29A at NSLS and Titus Boggon for valuable comments about the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914052107/DCSupplemental.
References
- 1.Olsson A-K, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling in control of vascular function. Nat Rev Mol Cell Biol. 2006;7(5):359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 2.Barleon B, et al. Mapping of the sites for ligand binding and receptor dimerization at the extracellular domain of the vascular endothelial growth factor receptor FLT-1. J Biol Chem. 1997;272(16):10382–10388. doi: 10.1074/jbc.272.16.10382. [DOI] [PubMed] [Google Scholar]
- 3.Shinkai A, et al. Mapping of the sites involved in ligand association and dissociation at the extracellular domain of the kinase insert domain-containing receptor for vascular endothelial growth factor. J Biol Chem. 1998;273(47):31283–31288. doi: 10.1074/jbc.273.47.31283. [DOI] [PubMed] [Google Scholar]
- 4.Wiesmann C, et al. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91(5):695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 5.Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312(5):549–560. doi: 10.1016/j.yexcr.2005.11.012. (in eng) [DOI] [PubMed] [Google Scholar]
- 6.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8(6):464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 7.Makinen T, et al. Inhibition of lymphangiogenesis with resulting lymphedema in transgenic mice expressing soluble VEGF receptor-3. Nat Med. 2001;7(2):199–205. doi: 10.1038/84651. [DOI] [PubMed] [Google Scholar]
- 8.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130(2):323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 9.Forbes S, et al. Cosmic 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105(22):7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238(4):528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 12.Hemmingsen JM, Gernert KM, Richardson JS, Richardson DC. The tyrosine corner: a feature of most Greek key beta-barrel proteins. Protein Sci. 1994;3(11):1927–1937. doi: 10.1002/pro.5560031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14(3):249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 14.Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97(6):727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 15.Meyer RD, Mohammadi M, Rahimi N. A single amino acid substitution in the activation loop defines the decoy characteristic of VEGFR-1/FLT-1. J Biol Chem. 2006;281(2):867–875. doi: 10.1074/jbc.M506454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochet C, et al. Demonstration of epidermal growth factor-induced receptor dimerization in living cells using a chemical covalent cross-linking agent. J Biol Chem. 1988;263(7):3290–3295. [PubMed] [Google Scholar]
- 17.Holden HM, Ito M, Hartshorne DJ, Rayment I. X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution. J Mol Biol. 1992;227(3):840–851. doi: 10.1016/0022-2836(92)90226-a. [DOI] [PubMed] [Google Scholar]
- 18.Stiegler AL, Burden SJ, Hubbard SR. Crystal structure of the agrin-responsive immunoglobulin-like domains 1 and 2 of the receptor tyrosine kinase MuSK. J Mol Biol. 2006;364(3):424–433. doi: 10.1016/j.jmb.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5:Unit 5 5. [DOI] [PubMed] [Google Scholar]
- 20.Cho NK, et al. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;108(6):865–876. doi: 10.1016/s0092-8674(02)00676-1. [DOI] [PubMed] [Google Scholar]
- 21.Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc Natl Acad Sci USA. 1999;96(24):13603–13610. doi: 10.1073/pnas.96.24.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satou Y, et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. V. Genes for receptor tyrosine kinase pathway and Notch signaling pathway. Dev Genes Evol. 2003;213(5-6):254–263. doi: 10.1007/s00427-003-0317-9. [DOI] [PubMed] [Google Scholar]
- 23.Shibuya M. Vascular endothelial growth factor receptor family genes: When did the three genes phylogenetically segregate? Biol Chem. 2002;383(10):1573–1579. doi: 10.1515/BC.2002.177. [DOI] [PubMed] [Google Scholar]
- 24.Grassot J, Gouy M, Perriere G, Mouchiroud G. Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Mol Biol Evol. 2006;23(6):1232–1241. doi: 10.1093/molbev/msk007. [DOI] [PubMed] [Google Scholar]
- 25.Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134(12):2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276(part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 30.Sawano A, et al. Flt-1, vascular endothelial growth factor receptor 1, is a novel cell surface marker for the lineage of monocyte-macrophages in humans. Blood. 2001;97(3):785–791. doi: 10.1182/blood.v97.3.785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.