Abstract
Infection of Escherichia coli by the T7 phage leads to rapid and selective inhibition of the host RNA polymerase (RNAP)—a multi-subunit enzyme responsible for gene transcription—by a small (∼7 kDa) phage-encoded protein called Gp2. Gp2 is also a potent inhibitor of E. coli RNAP in vitro. Here we describe the first atomic resolution structure of Gp2, which reveals a distinct run of surface-exposed negatively charged amino acid residues on one side of the molecule. Our comprehensive mutagenesis data reveal that two conserved arginine residues located on the opposite side of Gp2 are important for binding to and inhibition of RNAP. Based on a structural model of the Gp2-RNAP complex, we propose that inhibition of transcription by Gp2 involves prevention of RNAP-promoter DNA interactions required for stable DNA strand separation and maintenance of the “transcription bubble” near the transcription start site, an obligatory step in the formation of a transcriptionally competent promoter complex.
Keywords: gene protein 2, promoter melting , inhibitor
In all cellular organisms, gene transcription, the first and most regulated step during gene expression, is catalyzed by the highly conserved multi-subunit DNA-dependent RNA polymerase (RNAP). The activity of RNAP is tightly controlled to enable the rapid switching of gene expression patterns in response to various cues. The RNAP of Escherichia coli, the most-studied RNAP, consists of five polypeptides (α2ββ′ω), which constitute the catalytic core (E). Specificity is conferred on the core by a dissociable sigma (σ) subunit, which converts E to the holoenzyme (Eσ) capable of promoter-specific transcription initiation (reviewed in ref. 1). Most E. coli promoters with conserved sequences near positions −35 and −10 with respect to the transcription start site (the +1 site) are recognized by RNAP containing the major housekeeping σ factor, σ70, or a related σ70-family member. Under certain stress conditions, a small subset of E. coli promoters is used by the RNAP containing a major variant σ factor, σ54 (reviewed in ref. 2). In addition, the bacterial RNAP can be controlled by an array of transcription regulatory proteins and small ligands that repress, stimulate, or modulate its activity to fine-tune gene expression profiles and ultimately satisfy the requirements for cell survival. Not surprisingly, bacteriophages (phages) have evolved strategies to alter the activity of bacterial (host) RNAP during infection to shift host resources towards the production of viral progeny. This modulation can occur in two ways, either through covalent modifications, such as phosphorylation or ADP ribosylation of target sites in the RNAP, or through low–molecular weight (MW) phage-encoded proteins that bind to RNAP (3, 4).
Infection of E. coli by T7 phage provides paradigmatic examples of how posttranslational modifications and low-MW phage-encoded RNAP-binding proteins are used to modulate the activity of host RNAP (5). The gene expression program of T7 phage relies on both E. coli RNAP and the single-subunit T7 RNAP. Early T7 genes, including gene 0.7 (encoding Gp0.7) and gene 1 (encoding the T7 RNAP) are transcribed by E. coli RNAP; T7 RNAP subsequently transcribes middle T7 genes, including gene 2 (encoding Gp2), and the late T7 genes. Gp0.7 is a protein kinase that phosphorylates a specific threonine residue (T1068) in the evolutionarily variable region of the E. coli RNAP β′ subunit, called the β′ GNCD domain (6). Gp2 comprises a polypeptide of 64 amino acid residues that binds to the structurally conserved RNAP β′ jaw domain (Fig. 1A), a multifunctional domain that contributes to all stages of transcription (7 –9).
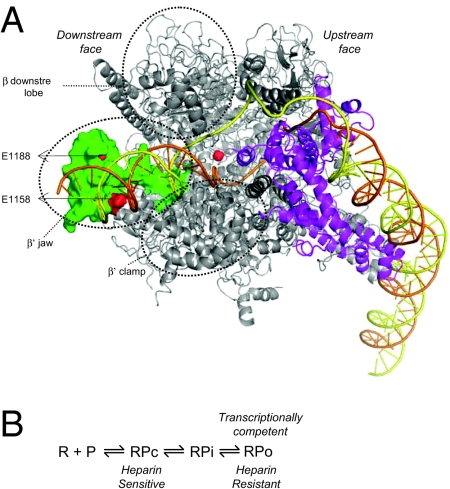
Fig. 1.
(A) Model of the RPo generated using the crystal structure of Thermus aquaticus RNAP (25) shown as a ribbon representation. Highlighted in green is the β′ jaw domain at the “downstream face” of the RNAP (E. coli residues 1149–1190), the deletion of which confers Gp2 resistance. The locations of amino acids E1158 and E1188 are indicated in red. The active site is indicated by the orange sphere. The location of the σ factor (magenta) at the “upstream face” of the RNAP and the path of the modeled DNA (orange, template strand; yellow, nontemplate strand) in the RPo is shown. Circled are the domains of the β and β′ subunit at the downstream face of the RNAP, which together with the β′ jaw contribute to the downstream DNA-binding channel. (The β′ GNCD domain is absent in the T. aquaticus RNAP.) (B) Schematic depiction of the steps leading to the transcriptionally competent RPo at the lacUV5 promoter (12). R, RNAP; P, promoter template; RPc, closed promoter complex; RPi, intermediate promoter complex; RPo, open promoter complex.
In the transcriptionally competent and competitor (heparin) resistant open promoter complex (RPo; Fig. 1A), the β′ jaw forms (together with the β downstream lobe, β′ clamp, and β′ GNCD domains) a trough at the downstream face of the RNAP (hereinafter called the downstream DNA-binding channel), which accommodates the double-stranded DNA exiting the active site (hereinafter called the downstream DNA). Structural models based on protein–DNA cross-linking (10), fluorescence energy transfer studies (11), and biochemical analyses of RNAP deleted for the β′ jaw (8) are consistent with the idea that the β′ jaw could have sequence-nonspecific interactions with bases of downstream DNA in the RPo and the transcribing complex (Fig. 1A). Deletion of the E. coli β′ jaw (amino acids 1149–1190) confers Gp2 resistance, but this mutant RNAP forms significantly destabilized RPo (8). Furthermore, charge-reversal point mutations in the β′ jaw (E1158K or E1188K) also confer resistance to Gp2 (Fig. 1A) (7). It is thus conceivable that the function of Gp2 is to prevent stable RPo formation by the host RNAP. However, an earlier study (7) reported that recombinant Gp2 prevents promoter recognition by Eσ70 and thus inhibits transcription by preventing formation of the early initiation intermediate RPc, an unstable closed promoter complex sensitive to heparin challenge (12) (Fig. 1B).
The finer details of RNAP recognition and inhibition by Gp2 remain unknown. Here we describe the high resolution structure of this low-MW phage-encoded RNAP inhibitor. Molecular modeling shows that two evolutionary conserved surface-exposed arginine residues of Gp2 are ideally positioned for direct electrostatic interaction with the β′ jaw glutamates (E1188 and E1158) known to be important for Gp2 binding. Indeed, in vitro binding and transcription assays using a library of single-alanine Gp2 mutants, as well as in vivo complementation assays, have confirmed the importance of these arginine residues for Gp2 function. By combining structural and mutagenesis data and the results of biochemical functional assays, we put forward a model of RNAP inhibition by Gp2. We provide experimental evidence in support of this model, which envisions that Gp2 inhibits late step(s) during RPo formation by antagonizing the β′ jaw–downstream DNA interactions required for the formation and stable maintenance of the RPo. In particular, the binding of Gp2 to Eσ70 seems to affect stable DNA strand separation near the +1 site.
Results
Structure Determination of T7 Gp2.
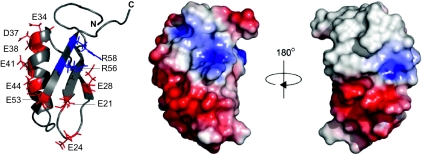
The solution structure of Gp2 was determined using standard heteronuclear NMR methods. Backbone Cα, Cβ, CO, N, and HN assignments were obtained from HNCACB/CBCA(CO)NH and HN(CA)CO/HNCO spectra, and side-chain assignments were obtained using HCCH total correlation spectroscopy (TOCSY) spectra. Using a combination of manual and automated NMR assignment methods using the ARIA program, a family of 20 structures was calculated (Fig. S1 and Table S1). The sequence of Gp2 folds into a compact globular domain comprising a three-stranded β sheet that packs against an α-helix in a β1β2α1β3 topology (Fig. 2). The central β2 strand is flanked by antiparallel β1 and β3 strands. A buried hydrophobic core can be defined by side chains of residues F16, A18, and V20 (β1); I31 and A33 (β2); A39 and A43 (α1); and V54 and V57 (β3). On the surface, a notable feature is the separation of negatively and positively charged residues on opposite sides of the molecule (Fig. 2). Although the invariant R56 and R58 (Fig. S2) at the C terminus are flanked by the side chains of E21 and E28, the remaining negative charges of E24, E34, D37, E38, E41, E44, and E53 form a contiguous strip of negative charges running the length of the α helix to the β1β2 loop (Fig. 2).
Fig. 2.
NMR-derived three-dimensional structure of Gp2. The left panel shows a ribbon representation of Gp2 indicating the side chains of the negatively charged amino acid residues and the conserved arginines at positions 56 (red) and 58 (blue). The middle and right panels show two views of the molecular surface of Gp2 (in the same orientation as the ribbon form) color-coded according to a basic electrostatic surface distribution, calculated using the vacuum electrostatics program in Pymol, version 0.99rc6.
Invariant Arginines 56 and 58 at the C Terminus Are Essential for the Inhibitory Function of Gp2.
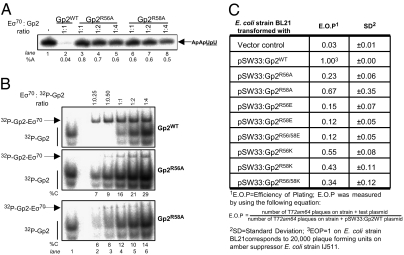
The structure of Gp2 reveals two prominent arginine side chains protruding from the edge of the β3 strand (Fig. 2). The two residues are conserved in all known Gp2-like proteins encoded by various T7 relatives (Fig. S2). Based on this observation, we speculated that invariant R56 and R58 might be important for function. To evaluate the contribution of these residues, the inhibitory activity of R56A and R58A Gp2 mutants on Eσ70 was measured in an in vitro transcription assay using a 65-bp DNA fragment containing the lacUV5 promoter sequence as the template. Under the conditions used here, this assay reports the ability of Eσ70 to bind to the promoter, initiate DNA strand separation, and synthesize a tetranucleotide RNA transcript, ApApUpU (SI Materials and Methods and Fig. S3). Incubation of Eσ70 with equimolar amounts of Gp2WT before the addition of promoter DNA effectively abolished the synthesis of ApApUpU (Fig. 3A, lanes 1 and 2). Gp2R56A and Gp2R58A, when present at equimolar amounts to Eσ70, inhibited the synthesis of ApApUpU by only ∼20% and ∼40%, respectively (Fig. 3A, lanes 3 and 6). Gp2R56A and Gp2R58A failed to fully inhibit Eσ70 even when present at ∼4-fold molar excess over Eσ70 (Fig. 3A, lanes 4, 5, 7, and 8). These findings indicate that residues R56 and R58 are important for Gp2 function.
Fig. 3.
Role of R56 and R58 in Gp2 function. (A) An autoradiograph of a 20% (wt/vol) denaturing gel showing synthesis of the transcript ApApUpU (indicated by the arrow; with the underlined nucleotides 32P-labeled) from lacUV5 by Eσ70 in the presence of increasing amounts of Gp2R56A and Gp2R58A. The percentage ApApUpU synthesized (%A) by Eσ70 in the presence of Gp2 with respect to reactions with no Gp2 are given at the bottom of the gels. (B) An autoradiograph of a 4.5% (wt/vol) native gel showing the binding of 32P-Gp2WT (Top), 32P-Gp2R56A (Middle), and 32P-Gp2R58A (Bottom) to Eσ70 are shown. The migration positions of 32P-Gp2 (lane 1) and the Eσ70-Gp2 complex (lanes 2–6) are indicated. Radioactivity in the mutant and WT Eσ70-Gp2 complexes was measured, and the Eσ70-binding activity of the R56A and R58A Gp2 mutants is expressed as the percentage of Gp2WT-binding activity (%C) for each corresponding ratio of Eσ70:32P-Gp2. At the bottom of the middle and bottom panels, the percentage of mutant 32P-Gp2 associated with Eσ70 compared with Gp2WT is given (%C). In A and B, the molar ratio of Gp2 present with respect to Eσ70 in each lane is shown at the top. (C) Plating efficiency of T72 AM64 phage on E. coli strain BL21 transformed with pSW33gp2 encoding mutant Gp2 proteins with the R to E and R to K substitutions at positions 56 and/or 58.
R56 and R58 Are Required for Gp2 Binding to Host RNAP.
We considered the possibility that Gp2R56A and Gp2R58A are ineffective RNAP inhibitors because they bind RNAP less effectively than Gp2WT. Toward this end, we conducted native-PAGE binding analysis of complexes formed between increasing amounts of 32P-labeled wild-type (WT) or mutant Gp2 proteins and fixed amounts of Eσ70. Our results reveal that Gp2R56A and Gp2R58A were significantly impaired for binding to Eσ70 (Fig. 3B). Semiquantitative analyses indicated that at equimolar amounts of 32P-Gp2 and Eσ70, there was 6-fold less Gp2R56A-Eσ70 complex and 8-fold less 32P-Gp2R58A-Eσ70 complex compared with the amounts of complex formed with Gp2WT. Even at ∼4-fold molar excess of 32P-Gp2, relatively weak binding of Gp2R56A and Gp2R58A to Eσ70 was detected (a 3- and 7-fold binding defect, respectively; Fig. 3B, lane 6). The impaired ability of Gp2R56A and Gp2R58A to bind Eσ70 closely correlates with the loss of inhibitory activity in the in vitro transcription assays. It seems that even though Gp2R58A showed some inhibition of Eσ70 in vitro (Fig. 3A), the complex formed between Gp2R58A and Eσ70 was less stable, because it did not survive native PAGE.
We also tested the functionality of Gp2 mutants in vivo. Toward this end, E. coli cells harboring pET-based expression plasmids containing WT or mutant gene 2 were infected with T7 phage harboring an am64 amber mutation in gene 2 (13). T72am64 infections of WT E. coli cells are not productive, because Gp2 is essential for phage development. We expected that infection of E. coli cells harboring a plasmid with WT gene 2 would be productive, because T7 RNAP synthesized during early stages of infection should transcribe the plasmid-borne gene 2 recall that genes cloned in pET plasmids are under the control of a T7 RNAP promoter. Indeed, E. coli BL21 cells harboring the pSW33:Gp2WT plasmid were productively infected by T72am64, as judged by the efficiency of plaque formation (EOP), calculated as the ratio of plaque observed on nonsuppressing hosts to plaque observed on suppressing hosts (Fig. 3C). As expected based on the in vitro results, plaque formation by T72am64 on lawns of E. coli BL21 harboring the pSW33:Gp2R56A and pSW33:R58A plasmids was less efficient (plating efficiencies 77% and 33% of the efficiency observed with cells harboring pSW33:Gp2WT; Fig. 3C). The reduced in vivo activity of Gp2R56A and Gp2R58A closely correlates with their reduced activity in vitro, where, when present at equimolar conditions with RNAP, they inhibit transcription by ∼20% and ∼40%, respectively (Fig. 3A, lanes 3 and 6). These findings confirm that arginine residues at positions 56 and 58 are important for Gp2 function both in vivo and in vitro.
Interaction Between Gp2 and Host RNAP Involves an Electrostatic Component.
Previous experiments identifying Gp2-resistant RNAP mutants containing charge-reversal substitutions at E1188K or E1158K within the β′ jaw (7) suggested that the interaction of Gp2 with host RNAP could have a significant electrostatic component. Our results showing that R56 and R58 are important for the binding of Gp2 to RNAP lend support to this idea. To evaluate the contribution of the charge at positions 56 and 58 to Gp2 function, we constructed R to E (charge-reversed) and R to K (charge-maintained) single mutants, as well as the corresponding double mutants R56/58E and R56/58K. As shown in Fig. 3C, plating efficiencies of T72am64 on lawns of E. coli cells expressing Gp2 mutants with charge-altering substitutions were reduced by ∼85%–90% compared with that on E. coli cells expressing Gp2WT. The plating efficiency on cells expressing R to K substitutions was less affected (a reduction of 3-fold or less). These findings are consistent with the view that the Gp2–RNAP interaction contains a significant electrostatic component, although introduction of lysines instead of Gp2 arginines at positions 56 and/or R58 affects the function as well.
Alanine Scanning of Gp2 Reveals Functional Contributions of Each Amino Acid Position.
To identify other amino acids involved in the interaction of Gp2 with Eσ70, we extended the in vitro transcription assay to a library of Gp2 mutants containing an alanine substitution at every position, excluding the starting methionine and six alanines found in the WT sequence. The results are summarized in Fig. S4A, where the ability of each mutant to inhibit Eσ70 at a 1:1 molar ratio is reported as percentage of activity relative to Eσ70 activity in the absence of Gp2. Gp2 mutants that inhibited Eσ70 by ≥90% were considered to have WT activity. Apart from residues R56 and R58, deleterious mutations were observed predominantly in residues buried within the hydrophobic core of Gp2 (Fig. S4 A and B). Alanine substitutions of buried residues F16, I31, V54, and V57 and partially exposed F52 could destabilize the structure of Gp2 by disrupting the hydrophobic core. The introduction of an alanine residue instead of G51 also likely affects Gp2 folding, because it could impair the ability of the polypeptide to form a turn connecting α1 and β3. Indeed, native gel migration properties of 32P-labeled versions of I31A, F16A, G51A, F52A, V54A, and V57A Gp2 mutants were markedly different from those of WT Gp2, indicating that alanine substitutions at these positions affect the overall structural integrity of Gp2 (Fig. S4C).
The proton (1H) NMR spectrum of Gp2WT is characteristic of a small, folded domain, as judged by the dispersion of resonances below ∼0.5 ppm (corresponding to Hγ1 and Hγ2 protons from V20 at −0.126 and 0.046 ppm, respectively; Fig. S4D). These resonances typically represent buried protons of methyls or methylenes in hydrophobic cores. Their frequencies are shifted by electronic ring currents from the plane of a neighboring aromatic side chain. Similarly, there was good dispersion of resonances in the amide and aromatic regions (∼6.5–9 ppm). These features were retained in the spectra of Gp2R56A and Gp2R58A (and at least in part in Gp2V57A), indicating that removal of these side chains does not abolish the native structure. In contrast, this pattern of resonances was lost in the spectrum of Gp2F16A, confirming that the structure is significantly disrupted by mutation (Fig. S4D).
Gp2 proteins with alanine substitutions at positions I31, F16, G51, F52, V54, and V57 were able to inhibit Eσ70 to some extent, indicating that a fraction of folded protein is retained in these mutants or that their folding is coupled to association with Eσ70. Overall, the functional screen of the alanine scan library revealed that Gp2 is rather resilient to site-specific mutations, with no strong correlation between positions showing a phenotype and the degree of sequence conservation in Gp2-like proteins. The results also confirm that arginine residues at positions 56 and 58 within the C-terminal region of Gp2 are the principal functional determinants for the binding and inhibition of host RNAP.
Gp2 Inhibits Transcription by Antagonizing RPo Formation.
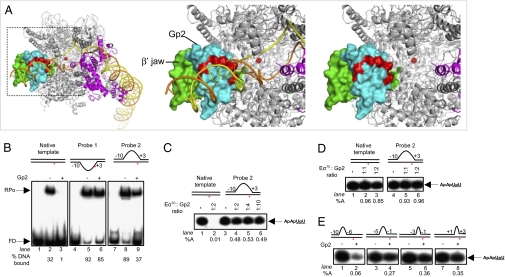
To better understand how Gp2 inhibits Eσ70, we developed a restraint-driven model of the Gp2-RNAP complex based on mutagenesis experiments, our NMR structure, and the earlier observation that binding of Gp2 does not induce large-scale structural changes in RNAP (14). In this model (Fig. 4A and SI Materials and Methods), Gp2 is bound to the β′ jaw such that it projects the negatively charged side chains of residues E21, E28, E34, D37, E38, E41, E44 and E53 into the downstream DNA-binding channel. We envisage that the negative charge introduced in the downstream DNA-binding channel repels the negatively charged DNA, thereby preventing efficient and stable RPo formation and leading to transcription inhibition. In addition, Gp2 also could sterically antagonize conformational rearrangements in the DNA-binding channel that are required for accommodation of downstream DNA and stable RPo formation (see earlier). Irrespective of a particular mechanism, the presence of Gp2 in the DNA-binding channel should shift the RPc–RPo equilibrium away from RPo. To test this view, we determined whether preopening of promoter DNA, which mimics the conformation of DNA in RPo, could allow Eσ70 to overcome inhibition by Gp2. We conducted experiments with linear lacUV5 templates containing a range of heteroduplex segments between positions −10 and +3 (with respect to the +1 site). The lacUV5 probe 1 contains a WT nontemplate strand sequence and the template strand with noncomplementary sequence from −10 to +3, whereas lacUV5 probe 2 contains a WT template strand sequence and a nontemplate strand with noncomplementary sequence from −10 to +3 (SI Materials and Methods). Because the consensus −10 sequence recognized by Eσ70 is partially disrupted in probe 2, we used a native gel shift assay to demonstrate that Eσ70 forms heparin-resistant RPo on both probes (Fig. 4B, lanes 5 and 8). In line with our expectations, complex formation on both probes was markedly more Gp2-resistant—∼85% and ∼40% more activity, respectively, in the presence of Gp2 than was observed with native lacUV5 template under the same conditions (∼2-fold molar excess of Gp2 over Eσ70; Fig. 4B, lanes 3, 6, and 9). Because probe 2 preserves the WT +1 site sequence, it was used in the in vitro transcription assay. Consistent with the native gel shift results, Eσ70 remained highly active on this probe even in the presence of ∼10-fold molar excess of Gp2 (Fig. 4C, lanes 4–6). The addition of Gp2 to preformed transcriptionally competent promoter complexes on heteroduplex lacUV5 probe 2 also had little effect on the amount of ApApUpU synthesized by Eσ70 (Fig. 4D). Additional data suggested that Gp2 was no longer bound to (and thus did not interact with) RNAP in transcriptionally competent promoter complexes formed on heteroduplex lacUV5 probe 2 (Fig. S5 and SI Experiment 1). Thus, preopening of promoter DNA facilitates displacement of Gp2, presumably by facilitating the interaction of downstream DNA with the β′ jaw and/or other RNAP elements of the downstream DNA-binding channel.
Fig. 4.
Gp2 inhibits step(s) leading to the RPo. (A) Model of the Gp2–RNAP complex. The boxed region in the image in the left panel is enlarged in the middle (with promoter DNA) and right panels (without promoter DNA) to emphasize the β′ jaw region. The RNAP is presented as in Fig. 1A. Gp2 is shown in cyan, and the negatively charged side chains of residues E21, E28, E34, D37, E38, E41, E44 and E53, which protrude into the DNA binding channel, are highlighted in red. (B) Autoradiograph of a 4.5% (wt/vol) native gel showing heparin-resistant RPo formation by Eσ70 in the absence (lanes 2, 5, and 8) and presence (lanes 3, 6, and 9) of ∼2-fold molar excess of Gp2 on 32P-labeled versions of the fully duplex (native) and heteroduplex probes 1 and 2 lacUV5 promoter templates (see text; FD, free DNA). The % template DNA in the RPo is shown at the bottom of the gel. (C) Autoradiograph of a 20% (wt/vol) denaturing gel showing synthesis of the transcript ApApUpU (indicated by the arrow) from the native (lanes 1 and 2) and heteroduplex probe 2 (lanes 3–6) lacUV5 promoter templates in the absence (lanes 1 and 3) and presence (lanes 2, 4, 5, and 6) of Gp2. The Gp2:Eσ70 molar ratio in each lane is shown at the Top. The percentage transcripts synthesized (%A) by Eσ70 in the presence of Gp2 with respect to reactions with no Gp2 are given at the bottom. (D) As in C, but Gp2 was added to the reactions after transcriptionally competent promoter complexes had formed on native (lanes 2 and 3) and heteroduplex probe 2 (lanes 5 and 6) lacUV5 promoter templates. (E) As in C, but assays were conducted with lacUV5 promoter templates containing heteroduplex segments of different lengths and at different positions with respect to the transcription start site (probes 3–6; see text). In B–E, the lacUV5 promoter templates used are shown schematically with the positions and lengths of the heteroduplex segment indicated with respect to +1 site (indicated by the red asterisk).
Because nucleation of DNA strand separation during RPo formation occurs around position −10 and then extends beyond the +3 position in fully formed RPo, we extended our analysis to determine the position and the length of the heteroduplex segment sufficient to overcome inhibition by Gp2. In vitro transcription assays were conducted with linear lacUV5 templates containing heteroduplex segments formed due to nonnative nontemplate strand sequences extending from −10 to −6 (probe 3), from −5 to −1 (probe 4), from −3 to −1 (probe 5), and from +1 to +3 (probe 6) (Fig. S3A). In the absence of Gp2, Eσ70 effectively synthesized ApApUpU from each of these templates (Fig. 4E, lanes 1, 3, 5, and 7). In the presence of ∼2-fold molar excess of Gp2 over Eσ70, complete inhibition of ApApUpU synthesis was seen only in reactions with probe 3 (Fig. 4E, lane 2). In contrast, Gp2 was able to inhibit ApApUpU synthesis by Eσ70 by only ∼70%–60% in reactions with templates containing +1 site-proximal heteroduplex segments (probes 4–6) (Fig. 4E, lanes 4, 6, and 8). Because Eσ70 can transcribe with varying degrees of efficiency from premelted promoter templates in the presence of Gp2, we conclude that Gp2 does not inhibit RPc formation (Discussion). Instead, Gp2 must be inhibiting Eσ70 at a step after RPc formation. The locations of premelted segments that allow Eσ70 to overcome Gp2 inhibition suggest that interactions antagonized by Gp2 occur at a step after the nucleation of promoter melting and are required for stable DNA strand separation near the +1 site.
Discussion
In bacteria, the regulation of RNAP activity is often accomplished by DNA-binding transcription regulatory factors. Gp2 is a phage T7-encoded, non–DNA-binding transcription factor that is a potent inhibitor of the E. coli Eσ70. Our analyses have revealed that two arginine residues (R56 and R58), which are conserved in all Gp2-like proteins, are essential for binding to and inhibition of Eσ70 and the interaction between Gp2 and the β′ jaw. Our results indicate that the interaction between Gp2 and the host RNAP likely involve a major electrostatic component, consistent with the view that the Gp2–RNAP complex is salt-sensitive (7). The striking charge separation on the surface of Gp2 is likely to be important for the binding of Gp2 to the β′ jaw and also for Gp2’s function as a transcription inhibitor. During RPo formation, interactions of promoter DNA with the catalytic cleft of the RNAP cause DNA to kink sharply in the −10 region, resulting in loading of the largely double-stranded DNA into the catalytic cleft of the RNAP, followed by nucleation of DNA strand separation (15). The transcription-competent and fully heparin-resistant RPo status is acquired when the “jaws” of the RNAP close onto the DNA and the DNA is unwound from approximately −10 to +3, thus creating a mature “transcription bubble.” In RPo, downstream DNA is secured in the RNAP downstream DNA-binding channel (Fig. 1A). In RPo formed by Eσ70, contacts with downstream DNA proximal to the +1 site (∼ +5 to +8) are likely to be made by the β lobe and β′ clamp domains, whereas distal contacts (∼ +10 to +20) with downstream DNA are likely to be made by the β′ jaw, β′ GNCD, and β′ clamp domains (Fig. 1A) (10, 11). Because Eσ70 lacking the β′ jaw, β′ GNCD, or β lobe domain, or containing deletions in the β′ clamp domain, form RPo with a markedly reduced half-life (8, 16–18), it is conceivable that interaction between the downstream DNA and the downstream DNA-binding channel contribute to stable propagation of DNA strand separation during RPo formation. Our results are consistent with this view and suggest that the binding of Gp2 to the β′ jaw could prevent interactions between the β′ jaw and downstream DNA and/or conformational changes in the downstream DNA-binding channel required for accommodation of downstream DNA. Both scenarios would antagonize formation and/or stable maintenance of RPo and thus lead to effective inhibition of transcription. The asymmetric distribution of charged residues in Gp2 and our structural model of the Eσ70–Gp2 complex suggest that electrostatic repulsion between the negatively charged DNA and the negatively charged surface of Gp2 (formed by residues E21, E28, E34, D37, E38, E41, E44, and E53), which is projected into the downstream DNA-binding channel, could be one basis for how Gp2 antagonizes RPo formation. Notably, the mutagenesis data suggest that removing any one of the negatively charged side chains at positions E21, E28, E34, D37, E38, E41, E44, or E53 has no detectable effect on the ability of Gp2 to inhibit Eσ70. This result is consistent with the view that Gp2 electrostatically repels downstream DNA during RPo formation, because substitution of any one negatively charged residue would not alter the surface electrostatic distribution of Gp2 significantly. However, we do not discount an alternate possibility, that the binding of Gp2 to the β′ jaw per se also sterically antagonizes, at some point during RPo formation, the conformational rearrangement in the downstream DNA-binding channel necessary to accommodate the downstream DNA.
The ability of Eσ70 to overcome inhibition by Gp2 on preopened (heteroduplex) promoter templates suggests that during the transition from RPc and RPo, preopening the promoter shifts the equilibrium toward RPo. Because Gp2 does not bind RPo [formed on native templates (7)], it ceases to be an effective inhibitor of transcription from heteroduplex templates and in fact is displaced from the β′ jaw (Fig. S5 and SI Experiment 1). The near-identical protection of DNA downstream of the +1 site in transcriptionally competent and heparin-resistant promoter complexes formed on heteroduplex lacUV5 probe 2 in the presence and absence of Gp2 (Fig. S6) further supports the idea that Gp2 is displaced on the formation of transcriptionally competent promoter complexes, and that downstream DNA follows the same path through RNAP in the absence and presence of Gp2. Apparently on native, fully double-stranded templates, there is insufficient binding energy to initiate DNA strand separation and displace Gp2, which are required for RPo formation (Fig. S5A).
The results indicate that Gp2 does not prevent promoter recognition by Eσ70 and thus does not inhibit RPc formation, but rather affects later steps en route to RPo. Indeed, experiments conducted with the native and heteroduplex probes 1 and 2 lacUV5 promoter templates at ∼4°C to determine whether Gp2 inhibits RPc formation indicated that the RPc formed on native and heteroduplex probes 1 and 2 lacUV5 promoter templates is resistant to inhibition by Gp2 (Fig. S7A and SI Experiment 2). The near-identical protection of the native lacUV5 promoter by Eσ70 in RPc in the presence and absence of Gp2 (Fig. S7B) further corroborates the view that Gp2 has no detectable effect on RPc formation. In addition, the observation that Gp2 does not inhibit RPc formation by a Gp2-sensitive form of Eσ54 (and can bind to Eσ54 in RPc) (19) is consistent with idea that Gp2 inhibits steps after RPc formation at most promoters.
The results with promoter templates containing heteroduplex segments of different lengths and locations with respect to the +1 site suggest that the step(s) inhibited by Gp2 relate to a late stage during RPo formation that is associated with stable opening of promoter DNA around the +1 site. Because the Gp2 binding site (β′ jaw) and the +1 site (at the active center of the RNAP) in the RPo are >30 Å apart, it seems that interactions between the β′ jaw and the downstream DNA are important for stable DNA strand separation around the +1 site. This conclusion is consistent with a previous result demonstrating that the half-life of RPo formed by Eσ70 (and Eσ54) lacking the β’ jaw was markedly improved on the lacUV5 and Sinorhizobium meliloti nifH promoter templates containing +1 site proximal heteroduplex segments compared with the native promoter template (9), and also with a study suggesting a genetic link between residues in the β′ jaw and the active site of the RNAP (20).
Each stage of the bacterial transcriptional cycle is targeted by phage-encoded host RNAP-binding regulatory proteins (4). Many phage transcription regulators function as alternative σ factors and redirect host RNAP to transcribe phage genes to support phage development and thereby affect a step associated with RPc formation at bacterial promoters. Other such regulators affect steps associated with transcription elongation by host RNAP (4). The inhibition of E. coli RNAP by T7 phage Gp2 provides a novel example of transcription regulation by a phage-encoded transcription regulator at the RPo formation step. Our results provide a structural and functional framework for unraveling the mechanism of inhibition of host RNAP by Gp2. Time-resolved analyses of RNAP–promoter DNA interactions, DNA strand separation, and RNAP–Gp2 interactions by rapid-footprinting methods are currently underway to precisely define the steps en route to the RPo inhibited by Gp2.
Materials and Methods
Proteins.
All of the proteins used in this work were prepared essentially as described previously (7, 19). Detailed descriptions are provided in SI Materials and Methods.
In Vitro Transcription Assays.
The 10-μL reactions were conducted using final concentrations of 75 nM Eσ70, 20 nM promoter DNA, 0.5 mM dinucleotide primer ApA, 100 μg/mL of heparin, 3 μCi of [α-32P]-UTP, and 0.5 μM UTP in buffer R [10 mM Tris-Cl (pH 8), 10 mM MgCl2, 1 mM DTT, and 100 mM NaCl]. Unless stated otherwise, Gp2 (at concentrations indicated in the figures and text) and Eσ70 were always preincubated before the promoter DNA was added to the reaction. A detailed description of the assay is provided in SI Materials and Methods.
Native Gel Mobility Assays.
All native mobility shift assays were conducted essentially as described previously (19, 21). Binding reactions (10 μL) were set up in buffer R at 37 °C and analyzed on a 4.5% (wt/vol) native polyacrylamide gel. The gel was run for 45–60 min at 100 V and then dried. Proteins or protein–DNA complexes were visualized and quantified using a Fuji PhosphorImager. For the experiments shown in Fig. 3B, 75 nM Eσ70 was incubated with 18.75–300 nM 32P-Gp2 for 5 min before electrophoresis. For the experiments shown Fig. S4C, 300 nM 32P-Gp2 was incubated for 5 min at 37 °C for buffer R before electrophoresis. For the experiments shown in Fig. 4B, 75 nM Eσ70 was incubated with 20 nM 32P-labeled promoter DNA fragment to allow promoter complex formation. Before electrophoresis, 100 μg/mL of heparin was added to the reaction for 5 min. The reaction products were resolved on a native gel kept at ∼37 °C in a water bath.
EOP Assays.
The EOP assays were conducted as described in detail in SI Materials and Methods.
NMR Spectroscopy and Structure Calculation.
Backbone and side-chain assignments were completed using standard double- and triple-resonance assignment methodology (22). Hα and Hβ assignments were obtained using HBHA(CBCACO)NH. The side-chain assignments were completed using HCCH TOCSY and (H)CC(CO)NH TOCSY. Three-dimensional 1H-15N/13C Nuclear Overhauser Effect Spectroscopy (NOESY)-Heteronuclear Multiple Quantum Coherence (HSQC) experiments (mixing time, 100 ms at 800 MHz) provided the distance restraints used in the final structure calculation. The ARIA protocol (23) was used for completion of the NOE assignment and structure calculation. A total of 1,037 NOE-derived distances (comprising 819 unambiguous restraints and 218 ambiguous restraints) were assigned from 13C- and 15N-edited spectra. Dihedral angle restraints derived from TALOS were implemented as well (24). The frequency window tolerances for assigning NOEs were ±0.03 ppm for direct proton dimensions and ±0.04 ppm for indirect proton dimensions, and ±0.5 ppm for nitrogen dimensions and ±1.2 ppm for carbon dimensions. The ARIA parameters p, Tv, and Nv were set to default values. The 20 lowest-energy structures had no NOE violations >0.5 Å and no dihedral angle violations >5°. The structural statistics are presented in Table S1.
Supplementary Material
Acknowledgments
We thank Seth Darst for providing the PDB coordinates for the RPo model. This project was funded by grants from the (BBSRC) (to S.W., S.M., E.C., and P.S.). S.W. is a recipient of a Biotechnology and Biological Sciences Research Council (BBSRC) David Philips Fellowship (BB/E023703). Work in the laboratory of K.S. was supported by National Institutes of Health Grant GM59295 and a grant from Russian Academy of Sciences Presidium Molecular and Cellular Biology program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NMR solution structure of Gp2. Assigned wwPDB ID code: 2wnm for the coordinate entry. NMR, atomic coordinates, chemical shifts, and restraints.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907908107/DCSupplemental.
References
- 1.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wigneshweraraj S, et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 3.Nechaev S, Severinov K. The elusive object of desire—interactions of bacteriophages and their hosts. Curr Opin Microbiol. 2008;11:186–193. doi: 10.1016/j.mib.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 5.Hesselbach BA, Nakada D. “Host shutoff” function of bacteriophage T7: Involvement of T7 gene 2 and gene 0.7 in the inactivation of Escherichia coli RNA polymerase. J Virol. 1977;24:736–745. doi: 10.1128/jvi.24.3.736-745.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Severinova E, Severinov K. Localization of the Escherichia coli RNA polymerase beta′ subunit residue phosphorylated by bacteriophage T7 kinase Gp0.7. J Bacteriol. 2006;188:3470–3476. doi: 10.1128/JB.188.10.3470-3476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nechaev S, Severinov K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J Mol Biol. 1999;289:815–826. doi: 10.1006/jmbi.1999.2782. [DOI] [PubMed] [Google Scholar]
- 8.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 9.Wigneshweraraj SR, Burrows PC, Severinov K, Buck M. Stable DNA opening within open promoter complexes is mediated by the RNA polymerase beta′ jaw domain. J Biol Chem. 2005;280:36176–36184. doi: 10.1074/jbc.M506416200. [DOI] [PubMed] [Google Scholar]
- 10.Korzheva N, et al. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 11.Mekler V, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 12.Buc H, McClure WR. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter: Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 13.Burck KB, Miller RC., Jr Marker rescue and partial replication of bacteriophage T7 DNA. Proc Natl Acad Sci USA. 1978;75:6144–6148. doi: 10.1073/pnas.75.12.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nechaev S, Yuzenkova Y, Niedziela-Majka A, Heyduk T, Severinov K. A novel bacteriophage-encoded RNA polymerase binding protein inhibits transcription initiation and abolishes transcription termination by host RNA polymerase. J Mol Biol. 2002;320:11–22. doi: 10.1016/S0022-2836(02)00420-5. [DOI] [PubMed] [Google Scholar]
- 15.Saecker RM, et al. Kinetic studies and structural models of the association of E. coli sigma(70) RNA polymerase with the lambdaP(R) promoter: large-scale conformational changes in forming the kinetically significant intermediates. J Mol Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 16.Nechaev S, Chlenov M, Severinov K. Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J Biol Chem. 2000;275:25516–25522. doi: 10.1074/jbc.M002511200. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett MS, Gaal T, Ross W, Gourse RL. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 18.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 19.Wigneshweraraj SR, et al. Regulated communication between the upstream face of RNA polymerase and the beta′ subunit jaw domain. EMBO J. 2004;23:4264–4274. doi: 10.1038/sj.emboj.7600407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ederth J, Mooney RA, Isaksson LA, Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 2006;356:1163–1179. doi: 10.1016/j.jmb.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 21.Wigneshweraraj SR, et al. Enhancer-dependent transcription by bacterial RNA polymerase: The beta subunit downstream lobe is used by sigma 54 during open promoter complex formation. Methods Enzymol. 2003;370:646–657. doi: 10.1016/S0076-6879(03)70053-6. [DOI] [PubMed] [Google Scholar]
- 22.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 23.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: Automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 25.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4-Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.