Abstract
Inhibitors of poly(ADP-ribose) polymerase (PARP) are in clinical trials for cancer therapy, on the basis of the role of PARP in recruitment of base excision repair (BER) factors to sites of DNA damage. Here we show that PARP inhibition to block BER is toxic to hypoxic cancer cells, in which homology-dependent repair (HDR) is known to be down-regulated. However, we also report the unexpected finding that disruption of PARP, itself, either via chemical PARP inhibitors or siRNAs targeted to PARP-1, can inhibit HDR by suppressing expression of BRCA1 and RAD51, key factors in HDR of DNA breaks. Mechanistically, PARP inhibition was found to cause increased occupancy of the BRCA1 and RAD51 promoters by repressive E2F4/p130 complexes, a pathway prevented by expression of HPV E7, which disrupts p130 activity, or by siRNAs to knock down p130 expression. Functionally, disruption of p130 by E7 expression or by siRNA knockdown also reverses the cytotoxicity and radiosensitivity associated with PARP inhibition, suggesting that the down-regulation of BRCA1 and RAD51 is central to these effects. Direct measurement of HDR using a GFP-based assay demonstrates reduced HDR in cells treated with PARP inhibitors. This work identifies a mechanism by which PARP regulates DNA repair and suggests new strategies for combination cancer therapies.
Keywords: DNA repair, hypoxia
Poly(ADP-ribose) polymerases (PARPs) comprise a family of enzymes that catalyze ADP ribosylation of a variety of cellular factors (1 –4). PARP-1 is thought to play a key role in DNA repair, primarily by modifying chromatin factors at sites of DNA damage and thereby recruiting repair factors. Inhibitors of PARP have attracted interest for cancer therapy because cancer cells deficient in BRCA1 or BRCA2 due to inactivating mutations are sensitive to PARP inhibition (5 –8). This has been attributed to the role of PARP in recruiting base excision repair (BER) factors that remove damaged bases and fix single-strand breaks (SSBs) (1). SSBs persisting into S-phase produce replication fork collapse, requiring BRCA1- and BRCA2-mediated homology-dependent repair (HDR) for resolution (5, 9, 10).
In prior work, we found that hypoxia suppresses HDR in human cells via transcriptional down-regulation of BRCA1 and RAD51 (11 –15). Hence, we hypothesized that cancer cells in hypoxia, with acquired deficiency in HDR, might have increased sensitivity to PARP inhibition. Work presented here confirms this hypothesis, showing that PARP inhibitors are more cytotoxic to hypoxic than to normoxic cells. Because hypoxia causes BRCA1 and RAD51 down-regulation by stimulating E2F4/p130 occupancy of the BRCA1 and RAD51 promoters, we asked whether disruption of p130 function via expression of human papillomavirus (HPV) E7 would reverse the sensitivity of hypoxic cells to PARP inhibition. We found that E7 expression, as predicted, does confer resistance to PARP inhibitors on hypoxic cells, but surprisingly, it also blocks the toxicity of PARP inhibition in normoxic cells.
As a basis for this effect, we present evidence that PARP inhibitors, themselves, cause BRCA1 and RAD51 down-regulation and do so at the transcriptional level via induction of E2F4/p130 binding to the BRCA1 and RAD51 promoters, a pathway that can be disrupted by HPV E7 expression or by siRNAs targeting p130. siRNAs that knock down PARP-1 expression also cause down-regulation of BRCA1. We also find that the radiosensitization caused by PARP inhibition, an effect previously observed but attributed to the direct role of PARP in BER, is partially reversed by E7 expression or knockdown of p130, suggesting that the down-regulation of BRCA1 and RAD51 has a role in the radiosensitizing effects of PARP inhibitors.
Results
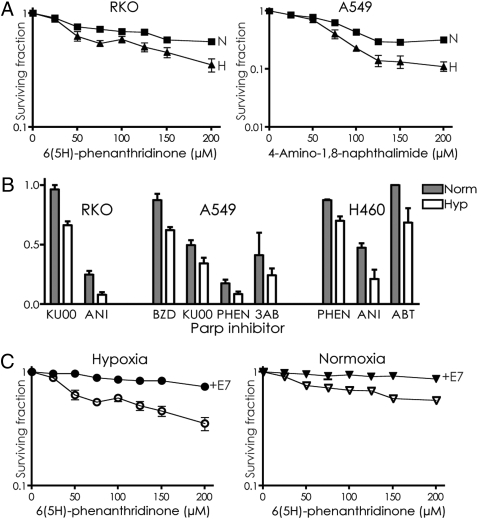
To test the impact of hypoxia on the cytotoxicity of PARP inhibition, a colon cancer cell line, RKO, was grown in normoxia or hypoxia for 2 days, exposed to the PARP inhibitor 6(5H)-phenanthridinone (PHEN), and assayed for cell survival by colony formation (Fig. 1A). The hypoxic cells were found to be more sensitive to PARP inhibition. Similar results were seen in another cell line, A549 (lung cancer derived), treated with the PARP inhibitor 4-amino-1,8-naphthalimide (ANI). We also tested the PARP inhibitors KU0058684 (KU00) and ANI on the RKO cells; benzamide (BZD), KU00, PHEN, and 3-aminobenzamide (3AB) on the A549 cells; and PHEN, ANI, and ABT-888 (ABT) on H460 cells (lung cancer line). In all cases, there was increased killing of cells under hypoxic compared with normoxic conditions (Fig. 1B).
Fig. 1.
Cytotoxicity of PARP inhibition in hypoxic and normoxic cancer cells. (A) Survival by colony formation of RKO colon cancer cells and A549 lung cancer cells under either hypoxic or normoxic conditions to increasing doses of the PARP inhibitors PHEN [6(5H)-phenanthridinone] and ANI (4-amino-1,8-naphthalimide). (B) Survival of RKO, A549, and H460 cells grown under hypoxic or normoxic conditions and exposed to the PARP inhibitors KU00 (100 μM), ANI (200 μM), BZD (5 mM), PHEN (200 μM), 3AB (20 mM), or ABT (4 μM). (C) Effect of E7 expression on survival of RKO cells treated with PHEN. Cells expressing E7 or not were placed in hypoxia or normoxia and treated with PHEN for 48 h in normoxia, followed by growth to allow colony formation. Error bars represent SEs based on three replicates in all cases.
To test the proposed role of BRCA1 and RAD51 down-regulation as a cause of the sensitization to PARP inhibition in hypoxic cells, RKO cells expressing the HPV E7 protein (RKO-E7) or just an empty vector control (RKO-neo) were tested. Previous work has shown that E7 prevents down-regulation of BRCA1 and RAD51 in hypoxia by disrupting p130/E2F4 binding to the respective promoters (11 –15). E7 expression was found to confer resistance to PARP inhibition in hypoxic cells (Fig. 1C), consistent with the proposed mechanism of sensitization. However, a surprising result was seen when the effect of E7 was evaluated in normoxic cells. Unexpectedly, the normoxic cells were also protected from the cytotoxic effects of PARP inhibition by E7 expression (Fig. 1C).
This finding was unexpected because the slight toxicity of PARP inhibition in normal conditions has been attributed just to a reduction in BER that occurs when PARP activity is blocked. With reduced BER, cells are less able to cope with the DNA damage that comes from the by-products of oxidative metabolism. However, the ability of E7 to reverse the toxicity, instead, raised the possibility that a second mechanism was being invoked by inhibition of PARP.
As a putative second mechanism, we hypothesized that the PARP inhibitors might be causing BRCA1 and/or RAD51 suppression and might do so via an E2F4/p130-dependent pathway. In this model, PARP inhibitors would produce toxicity in two ways: (i) the consequences of reduced BER, as discussed above; and (ii) the suppression of BRCA1 or RAD51, or both, by PARP inhibition, leading to reduced HDR. The concurrent inhibition of BER would yield unrepaired SSBs and stalled replication forks. Such stalled forks require HDR for resolution, but the reduction of BRCA1 or RAD51 by PARP inhibition would render cells unable to cope with the lesions, causing increased cell death.
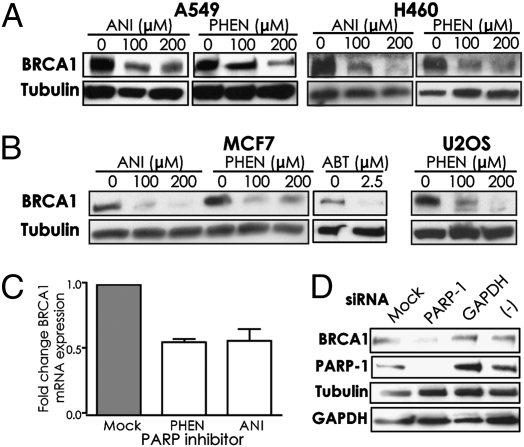
To test this, we asked whether the actual levels of BRCA1 are altered in the PARP inhibitor-treated cells. We found that A549 or H460 cells exposed to PHEN or ANI showed substantially reduced levels of BRCA1 protein (Fig. 2A). MCF7 cells (breast cancer line) also showed reduced levels of BRCA1 when exposed to ANI or to PHEN, as well as to ABT, and U2OS cells, a sarcoma line, showed reduced BRCA1 levels in response to PHEN (Fig. 2B). These results were obtained after exposure of cells to the PARP inhibitors for 72 h. Examination of BRCA1 levels at earlier time points of exposure revealed slight suppression at 6 h to 12 h but substantial suppression by 24 h (Fig. S1A) and 48 h (Fig. S1B). Suppression of BRCA1 at the mRNA level by both PHEN and ANI was seen in A549 cells by quantitative real-time PCR analyses (Fig. 2C). Inhibition of PARP-1 catalytic activity was confirmed in cells treated with the concentrations of PHEN and ANI used in the above experiments (Fig. S1C).
Fig. 2.
Inhibition or knockdown of PARP causes decreased levels of BRCA1 protein and BRCA1 mRNA in human cells. (A) A549 or H460 cells were exposed to the PARP inhibitors PHEN or ANI and harvested at 72 h for analysis of BRCA1 levels by immunoblot. (B) MCF7 and U2OS cells treated with ANI, PHEN, or ABT were analyzed for BRCA1 expression. (C) Analysis of BRCA1 mRNA levels by quantitative real-time RT-PCR in A549 cells after exposure to PARP inhibitors. (D) MCF7 cells were transfected with siRNAs targeting either PARP-1 or GAPDH or with a negative control siRNA pool. After 72 h cells were harvested for immunoblot analysis of the indicated proteins.
Knockdown of PARP-1 by siRNAs also produced down-regulation of BRCA1, whereas neither control siRNAs directed at GAPDH mRNA nor pooled negative control siRNAs had an effect on BRCA1 levels in MCF7 cells (Fig. 2D). These results indicate that three different chemical inhibitors of PARP, as well as siRNAs targeting PARP-1, all yield BRCA1 down-regulation.
Prior work had shown that expression of RAD51 is regulated in response to hypoxic stress in a manner parallel to the regulation of BRCA1 (14). We therefore tested whether the levels of RAD51 are similarly down-regulated upon PARP inhibition. We found that RAD51 levels are reduced in A549, H460, and U2OS cells treated with PARP inhibitors for 72 h (Fig. 3A) and are reduced as early as 48 h in A549 and H460 cells (Fig. S2). RAD51 mRNA levels are also suppressed by PARP inhibition (Fig. 3B), suggesting regulation at the level of transcription, as with BRCA1. siRNA knockdown of PARP-1 in A549 cells was also found to produce down-regulation of RAD51 (Fig. 3C), showing that disruption of PARP-1 by either chemical inhibition or knockdown reduces RAD51 expression.
Fig. 3.
PARP inhibition or knockdown suppresses RAD51 expression. (A) A549, H460, or U2OS cells were treated with PARP inhibitors and harvested at 72 h for analysis of RAD51 levels by immunoblot. (B) Analysis of RAD51 mRNA levels by quantitative real-time RT-PCR in A549 cells after PARP inhibition. (C) A549 cells were transfected with siRNAs targeting either PARP-1 or GAPDH or with a negative control siRNA pool. After 72 h cells were harvested for immunoblot analyses of the indicated proteins. (D) A549 cells were exposed to either hypoxia or PHEN treatment, or both, as indicated, and analyzed for BRCA1 and RAD51.
We next tested the combined effect of PARP inhibition and hypoxia compared with each individual treatment (Fig. 3D). PARP inhibition and hypoxia both yielded substantial down-regulation of BRCA1 and RAD51 in A549 cells. The combined treatments had only a slightly larger effect (detectable only upon longer exposure of the Western blot; Fig. 3D).
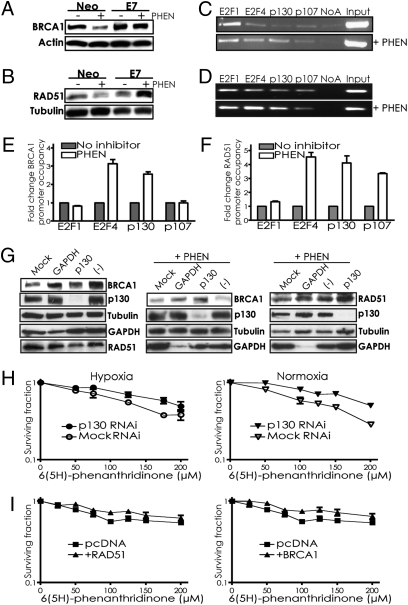
To further probe the mechanism by which PARP inhibition suppresses BRCA1 and RAD51, we examined BRCA1 and RAD51 levels in RKO cells expressing either E7 or an empty vector (Fig. 4 A and B). The RKO-neo cells display down-regulation of BRCA1 and RAD51 when treated with PHEN, consistent with the results in Figs. 2 and 3. However, the isogenic cells expressing E7 (RKO-E7) show no down-regulation of BRCA1 or RAD51 with PHEN exposure (Fig. 4 A and B). These results point to a role for E2F4/p130 complexes in suppression of BRCA1 and RAD51 in response to PARP inhibition, just as in response to hypoxia.
Fig. 4.
PARP inhibitor–mediated suppression of BRCA1 and RAD51 is reversed by E7 expression or p130 knockdown, occurs via E2F4 and p130 occupancy of the respective promoters, and contributes to the cytotoxicity of PARP inhibition. (A and B) RKO cells expressing either E7 or a vector control (Neo) were treated or not with 200 μM PHEN and assayed by immunoblot for (A) BRCA1 or (B) RAD51 expression. ChIP assays of (C) BRCA1 or (D) RAD51 promoter occupancy were performed using antibodies to the indicated factors with lysates from A549 cells treated or not with 200 μM PHEN. Representative agarose gels containing BRCA1 or RAD51 promoter region PCR amplification products are shown. (E) Quantification by real-time PCR of BRCA1 or (F) RAD51 promoter occupancy by the indicated factors is shown, based on three independent ChIP assays, with error bars based on SEs. Promoter occupancy is expressed as the fold change relative to that observed in untreated cells. (G) BRCA1 and RAD51 levels were determined by Western blot in A549 cells that were treated or not with PHEN after transfection with either siRNAs targeting p130, GAPDH, nontargeting negative control pool, or no siRNAs (Mock). Note that the order of the lanes in the third panel is different from in the first and second panels. (H) Effect of p130 knockdown by siRNAs on survival of A549 cells treated with PHEN. Cells transfected or not with siRNAs targeting p130, as indicated, were placed in hypoxia or normoxia and then treated with increasing doses of PHEN for 72 h under normoxic conditions, followed by growth to allow colony formation. Error bars represent SEs based on three replicates. (I) Effect of forced BRCA1 or RAD51 expression on survival of MCF7 cells treated with PHEN. Cells transfected with cDNA expression vectors for BRCA1 or RAD51, or an empty vector control, were treated with increasing doses of PHEN for 72 h under normoxic conditions, followed by growth to allow colony formation. Error bars as above.
ChIP assays of transcription factor occupancy at the BRCA1 and RAD51 promoters (Figs. 4 C–F) reveal increased binding of both promoters by E2F4 and p130 in response to PHEN. Similarly, siRNA knockdown of p130 blocks down-regulation of BRCA1 and RAD51 levels by PHEN (Fig. 4G). p130 knockdown also reverses the sensitivity of normoxic and hypoxic cells to PARP inhibition (Fig. 4H), similar to the effect of E7 expression (Fig. 1C). Hence, the roles of E2F4 and p130 in mediating the effects of PARP inhibition on BRCA1 expression provide an explanation for our initial observation of the impact of E7 (Fig. 1).
MCF7 cells were also transfected with cDNA expression vectors to force expression of either BRCA1 or RAD51 from heterologous promoters in cells exposed to PHEN. The cytotoxicity of PHEN was reduced by forced expression of either BRCA1 or RAD51 (Fig. 4I), further linking BRCA1 and RAD51 levels to the sensitivity of cells to PARP inhibition. Forced expression was confirmed by immunoblot (Fig. S3).
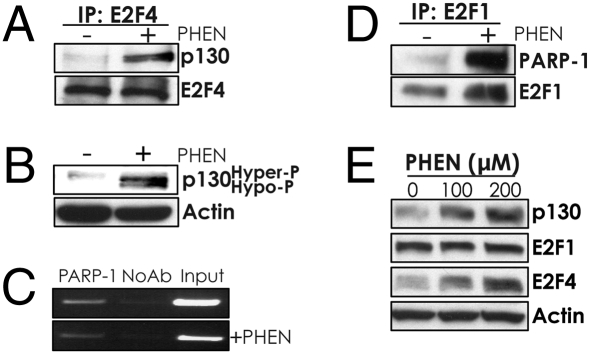
Next, we found that PARP inhibition promotes E2F4/p130 complex formation, as judged by increased coimmunoprecipitation of these two factors from lysates of PHEN-treated cells (Fig. 5A). Further, PHEN treatment induces increased levels of the hypo-phosphorylated form of p130, which is the form that binds to E2F4 (Fig. 5B).
Fig. 5.
PARP inhibition stimulates interaction between p130 and E2F4, whereas PARP-1 binds to the BRCA1 promoter and physically interacts with E2F1. (A) Analysis of the association between p130 and E2F4 by coimmunoprecipitation using an E2F4 antibody in extracts from A549 cells treated or not with PHEN, followed by immunoblot with antibodies to either p130 or E2F4. (B) Western blot analysis of phosphorylation status of p130, as judged by gel mobility, after treatment or not with 200 μM PHEN for 72 h. (C) ChIP assay of BRCA1 promoter occupancy by PARP-1 in A549 cells treated or not with 200 μM PHEN. (D) Analysis of the association between PARP-1 and E2F1 by coimmunoprecipitation using an antibody specific to E2F1 in extracts from MCF7 cells treated or not with PHEN, followed by immunoblot with an antibody to PARP-1. (E) Immunoblot analysis of expression levels in A549 cells of p130, E2F1, and E2F4 after treatment or not with PHEN.
Using promoter–luciferase constructs (11), we found that disruption of either or both of the E2F sites in the BRCA1 promoter attenuates the suppressive effects of PARP inhibition on expression from the promoter (Fig. S4), providing further evidence linking E2F-related factors to regulation of BRCA1 by PARP.
Reports indicate that PARP-1, itself, can interact with gene promoters (16 –18), and so we asked whether PARP-1 could be detected at the BRCA1 promoter by ChIP. We were able to detect association of PARP-1 with the BRCA1 promoter in untreated A549 cells (Fig. 5C), and we found that this association decreases slightly upon PHEN treatment [and does so to a degree similar to the slight reduction in E2F1 binding to the BRCA1 promoter upon PHEN treatment (compare Fig. 5C with Fig. 4C)].
One prior study had also suggested that PARP-1 and E2F1 can physically interact (19). We therefore tested for coimmunoprecipitation of PARP-1 with E2F1 in MCF7 cells treated or not with PHEN (Fig. 5D). There was some detectable interaction between E2F1 and PARP-1 in untreated cells, but this interaction was increased in treated cells. These results suggest that PARP-1 may regulate E2F1 activity through physical interaction and possibly sequestration from promoter sites. The levels of E2F1 are not substantially altered in PHEN-treated cells (Fig. 5E), so the increased association of E2F1 with PARP-1 is not simply due to increased E2F1 levels. Interestingly, E2F4 and p130 levels seem to be increased in PHEN-treated cells (Fig. 5E), and this elevation may contribute to their increased occupancy at the BRCA1 promoter. No interaction was detected between PARP-1 and either E2F4 or p130 (Fig. S5).
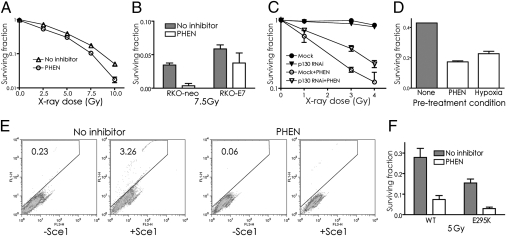
It has been reported that PARP inhibitors can sensitize cells to ionizing radiation (20, 21), a result that we were able to reproduce (Figs. 6 A–C). However, the radiosensitization of RKO cells by PHEN is partially reversed by expression of E7 (Fig. 6B). Radiosensitization by PHEN is also partially reversed by siRNA knockdown of p130 (Fig. 6C). Hence, preventing the down-regulation of BRCA1 and RAD51 caused by PARP inhibition blocks a substantial portion of the radiosensitizing effect of PARP inhibitors. Interestingly, pretreatment of cells with hypoxia also produces subsequent radiosensitization (if the cells are irradiated in oxic conditions), and this effect has also been linked to down-regulation of HDR (22).
Fig. 6.
PARP inhibitor treatment sensitizes cells to ionizing radiation in a pathway dependent on p130 and suppresses HDR of DSBs. (A) Survival of A549 cells exposed to ionizing radiation. Cells were pretreated or not with PHEN at 200 μM for 48 h. (B) Radiosensitization by PARP inhibition is partially reversed by E7 expression. Survival of RKO cells with or without E7 expression to 7.5 Gy of ionizing radiation. Cells were pretreated or not with PHEN (200 μM for 48 h). (C) Radiosensitization by PARP inhibition is partially reversed by siRNA knockdown of p130. Cells transfected or not (Mock) with siRNAs targeting p130 were treated or not with PHEN for 48 h and exposed to ionizing radiation, followed by growth to allow colony formation. Error bars represent SEs based on three replicates. (D) Survival of A549 cells pretreated or not with PHEN (200 μM for 48 h) or pretreated with exposure to hypoxia (0.1% for 48 h), followed by treatment with PHEN (100 μM for 24 h), as indicated. Error bars as above. (E) Impaired HDR in MCF7 cells after treatment with PHEN, as detected using the DR-GFP recombination substrate. MCF7 cells containing the DR-GFP substrate were exposed or not to PHEN for 72 h, followed by transfection with either an I-SceI expression vector (+SceI) to induce a site-specific DSB or with an empty vector (-SceI) as a control. Cells were assayed 72 h later by FACS for HDR of the I-SceI-induced DSB, as indicated by production of GFP+ cells. Percentages of GFP+ cells are shown in each panel. (F) PARP inhibition produces radiosensitization even in cells already defective in BER. Mouse embryonic fibroblasts expressing wild-type or a dominant negative variant (E295K) of DNA polymerase β were treated or not with PHEN for 48 h and then exposed to 5 Gy of ionizing radiation. Survival was determined by colony formation, with SEs calculated on the basis of three replicates.
As a corollary to the radiosensitization by PARP inhibitors via BRCA1 suppression, we hypothesized that pretreatment with a PARP inhibitor would also sensitize cells to a subsequent exposure to the inhibitor. We tested this by comparing the effects on cell survival of pretreatment of A549 cells either with hypoxia (which we know from the results in Fig. 1 sensitizes cells to subsequent PARP inhibition) or with PHEN, followed by further exposure to PHEN. As shown (Fig. 6D), both pretreatments led to reduced survival in response to the follow-up treatment with PHEN compared with the non-pretreated cells.
To directly test whether PARP inhibition causes a functional change in HDR of double-strand breaks (DSBs), we used a GFP-based, chromosomal assay in which a DSB is generated by expression of the I-SceI endonuclease, whose recognition site is integrated in the GFP gene such that it disrupts the gene (23). Repair of the I-SceI–mediated DSB by HDR using a downstream GFP fragment as a template gives rise to a functional GFP gene, which is quantified by FACS (Fig. 6E). HDR activity was found to be reduced by PARP inhibition compared with untreated cells (0.20% vs. 3.26% GFP-positive cells, respectively; Fig. 6E).
Because the above experiments show that PARP inhibition reduces HDR in addition to its known inhibitory effect on BER, we sought to determine the relative contributions of BER inhibition vs. HDR inhibition to the radiosensitivity conferred by PARP inhibitors. We used a matched pair of cell lines expressing either wild-type or a dominant negative version of polymerase β (E295K), the primary polymerase in short patch BER (24). The cells expressing the E295K are already somewhat radiosensitive relative to the wild type (consistent with the deficiency in BER), but these cells are even further radiosensitized by PARP inhibition (Fig. 6F). These results suggest that the impact of PARP inhibition on radiation response goes beyond an effect on BER and reflects an additional effect on HDR. Hence, the radiosensitization by PARP inhibitors reflects both inhibition of BER and suppression of HDR. Consistent with this, siRNA knockdown of BRCA1 or PARP-1 does not add much to the radiosensitizing effect of PHEN treatment (Fig. S6).
Discussion
DNA repair pathways have emerged as promising targets for cancer therapy, in part because many cancers arise in the setting of repair abnormalities and in part because many cancer therapies act via DNA damage. PARP inhibitors have attracted attention as potential antineoplastic agents because they inhibit an important enzyme in the BER pathway. The work reported here shows that PARP inhibitors can also suppress the expression of BRCA1 and RAD51, two key molecules in the HDR pathway. In particular, the experiments demonstrate that suppression of BRCA1 and RAD51 in response to PARP inhibition occurs in a pathway mediated by repressive E2F4/p130 complexes acting on the BRCA1 and RAD51 promoters. This is supported by ChIP assays showing increased occupancy of the respective promoters by E2F4 and p130 after PARP inhibition and by the reversal of BRCA1 and RAD51 suppression by HPV E7 expression, which serves as a molecular tool to block p130 activity, or by direct siRNA-mediated p130 knockdown. Functionally, cells treated with PARP inhibitors are sensitized to ionizing radiation, an effect partially reversed by expression of E7 or by p130 knockdown, thereby linking a portion of the induced radiosensitivity to the down-regulation of BRCA1 and RAD51. Direct measurement of DSB repair using a GFP-based assay provides further evidence that PARP inhibition suppresses HDR of DSBs, consistent with down-regulation of BRCA1 and RAD51.
In prior work, we had shown that exposure of cells to hypoxia can lead to suppression of BRCA1 and RAD51 and that this occurs in a coordinated fashion via stimulation of E2F4/p130 binding to the respective promoters (11, 14). It is interesting that PARP inhibition also invokes this pathway. Hence, both hypoxia and PARP inhibition suppress the HDR factors BRCA1 and RAD51, and accordingly, both treatments lead to increased susceptibility of cells to killing by subsequent treatment with a PARP inhibitor.
With respect to the radiosensitization produced by PARP inhibitors, we show that E7 expression or p130 knockdown can reverse a portion of this effect. Because E7 expression or p130 knockdown both serve to maintain BRCA1 and RAD51 levels despite PARP inhibition, our results suggest that manipulation of BRCA1 and RAD51 activity is an important factor in the ability of PARP inhibitors to modify the radiation response.
In addition, radiosensitization by PARP inhibition was seen even when BER was already inhibited in cells by expression of a dominant negative polymerase β variant. This result provides additional evidence that the radiation sensitivity produced by PARP inhibition is not just due to its known effect on BER.
However, inhibition of BER by PARP inhibitors may still play an important role because some residual radiosensitization can still be detected even in the presence of E7 or in the setting of p130 knockdown. By blocking BER, PARP inhibition perpetuates toxic intermediates arising from attempted repair of otherwise nonlethal (but mutagenic) base damage. In addition, there is evidence that PARP may also participate in a minor subpathway of nonhomologous end joining for DSB repair (25, 26), although the influence of this pathway on radiosensitivity is not clear.
In the case of hypoxia, it is well established that low oxygen is radioprotective if the irradiation occurs when the cells are hypoxic; however, in the posthypoxic period, when oxygen is present, there is increased sensitivity of cells to radiation. This effect was reported almost 20 years ago (27), and it was recently shown to correlate with the suppression of HDR via down-regulation of BRCA1 and RAD51 by hypoxia (22).
Our finding that PARP plays a regulatory role in the expression of BRCA1 and RAD51 fits with an accumulating body of work indicating that PARP can influence transcription (2, 28). Several mechanisms have been proposed to account for the effects of PARP on gene expression. PARP can catalyze addition of poly(ADP-ribose) chains to histones, loosening chromatin structure. PARP can also serve as a coactivator of transcription by interaction with other transcription factors, such as NF-κB (28) and c-Myb (2). However, the mechanism by which PARP influences the activity of these factors at specific promoters has not been worked out. In the cases of NF-κB and c-Myb, the catalytic activity of PARP is not required (2), and so some sort of scaffolding or allosteric function has been proposed.
PARP-1 has also been found to physically interact with E2F1, on the basis of both in vitro binding and coimmunoprecipitation (19). Prompted by this prior observation, we tested for interactions of PARP-1 with E2F1, E2F4, or p130. We did detect coimmunoprecipitation of PARP-1 and E2F1, and we found that the extent of interaction was increased by treatment of cells with PARP inhibitors, suggesting that PARP-1 may regulate E2F1 activity via differential physical interactions. However, we could not detect physical interaction of PARP-1 with either E2F4 or p130.
Using ChIP assays, we found that PARP inhibition causes a slight decrease in BRCA1 promoter occupancy by E2F1 (and by PARP-1 itself) along with a marked increase in promoter occupancy by E2F4 and p130. This increased binding by E2F4 and p130 to the BRCA1 promoter is associated with modulation of p130 phosphorylation status and thereby induction of its interaction with E2F4, leading to formation of repressive complexes. In addition, treatment of cells with PARP inhibitors was also seen to cause an elevation in the overall levels of p130 and E2F4, providing a further mechanism that may contribute to increased BRCA1 promoter occupancy by these factors.
Initially, this work was prompted by our prior findings that BRCA1 and RAD51 are down-regulated in hypoxic cells (11, 14), leading us to propose the hypothesis that hypoxic cells would therefore be sensitive to PARP inhibition. The results reported here confirm this sensitivity in hypoxic cells, indicating that PARP inhibitors may be useful as therapeutic agents for cancers with extensive hypoxic fractions. However, our unexpected finding that PARP inhibition, itself, can suppress BRCA1 and RAD51 expression identifies a previously uncharacterized mechanism of action for PARP inhibitors and may provide the basis for new therapeutic strategies that combine PARP inhibition with agents that exploit decreased HDR. It also suggests that proposed strategies to use PARP inhibitors for cancer chemoprevention may be counterproductive (8).
Materials and Methods
Cell Culture and Hypoxia.
A549, H460, MCF7, and U2OS cells were obtained from the ATCC. RKO-neo and RKO-E7 cell lines were provided by Dr. Kathleen Cho (University of Michigan). The MCF7 DR-GFP cell line was provided by Dr. Maria Jasin (Memorial Sloan-Kettering Cancer Center). Wild-type polymerase β and E295K polymerase β dominant negative cells were provided by Dr. Joann Sweasy (Yale University School of Medicine). Refer to SI Materials and Methods for cell culture growth conditions. For hypoxic culture conditions, cells received a continuous flow of a humidified mixture of 95% N2 and 5% CO2 gas certified to <10 ppm O2 for 48 h at 37 °C, as previously described (29).
PARP Inhibitors.
KU0058684 was provided by Graeme C. M. Smith (KuDOS Pharmaceuticals). BZD, ANI, PHEN, and 3AB were from Sigma-Aldrich. ABT-888 was from Alexis Biochemicals.
Clonogenic Survival Assays.
Clonogenic survival was determined by colony formation, as previously described (30). Details are provided in SI Materials and Methods.
Immunoblotting.
Cells were either treated with PARP inhibitors or transfected with siRNA or protein expression vector for the specified length of time, and harvested and lysed as previously described (31). Western blotting was then performed as previously described (32). The primary antibodies used are described in SI Materials and Methods. Proteins were visualized with HRP-conjugated anti-mouse or anti-rabbit IgG and the SuperSignal West Pico Chemiluminescent Substrate detection system (Thermo Scientific). Immunoprecipitations were performed as previously described (14), and the antibodies used are listed in SI Materials and Methods.
Quantitative Real-Time PCR Analysis.
Quantitative real-time PCR analyses were performed as previously described (11, 12). Details are given in SI Materials and Methods.
RNA Interference.
siRNAs directed against PARP-1, GAPDH, p130, BRCA1 (ON-TARGETplus SMARTpool reagents; Dharmacon) and a negative control (ON-TARGETplus siCONTROL Nontargeting Pool reagent; Dharmacon) were transfected into MCF7 or A549 cells using DharmaFECT1 transfection reagent (Dharmacon) according to the manufacturer’s instructions. Cells were lysed after 48–72 h, and Western blotting was performed to ascertain protein knockdown. The antibodies used are described in SI Materials and Methods.
Chromatin Immunoprecipitation.
A549 cells were treated or not with 200 μM PHEN for 72 h, and ChIP assays were done as previously described (11, 12). The primer sequences for the BRCA1 and RAD51 promoter regions have also been reported (11, 12). Antibodies for E2F1, E2F4, and p130 are listed in SI Materials and Methods; others were: p107 (C-18; Santa Cruz Biotechnology) and PARP-1 (p716/p25; Millipore).
Plasmids.
For the BRCA1 and RAD51 expression vectors cDNAs were cloned into the expression vector pcDNA3.1. Transfections were done in MCF7 cells using the FuGENE 6 Transfection Reagent (Roche) according to the manufacturer’s instructions.
Functional Assay for Homology-Dependent Repair.
The assay to measure HDR of I-SceI-induced DSBs using the DR-GFP reporter construct in MCF7 cells has been previously described (11, 23). Briefly, MCF7 DR-GFP cells were treated or not with 200 μM PHEN for 72 h and then transfected or not with a vector expressing I-SceI by electroporation, as described previously (11). The cells were analyzed by FACS for GFP expression 72 h later, as described previously (11). The PHEN-treated cells were maintained in medium containing PHEN postelectroporation.
Supplementary Material
Acknowledgments
We thank L. Cabral and B. Johnson for their help and J. Sweasy, K. Cho, and M. Jasin for providing cell lines. This work was supported by National Institutes of Health Grants R01ES005775 and P01CA129186 (to P.M.G.) and by American Cancer Society Grant PF-07-263-01 (to M.E.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904783107/DCSupplemental.
References
- 1.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 3.Bürkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 4.Ivana Scovassi A, Diederich M. Modulation of poly(ADP-ribosylation) in apoptotic cells. Biochem Pharmacol. 2004;68:1041–1047. doi: 10.1016/j.bcp.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 5.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.McCabe N, et al. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-ribose) polymerase: An issue of potency. Cancer Biol Ther. 2005;4:934–936. doi: 10.4161/cbt.4.9.2141. [DOI] [PubMed] [Google Scholar]
- 8.Hay T, et al. Efficient deletion of normal Brca2-deficient intestinal epithelium by poly(ADP-ribose) polymerase inhibition models potential prophylactic therapy. Cancer Res. 2005;65:10145–10148. doi: 10.1158/0008-5472.CAN-05-1186. [DOI] [PubMed] [Google Scholar]
- 9.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31:4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helleday T, Bryant HE, Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4:1176–1178. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- 11.Bindra RS, et al. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 12.Bindra RS, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindra RS, Glazer PM. Basal repression of BRCA1 by multiple E2Fs and pocket proteins at adjacent E2F sites. Cancer Biol Ther. 2006;5:1400–1407. doi: 10.4161/cbt.5.10.3454. [DOI] [PubMed] [Google Scholar]
- 14.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 15.Bindra RS, Glazer PM. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat Res. 2005;569:75–85. doi: 10.1016/j.mrfmmm.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Chu S, Xu H, Ferro TJ, Rivera PX. Poly(ADP-ribose) polymerase-1 regulates vimentin expression in lung cancer cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1127–L1134. doi: 10.1152/ajplung.00197.2007. [DOI] [PubMed] [Google Scholar]
- 17.Soldatenkov VA, et al. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277:665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 18.Zampieri M, et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simbulan-Rosenthal CM, et al. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene. 2003;22:8460–8471. doi: 10.1038/sj.onc.1206897. [DOI] [PubMed] [Google Scholar]
- 20.Brock WA, et al. Radiosensitization of human and rodent cell lines by INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase. Cancer Lett. 2004;205:155–160. doi: 10.1016/j.canlet.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 21.Küpper JH, et al. trans-dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against gamma-irradiation and N-methyl-N’-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol Cell Biol. 1995;15:3154–3163. doi: 10.1128/mcb.15.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan N, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–614. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 23.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol Cell Biol. 2007;27:5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miwa M, Masutani M. PolyADP-ribosylation and cancer. Cancer Sci. 2007;98:1528–1535. doi: 10.1111/j.1349-7006.2007.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok TT, Sutherland RM. The radiation response of cells recovering after chronic hypoxia. Radiat Res. 1989;119:261–267. [PubMed] [Google Scholar]
- 28.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–5757. [PubMed] [Google Scholar]
- 30.Zeng M, et al. Ionizing radiation-induced apoptosis via separate Pms2- and p53-dependent pathways. Cancer Res. 2000;60:4889–4893. [PubMed] [Google Scholar]
- 31.Peterson-Roth E, Reynolds M, Quievryn G, Zhitkovich A. Mismatch repair proteins are activators of toxic responses to chromium-DNA damage. Mol Cell Biol. 2005;25:3596–3607. doi: 10.1128/MCB.25.9.3596-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mihaylova VT, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.