Abstract
Fishery genetics have greatly changed our understanding of population dynamics and structuring in marine fish. In this study, we show that the Atlantic Bluefin tuna (ABFT, Thunnus thynnus), an oceanic predatory species exhibiting highly migratory behavior, large population size, and high potential for dispersal during early life stages, displays significant genetic differences over space and time, both at the fine and large scales of variation. We compared microsatellite variation of contemporary (n = 256) and historical (n = 99) biological samples of ABFTs of the central-western Mediterranean Sea, the latter dating back to the early 20th century. Measures of genetic differentiation and a general heterozygote deficit suggest that differences exist among population samples, both now and 96–80 years ago. Thus, ABFTs do not represent a single panmictic population in the Mediterranean Sea. Statistics designed to infer changes in population size, both from current and past genetic variation, suggest that some Mediterranean ABFT populations, although still not severely reduced in their genetic potential, might have suffered from demographic declines. The short-term estimates of effective population size are straddled on the minimum threshold (effective population size = 500) indicated to maintain genetic diversity and evolutionary potential across several generations in natural populations.
Keywords: ancient DNA, effective population size, genetic structure, large pelagic fishes, Thunnus thynnus
With an impressive amount of data accumulated in the last 10–15 years, genetic studies have greatly changed our understanding of the ecology and evolution of marine fish populations by showing evidence of complex genetic structure at the spatial and temporal scales of variation. This is especially true for fish species with high mobility, high potential for dispersal during egg and larval stages, and large population sizes (1, 2). Accurate estimates of genetic variation in fish populations, both for neutral and adaptive loci, can also suggest unique strategies for stock management and conservation of evolutionary potential in severely exploited fishery resources (3–5). The assessment of population dynamics, environmentally driven changes, and levels of exploitation in large pelagic fish is crucial for both stock management and conservation of marine ecosystems dominated by these oceanic top predators (6). Although fisheries still remain the chief source of data for assessing spatiotemporal population dynamics of these fish (7–10), whenever possible, their findings should be compared with those of fishery-independent approaches, such as genetic, electronic, and microchemical tagging experiments (11–13), to have more accurate data on the key features of population dynamics, such as spawning and migrations (10).
The Atlantic Bluefin tuna (ABFT, Thunnus thynnus) is a large top-predator fish exploiting the pelagic ecosystems of the North Atlantic Ocean and Mediterranean Sea. Much like the other large tunas, the ABFT displays highly migratory behavior, with documented transoceanic and large-scale movements for feeding and reproduction, high fecundity, and high mortality of larval stages (reviewed in 14, 15). Nonetheless, tagging experiments and fishery data analyses have unraveled an unexpected and complex interplay of ecological, behavioral, and reproductive factors that could affect the spatial and temporal population dynamics at the large scale (11, 16). Microchemical signatures in otoliths of yearlings (i.e., young-of-the-year) unequivocally identified two main spawning areas [i.e., the Mediterranean Sea for the Eastern Atlantic population, the Gulf of Mexico for the Western Atlantic population (13)]. Because of high rates of natal homing of spawning adults to their native areas (95.8% for the Mediterranean Sea and 99.3% for the Gulf of Mexico) and limited and more complex movements in sexually mature ABFTs (11, 17), the two populations show significant genetic divergence at both microsatellite (12) and mtDNA (18) loci. The two ABFT populations differ in size, with the eastern population approximately 10 times as large as the western population (19). Both are suffering from overfishing and are considered depleted, consistent with the continuous decline of the spawning stock biomass (which, in the eastern stock, is now at 40% of the 1970s levels), with the high fishing mortality rate of age class 8 and older individuals, and with modeling predictions (16, 19, 20).
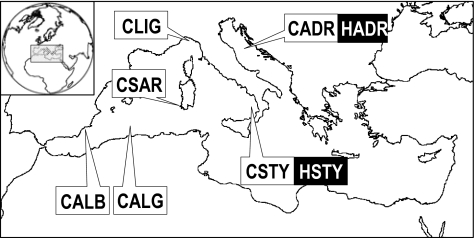
The complex population dynamics exhibited by ABFTs at the large scale is also apparent at a finer scale in the Mediterranean Sea (16). Size-dependent movements and spawning in different areas and periods have been documented (15, 21–23). Genetic variation among three geographical samples suggested that at least two subpopulations inhabit and persist over short time periods (3 years) in the western and eastern Mediterranean Sea, yielding Wright’s genetic variance (FST) values within the Mediterranean Sea between 0.0007 and 0.0087 (12, 24). In this study, we started from a more thorough sampling of Mediterranean ABFTs in space and time. Along with an extensive spatial sampling of contemporary ABFTs carried out from 1999 to 2007 in the central-western Mediterranean (CWM), we also had access to historical ABFT specimens collected by one of the authors (M.S.) in the CWM tuna traps between 1911 and 1926 (Fig. S1). We could thus compare the genetic composition of historical and contemporary ABFT samples, respectively, collected in the CWM before and after intense stock exploitation to address three main questions, namely: (i) Is there any genetic evidence of population structuring, (ii) If so, is such population structuring persistent over several decades and generations, and (iii) Have the Mediterranean ABFT populations suffered significant and consistent genetic bottlenecks?
Results
We surveyed genetic variation in historical (H, n = 219) and contemporary (C, n = 256) ABFTs at eight microsatellite loci (Fig. 1 and Tables S1 and S2). However, a historical sample (HCWM, n = 111) showed a strong excess of heterozygotes at many loci, which we could not logically justify (SI Materials and Methods and Table S2). For this reason, these data were disregarded in all successive analyses. After this removal and according to the sampling design, we analyzed ABFT genetic diversity and differentiation in contemporary population samples collected from six locations of the CWM and in two temporal replicates from the southern Tyrrhenian (STY) and Adriatic (ADR) Seas (Fig. 1 and Table S2).
Fig. 1.
Sampling locations of historical (H, full boxes) and contemporary (C, white boxes) T. thynnus samples in the CWM. Sampling data are detailed in SI Text and Table S1. Because the HCWM sample lacks data on the geographical sampling area and it was excluded from the data analysis (SI Text), it is not reported.
All samples but the CSTY sample showed deviations of allele frequencies across loci from Hardy–Weinberg equilibrium, and most inbreeding coefficient (FIS) values, Wright’s measure of heterozygote deficit, were positive and significant (Table 1). Exceptions were the contemporary Alboran Sea (CALB) sample, in which the departure from equilibrium frequencies did not reach significance, and the CSTY sample, in which there was a moderate but insignificant heterozygote excess. Single-locus effect was ruled out by jackknifing over loci, estimating the FIS value each time. The presence of null alleles was inspected by applying Microchecker (25); after correction of allele frequencies, only four initially significant single-locus tests (detected in the two historical samples) (Table S2) lost their significance. Allelic richness and gene diversity across contemporary and historical samples were not significantly different (Table 1).
Table 1.
Gene diversity, Hardy–Weinberg equilibrium deviation test, and FIS estimates of T. thynnus samples
| Gene diversity |
|||||||
| Sample | n | aR* | He* | Ho* | PHW | FIS | PF |
| HADR | 69 | 8.7 | 0.73 | 0.64 | 0.0000 | 0.119 | 0.0005 |
| HSTY | 39 | 9.3 | 0.73 | 0.62 | 0.0000 | 0.140 | 0.0001 |
| CADR | 73 | 10.0 | 0.74 | 0.66 | 0.0000 | 0.099 | 0.0007 |
| CSTY | 39 | 9.4 | 0.71 | 0.74 | NS | −0.057 | NS |
| CLIG | 36 | 9.2 | 0.76 | 0.69 | 0.0000 | 0.087 | 0.0033 |
| CSAR | 29 | 8.5 | 0.70 | 0.63 | 0.0025 | 0.088 | 0.0113 |
| CALG | 39 | 9.5 | 0.73 | 0.69 | 0.0015 | 0.054 | 0.0369 |
| CALB | 40 | 8.5 | 0.71 | 0.67 | 0.0026 | 0.046 | NS |
aR, mean allelic richness; He, mean expected heterozygosity; Ho, mean observed heterozygosity; NS, not significant; PHW, P value of the Hardy–Weinberg equilibrium deviation test; PF, P value of FIS calculated after 10,100 permutations.
*All Mann–Whitney U test values across ancient and modern samples were not significant.
Most pairwise differences between contemporary samples, as estimated by FST appeared significantly greater than 0, with three exceptions, CADR-contemporary Ligurian (CLIG) Sea samples, central Sardinian (CSAR) Sea-CLIG samples, and CSTY-contemporary Algerian (CALG) Sea samples (Table 2). After sequential Bonferroni correction for multiple tests, all these differences remained significant except two, CALB-CSTY and CSAR-CSTY samples. Nonsignificant FSTs were detected only among contemporary ABFT samples. The overall level of genetic differentiation was of the same extent among contemporary samples (FST = 0.014, P < 0.0001) and between the two historical samples (FST = 0.020, P < 0.0001), suggesting this is not a transient phenomenon. The limited but significant differentiation in space and time among all samples was confirmed by the analysis of molecular variance (AMOVA; 1.70% of the total variance; P < 0.0001). Moreover, differences were significant both within groups, namely, among contemporary and historical samples (1.37%; P < 0.0001), and in the overall comparison of contemporary vs. historical samples (0.50%; P = 0.033). On the other hand, we tried to assign genotypes to K = 1, 2, 3...10 clusters using STRUCTURE software (26) and found that the most likely clustering was in a single group. For any K > 1, the individuals’ posterior assignment probability was essentially the same for each specified cluster, showing that there is no detectable population structuring, at least for the number of loci and sample sizes we are considering.
Table 2.
Pairwise FSTs (below the diagonal) and P values (above the diagonal) among T. thynnus samples
| HADR | HSTY | CADR | CSTY | CLIG | CSAR | CALG | CALB | |
| HADR | <0.0001 | <0.0001 | 0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| HSTY | 0.020 | <0.0001 | 0.0018 | 0.0019 | 0.0001 | 0.0009 | 0.0008 | |
| CADR | 0.015 | 0.016 | <0.0001 | 0.0996 | 0.0077 | <0.0001 | <0.0001 | |
| CSTY | 0.022 | 0.015 | 0.015 | 0.0001 | 0.0030 | 0.1217 | 0.0032 | |
| CLIG | 0.019 | 0.017 | 0.005 | 0.018 | 0.0851 | 0.0014 | <0.0001 | |
| CSAR | 0.028 | 0.027 | 0.011 | 0.014 | 0.008 | 0.0005 | 0.0004 | |
| CALG | 0.024 | 0.020 | 0.016 | 0.004 | 0.016 | 0.021 | 0.0007 | |
| CALB | 0.026 | 0.019 | 0.015 | 0.011 | 0.019 | 0.021 | 0.015 |
Significance was obtained on 10,100 permutations. Values that lost significance after the Bonferroni sequential correction (α = 0.0029) are presented in boldface.
Values of short-term effective population size (Ne) were estimated by two methods: a pseudolikelihood method for estimating effective population size implemented in MLNE v.1.1 software (27) and a likelihood-based method implemented in TM3 v.1 software (28). Both methods estimated Ne by comparing historical and contemporary samples from the same area and assuming a given generation time (tg). We initially used for the CWM ABFT a tg of 8 years, which approximately corresponds to the length mode of ∼200+ cm observed in the catch composition from this area (22). These methods assume discrete generations, which was not the case here. However, the time distance between our historical and contemporary samples appears sufficient to minimize any bias attributable to age structure (29). In the ADR samples, effective population size estimates [MLNE: 752, confidence interval (95% CI): 429–1,853; TM3: 682, 95% CI: 363–1,575] was marginally higher than in the STY (MLNE: 326, 95% CI: 193–695; TM3: 405, 95% CI: 221–974). In the STY samples, the TM3 coalescent-based method gave a slightly higher Ne than the likelihood-based MLNE method, but confidence intervals overlap. Using a tg longer than 8 years, the short-term Ne estimates will be reduced, because the genetic drift effect would be obtained in a smaller number of generations. In fact, slightly lower Ne values (ADR samples, MLNE: 537 95% CI: 307–1,330, TM3: 582, 95% CI: 306–1,587; STY samples, MLNE: 222, 95% CI: 132–477, TM3: 353, 95% CI: 186–1,664) were obtained for both populations applying an ABFT tg of 13 years, estimated according to Kell et al. (30) and Fromentin and Kell (31) using International Commission for the Conservation of Atlantic Tuna (ICCAT) data on ABFT life history traits and considering natural mortality alone. This value could be an overestimation of the contemporary tg either for the western Mediterranean ABFT, for which the mean age of spawning stock is ∼8 years (∼200+ cm) or for the eastern Mediterranean ABFT, whose length mode appears to be even lower (∼75 cm) (22). However, tg = 13 years could be a better choice for the historical samples and for the period of our temporal sampling.
We then tried to infer Ne from levels of genetic diversity within each population sample (long-term estimates) by a coalescent-based genealogical analysis (32). The estimates of θ (4Neμ, where μ is the mutation rate expressed as the number of mutations per generation per locus) in the various samples ranged between 21.9 (in the CALB sample) and 39.9 (in the HSTY sample) (Table S3) and were of the same order of magnitude across historical and contemporary population samples. Solving for Ne based on a mutation rate, μ = 10−4 (intermediate between two widely used estimates: 10−3 and 10−5) (33), the long-term Ne values appeared much higher than their short-term counterparts (i.e., between 54,750 and 99,670). Even using the extremely high mutation rate of 10−3 suggested by Fraser et al. (34), the long-term Ne estimates are almost 10-fold as high as short-term estimates. By the same method, we could also obtain estimates of the growth rate. Point estimates were negative for all contemporary and historical samples, even if CIs included 0 (Table S3).
When looking for signatures of population bottlenecks, one reasonable assumption is that rare alleles have a greater chance of being lost than common alleles. This should result in a faster decline of allele number than of heterozygosity (35) and the range in microsatellite allele size (36). Thus, we compared these quantities in two tests for demographic bottlenecks. For the first of these approaches, it is also necessary to define a mutation model, and we chose three, namely, the infinite alleles model (IAM), single-step stepwise model (SMM), and multiple-step stepwise model (TPM) (SI Materials and Methods and Table S4). We detected a significant heterozygote excess in all samples except the CALG samples under the IAM mutation model, potentially corresponding to a bottleneck signal. However, it is controversial to what extent the IAM model faithfully describes the process of microsatellite mutation (37); indeed, the alternative mutation models gave radically different results. In fact, 10 of the 16 tests (2 for each population sample) performed with SMM and TPM gave significant results, and in all cases, this was attributable to a heterozygote deficit, which does not support population bottlenecks. No departure from null expectations was observed in the CLIG and CALB samples under both the SMM and TPM models and in the HSTY and CALG samples under the TPM model.
The second approach is an M-ratio bottleneck test (36) that requires a prior definition of the θ value. We tried three values, namely, those estimated for each sample as long-term Ne and two additional values often reported for natural fish populations (SI Text): 0.5 and 10. For the CADR and CSTY samples, we also had available an estimate of θ derived from the short-term Ne values (0.29 and 0.15, respectively), which we incorporated in the analysis (Table 3). The M-ratio bottleneck test proved to be rather sensitive to the choice of θ. The M ratios in the ABFT Mediterranean samples ranged from 0.73 to 0.84 (Table 3). All samples but the HADR and CADR samples showed M ratios below the 0.82 cutoff value empirically obtained for stable natural populations (36). On the other hand, all ABFT samples showed M > 0.70, which is a diagnostic value of genetic bottlenecks (36). Only seven M values reached statistical significance, and these appeared to be affected by the choice of θ. In fact, most significant M-ratio values were obtained only using values of θ as low as 0.5, except for the HADR and CADR samples. Statistical significance was also detected in the CSTY sample, with the short-term estimate of θ. With higher values of θ (10 and long-term inferences), we did not find evidence for a genetic bottleneck.
Table 3.
M-ratio analysis in the T. thynnus samples
| Sample | θ | M | Mcrit | P value |
| HADR | 29.7 | 0.82 | 0.69 | 0.465 |
| 0.5 | 0.82 | 0.79 | 0.056 | |
| 10 | 0.82 | 0.70 | 0.278 | |
| HSTY | 39.9 | 0.73 | 0.60 | 0.490 |
| 0.5 | 0.73 | 0.79 | 0.004 | |
| 10 | 0.73 | 0.66 | 0.095 | |
| CADR | 35.0 | 0.84 | 0.69 | 0.634 |
| 0.29* | 0.84 | 0.82 | 0.065 | |
| 0.5 | 0.84 | 0.79 | 0.090 | |
| 10 | 0.84 | 0.70 | 0.392 | |
| CSTY | 28.0 | 0.76 | 0.65 | 0.287 |
| 0.15* | 0.76 | 0.83 | 0.005 | |
| 0.5 | 0.76 | 0.79 | 0.011 | |
| 10 | 0.76 | 0.68 | 0.112 | |
| CLIG | 31.3 | 0.79 | 0.64 | 0.660 |
| 0.5 | 0.79 | 0.79 | 0.030 | |
| 10 | 0.79 | 0.67 | 0.300 | |
| CSAR | 20.8 | 0.75 | 0.63 | 0.320 |
| 0.5 | 0.75 | 0.75 | 0.008 | |
| 10 | 0.75 | 0.75 | 0.148 | |
| CALG | 29.3 | 0.79 | 0.79 | 0.523 |
| 0.5 | 0.79 | 0.79 | 0.024 | |
| 10 | 0.79 | 0.79 | 0.239 | |
| CALB | 21.9 | 0.80 | 0.80 | 0.521 |
| 0.5 | 0.80 | 0.80 | 0.030 | |
| 10 | 0.80 | 0.80 | 0.317 |
For each sample, we performed M estimates using three tentative θ values based on the long-term θ estimated by LAMARC (first row), θ = 0.5 (second row), and θ = 10 (third row), except for CADR and CSTY samples. In these samples, an estimate of M was also calculated using a short-term θ value (*) computed by the temporal method. Significant P values are presented in boldface.
Discussion
With this study, we wanted to address three general questions on spatial and temporal population dynamics of ABFT. The first was whether there is solid genetic evidence for population structuring in the Mediterranean Bluefin tuna. Our data show that there are statistically significant differences among population samples, confirming and strengthening previous results obtained with the same approach on a few samples (12, 24). Despite the documented individual tendency to disperse in various life stages, different samples of ABFT show genetic differences over space and time, both at the fine and large scales of variation. Our FST estimates among adult and juvenile ABFT samples in the CWM were greater than those previously estimated at a much larger geographical scale using yearlings (12, 24) and in another tuna species, Thunnus obesus (38). We also asked whether such genetic structuring of Mediterranean ABFTs has been persistent over several decades and generations. Our analysis shows that genetic differences between ABFT populations were present long before the development of industrial fisheries and apparently persisted across approximately the past century and several generations (i.e., 10–12 and 6–7 generations estimated using tgs of 8 and 13 years, respectively).
As a consequence, taking into account the ecological and reproductive traits of ABFTs (reviewed in 14, 15), these levels of genetic structuring led us to reject the hypothesis that the analyzed samples come from a single panmictic population. The general deficit of heterozygotes, observed consistently across loci and population samples, can indeed be interpreted as a form of Wahlund effect or as an effect of inbreeding related to the variance of reproductive success in ABFTs (i.e., only a few spawners contributing to the next generation).
No clear genetic structuring was recognized in this dataset by the method implemented using STRUCTURE software; this inconsistency with respect to the FST results may have to do with a different sensitivity of the statistics chosen to describe the phenomenon. It has been shown that the performance of STRUCTURE to detect the true number of clusters within a given dataset correctly increases with FST, and the assignment failed at FST values of 0.01–0.02 (39). In other terms, by FST analysis in the modern and ancient samples, we identified a low yet apparently stable structuring, which is not evident using less sensitive measures of genetic diversity. Given the recent recolonization (likely after the Last Glacial Maximum) of the Mediterranean Sea by ABFTs (18, 40) and the levels of genetic differentiation detected by FST analysis, it is unlikely that there would be relevant gene flow among these genetically structured populations (18).Our results are consistent with genetic subdivision which could suggest independent breeding populations exist within the CWM.
A general difficulty with highly mobile species such as the ABFT is that it is hard to define population units unequivocally. The necessary assumptions are not straightforward, and species-specific ecological features are also important (41). However, if we consider the heterogeneity of spatial genetic patterns detected within the Mediterranean Sea (present work and refs. 12, 24) and the variation detected in the demographic pattern retrieved both from current genetic variation and from comparisons between historical and contemporary samples (see below), we have reason to suspect that the basin is inhabited by distinct geographical populations, with each of them showing a unique demographic history. Tendency to demographic independence in the Mediterranean ABFTs (more or less pronounced) might also be supported by the occurrence of multiple environmentally suitable spawning areas in CWM [e.g., around the Balearic Islands and in the Southern Tyrrhenian (14)], and by recent fishery (16) and tagging (11) data.
Given the evidence that geographical structuring has persisted through time in the CWM ABFTs, we looked for evidence of genetic bottlenecks in the Mediterranean samples. Our analyses did not identify coherent and significant signals of genetic erosion, although the Eastern Atlantic ABFT stock is known to have declined for a long time and the spawning stock biomass is presently at its lowest on record, with the steepest phase of decline in past 5–10 years (14, 19, 42). Population samples did not show significant heterozygosity excess, and none of the M ratios were below the diagnostic value for genetic bottlenecks (0.70) proposed by Garza and Williamson (36). Although some individual M-ratio tests reached nominal significance, that result was clearly influenced by the choice of the parameter θ. Indeed, using the long-term estimates of θ, there is no evidence for the reduction in genetic diversity. Genetic bottleneck signals would appear only if the real value of θ was 0.5, that is, between 40 and 60 times as low as estimated from our data. By contrast, evidence of genetic bottlenecks is considered reliable only when consistent results emerge regardless of the parameter θ (43).
However, in interpreting these findings, one should keep in mind that the genetic methods to detect bottlenecks are sensitive to downward demographic fluctuations, resulting in a radical loss of genetic diversity. Therefore, what these methods fail to detect as a significant episode at the genetic level may well represent a catastrophe in terms of conservation biology. Estimating the magnitude of the detectable bottlenecks means asking how seriously population sizes might decrease without the allele number being significantly affected. The answer is not straightforward because it depends on the sample size, mutation rates, and gene diversities before the bottleneck (44), and only the first variable can really be measured. For the other two variables, we only have a range of estimates which, as we have seen, lead to different results.
At any rate, the negative growth rates (Table S3) and M ratios <0.82 [a cutoff value for selecting demographically stable natural populations (36)] detected in most historical and contemporary samples likely suggest that CWM ABFT populations, although not severely reduced on their genetic potential, might have suffered from demographic declines. Although we did not find evidence of dramatic losses of genetic diversity, the aforementioned signals suggest that recent demographic declines may well have occurred. Given the current concern for the ABFT fishery and conservation, these findings might represent alarming signals that should not be overlooked.
All in all, our analyses clearly illustrate that structured CWM ABFT populations retain a high level of genetic diversity across space and time, as was not suggested by previous studies (reviewed in 20). This retention has been shown despite the fact that population sizes might indeed be decreasing, primarily because of overexploitation and changes of the population age structure and reproductive demographics (42). These and combined effects can increase the populations’ vulnerability to ecosystem changes (20).
From this intriguing scenario, two downstream questions arise concerning the management and evolutionary aspects of ABFT conservation:
(i) Do our temporal genetic data provide previously undescribed perspectives for ABFT management? To forecast ABFT biomass trajectories reliably, it is crucial to infer the ABFT recruitment–spawner biomass relation (16, 20). The Ne can represent a reliable descriptor of recruitment dynamics in marine fish populations (2). Several studies have shown that in marine fish species and populations, the Ne ranges from the 100s to the low 1,000s; that its relation to the census size can be approximated, on average, by Ne/N = 10−4; and that Ne/N decreases with increasing population size (2). The short-term Ne estimates we obtained (never previously calculated for ABFT populations) are within the range documented in marine fish and they straddle on the minimum threshold Ne value of 500 indicated to maintain genetic diversity and evolutionary potential across several generations in natural populations (45). In addition, they were not much higher than those obtained for bottlenecked populations of Atlantic cod and New Zealand snapper (46, 47). The “mean-marine-fish” 10−4 ratio between Ne and N (2) is a very rough figure, probably an underestimate, and one that should not be generalized if more precise estimates become available. However, in the absence of better estimates, the Ne values inferred from our data suggest that the census ABFT population size might be close to 106 and even higher if we accept a lower ratio between Ne and N, as seems reasonable for large populations (2). This estimate of N appears to be quite reliable because it is entirely consistent with the results of recent ICCAT stock assessments (42), and this issue gives relevance to our choice of a genetic approach for management purposes.
(ii) How then, has genetic diversity been retained in the depleted Eastern ABFT? Genetic responses to demographic bottlenecks can vary (36), and retention of diversity in the face of demographic instability is not unprecedented in bottlenecked populations of marine fish (48–50). In the Eastern ABFT, it is likely that different populations might have exploited the Mediterranean habitats and might have become dominant demographically in space and time (16). According to this hypothesis, the contemporary CWM ABFTs might be composed of distinct reproductive units, with each displaying a somewhat different demographic history. ABFTs with rather resident behavior spawning preferentially in the central-eastern Mediterranean Sea have probably been dominating and expanding since the 1980s (16), and they likely include our ADR sample. Currently, they would be intermingled with more mobile ABFTs spawning in the CWM that have been demographically declining since the 1950s and 1960s (16), including our STY and other western Mediterranean samples. Therefore, even limited levels of population subdivision might have contributed to the maintenance of genetic diversity among ABFT populations. Additional factors might include the increase in size of some populations attributable to a positive response to environmental changes (16), overlapping generations (51), and a recent bottleneck too recent indeed to have fully manifested its genetic effects.
Materials and Methods
We analyzed microsatellite loci variation in 256 contemporary ABFTs collected between 1999 and 2007 from traps, long-lines, and purse-seines of the CWM (Fig. 1 and Table S1). In addition, we extracted and analyzed DNA from dried vertebrae of 219 historical ABFTs from the collection of one of the authors (M.S.), which were caught between 1911 and 1927 in CWM traps (Fig. 1, SI Materials and Methods, Fig. S1 and Table S1). Protocols for DNA extraction and amplification of historical and contemporary DNAs as well as details of the eight scored microsatellites are illustrated in SI Materials and Methods.
Allele and genotype frequencies were estimated using FSTAT version 2.9.3.2 (52). Expected (He) and observed (Ho) heterozygosities, leading to an exact test for Hardy–Weinberg equilibrium, were calculated by Arlequin version 3.1 (53) after 100,000 steps of Markov chains and 1,000 dememorization steps. The GENEPOP 3.1 (54) software package was used to test for the global significance over loci of the estimated statistics (10,000 dememorization steps, 100 batches, and 5,000 iterations per batch).
The Arlequin software package was also used to estimate Wright’s F-statistics, but global FST values were estimated by FSTAT. We tested for FIS significance using 10,000 permutations of the data. Null allele contribution was tested either by jackknifing over loci or by applying Microchecker (25). Pairwise FST values were calculated from the number of different alleles, and the null distribution of pairwise FST under the hypothesis of no difference between subpopulations was obtained with 10,100 permutations. With the same program, we carried out AMOVA so as to quantify genetic variation among samples through both place and time (significance evaluated by running 10,100 permutation tests). Significance values for multiple comparisons were adjusted by Bonferroni sequential correction (55).
Individual genotypes were clustered by an Monte Carlo Markov Chain (MCMC) approach implemented in the package STRUCTURE v2.2 (26). The program estimates the probability that genotypes fall into K clusters (for K = 1, 2, 3…n), each of them at Hardy–Weinberg equilibrium, and calculates the membership coefficient (i.e., the probability of each individual belonging to each cluster). The model we chose allows for admixture and correlation among allele frequencies. Ten independent analyses (1 ≤ K ≤ 10) were performed, with 30,000 burn-in steps and 10,000 MCMC iterations.
Ne was inferred in three ways. Two methods were based on comparisons of contemporary and historical data and led to the estimation of short-term values reflecting demographic phenomena occurring between the two samplings. The third method considered individual samples and yielded a long-term Ne estimate. In this context, long-term Ne reflects the degree of genetic diversity observed in a population and interpreted as a consequence of its whole demographic and evolutionary history. Short-term Ne values were estimated for STY and ADR samples, for which we had matching pairs of historical and contemporary data. Under a model of genealogical coalescence, the method of Berthier et al. (28) incorporates Bayesian prior information about the maximal Ne value (Nemax) and is implemented in the program TM3. Conversely, Wang’s pseudolikelihood method (27) is implemented in the MLNE v1.1 program. We assumed for the CWM ABFT two estimates of tg: 8 years (11, 22) and 13 years (31), which correspond to time lags between historical and contemporary samples of 12/7 generations in the STY and 10/6 generations in the ADR, respectively. For TM3 estimates, we used 300,000 MCMC iterations and Nemax = 5,000. Using the method of Berthier et al. (28), three different values of Nemax (5,000, 10,000, and 50,000) were preliminarily tested to evaluate the robustness of the estimate. However, we did not observe any marked difference in the Ne estimates. Convergence was evaluated using Tracer v1.4 (56).
Bayesian long-term estimates of Ne were obtained, together with estimates of the demographic growth rate scaled by the mutation rate, using an MCMC coalescent genealogy sampler (LAMARC v2.1.2b) (32). Two replicates were run for each population under an exponential growth model; in each replicate, 1 billion genealogies were generated and 1 of every 10,000 genealogies was sampled, with a burn-in of 1,000 iterations. Mutation was modeled by a Brownian-motion approximation to the stepwise model (57). Prior values for g (units for g are 1/μ generations) (58) and for θ (4Neμ) were drawn from uniform distributions (1−10 ≤ θ ≤ 100, −500 ≤ g ≤ 100). Tracer v1.4 (56) was used to check the convergence of the chains.
Finally, we tested in two ways for the effects of a population bottleneck resulting in an impoverishment of genetic diversity. First, assuming that the allele number decreases faster than heterozygosity after a bottleneck (35), we compared the expected heterozygosities (Hes) with their values (Heqs) estimated from the allele number. He > Heq means a heterozygosity excess and suggests a bottleneck; demographic growth has the opposite effect, and He < Heq (35). Second, we calculated the M ratio between the number of alleles (k) and the range in allele size (r), assuming that k (and hence M) decreases faster than r during a bottleneck (36). Methods and software used in these simulations are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the editor and the two anonymous reviewers for valuable comments that greatly improved the manuscript. We also thank R. Waples, J.-M. Fromentin, and L. Chikhi for critical reading of previous versions of this manuscript; E. Ciavaglia and C. Piccinetti at the University of Bologna, G. De Metrio at the University of Bari, A. Cau at the University of Cagliari, L. Orsi at the University of Genoa, and F. Hemida at the Ecole Supérieure des Sciences de la Mer et de l’Aménagement du Littoral of Algiers for providing tuna samples; and Piero Addis at the University of Cagliari for providing and managing biological data. Financial support was from the Italian Ministry of University and Scientific Research and from the Universities of Bologna and Ferrara. This work was part of the Italian PRIN 2005 TUNING “Tuna’s changing” coordinated by F.T.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908281107/DCSupplemental.
References
- 1.Patarnello T, Volckaert F, Castilho R. Pillars of Hercules: Is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol. 2007;16:4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- 2.Hauser L, Carvalho GR. Paradigm shifts in marine fisheries genetics: Ugly hypotheses slain by beautiful facts. Fish and Fisheries. 2008;9:333–362. [Google Scholar]
- 3.Frankham R. Effective population size/adult population size ratios in wildlife: A review. Genet Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- 4.Frankham R. Stress and adaptation in conservation genetics. J Evol Biol. 2005;18:750–755. doi: 10.1111/j.1420-9101.2005.00885.x. [DOI] [PubMed] [Google Scholar]
- 5.Waples RS, Punt AE, Cope JM. Integrating genetic data into management of marine resources: How can we do it better? Fish and Fisheries. 2008;9:423–449. [Google Scholar]
- 6.National Research Council. Dynamic Changes in Marine Ecosystems: Fishing, Food Webs and Future Options. Washington, DC: National Academy Press; 2006. [Google Scholar]
- 7.Hampton J, et al. Fisheries: Decline of Pacific tuna populations exaggerated? Nature. 2005;434:E1–E2. doi: 10.1038/nature03581. [DOI] [PubMed] [Google Scholar]
- 8.Sibert J, Hampton J, Kleiber P, Maunder M. Biomass, size, and trophic status of top predators in the Pacific Ocean. Science. 2006;314:1773–1776. doi: 10.1126/science.1135347. [DOI] [PubMed] [Google Scholar]
- 9.Safina C, Klinger DH. Collapse of bluefin tuna in the Western Atlantic. Conserv Biol. 2008;22:243–246. doi: 10.1111/j.1523-1739.2008.00901.x. [DOI] [PubMed] [Google Scholar]
- 10.Rouyer T, et al. Complex interplays among population dynamics, environmental forcing, and exploitation in fisheries. Proc Natl Acad Sci USA. 2008;105:5420–5425. doi: 10.1073/pnas.0709034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block BA, et al. Electronic tagging and population structure of Atlantic bluefin tuna. Nature. 2005;434:1121–1127. doi: 10.1038/nature03463. [DOI] [PubMed] [Google Scholar]
- 12.Carlsson J, McDowell JR, Carlsson JE, Graves JE. Genetic identity of YOY bluefin tuna from the eastern and western Atlantic spawning areas. J Hered. 2007;98:23–28. doi: 10.1093/jhered/esl046. [DOI] [PubMed] [Google Scholar]
- 13.Rooker JR, et al. Natal homing and connectivity in Atlantic bluefin tuna populations. Science. 2008;322:742–744. doi: 10.1126/science.1161473. [DOI] [PubMed] [Google Scholar]
- 14.Fromentin JM, Powers JE. Atlantic bluefin tuna: Population dynamics, ecology, fisheries and management. Fish and Fisheries. 2005;6:281–306. [Google Scholar]
- 15.Rooker JR, et al. Life history and stock structure of Atlantic Bluefin Tuna (Thunnus thynnus) Rev Fish Sci. 2007;15:265–310. [Google Scholar]
- 16.Fromentin JM. Lessons from the past: Investigating historical data from bluefin tuna fisheries. Fish Fish. 2009;10:197–216. [Google Scholar]
- 17.Teo SLH, et al. Annual migrations, diving behavior, and thermal biology of Atlantic bluefin tuna, Thunnus thynnus, on their Gulf of Mexico breeding grounds. Mar Biol (Berlin) 2007;151:1–18. [Google Scholar]
- 18.Boustany AM, Reeb CA, Block BA. Mitochondrial DNA and electronic tracking reveal population structure of Atlantic bluefin tuna (Thunnus thynnus) Mar Biol (Berlin) 2008;156:13–24. [Google Scholar]
- 19.ICCAT. Report of the 2006 Atlantic Bluefin Tuna Stock Assessment Session (SCRS/2006/013). International Commission for the Conservation of Atlantic Tuna. Madrid: ICCAT; 2007. [Google Scholar]
- 20.MacKenzie BR, Mosegaard H, Rosenberg NA. Impending collapse of bluefin tuna in the northeast Atlantic and Mediterranean. Conserv Lett. 2009;2:25–34. [Google Scholar]
- 21.Karakulak S, et al. Evidence of a spawning area for the bluefin tuna (Thunnus thynnus L.) in the eastern Mediterranean. J Appl Ichthyol. 2004;20:318–320. [Google Scholar]
- 22.Heinisch G, et al. Spatial-temporal pattern of bluefin tuna (Thunnus thynnus L. 1758) gonad maturation across the Mediterranean Sea. Mar Biol (Berlin) 2008;154:623–630. [Google Scholar]
- 23.De Metrio G, et al. Movements of bluefin tuna (Thunnus thynnus L.) tagged in the Mediterranean Sea with pop-up satellite tags. ICCAT Collective Volume of Scientific Papers. 2005;58:1337–1340. [Google Scholar]
- 24.Carlsson J, et al. Microsatellite and mitochondrial DNA analyses of Atlantic bluefin tuna (Thunnus thynnus thynnus) population structure in the Mediterranean Sea. Mol Ecol. 2004;13:3345–3356. doi: 10.1111/j.1365-294X.2004.02336.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Oosterhout C, Hutchinson WF, Willis DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- 26.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J. A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet Res. 2001;78:243–257. doi: 10.1017/s0016672301005286. [DOI] [PubMed] [Google Scholar]
- 28.Berthier P, Beaumont MA, Cornuet JM, Luikart G. Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: a genealogical approach. Genetics. 2002;160:741–751. doi: 10.1093/genetics/160.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waples RS, Yokota M. Temporal estimates of effective population size in species with overlapping generations. Genetics. 2007;175:219–233. doi: 10.1534/genetics.106.065300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kell LT, et al. An evaluation of management strategies for Atlantic tuna stocks. Scientia Marina. 2003;67(Suppl 1):353–370. [Google Scholar]
- 31.Fromentin JM, Kell LT. Consequences of variations in carrying capacity or migration for the perception of Atlantic bluefin tuna (Thunnus thynnus) population dynamics. Can J Fish Aquat Sci. 2007;64:827–836. [Google Scholar]
- 32.Kuhner MK. LAMARC 2.0: Maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- 33.Waldick RC, Kraus S, Brown M, White BN. Evaluating the effects of historic bottleneck events: An assessment of microsatellite variability in the endangered, North Atlantic right whale. Mol Ecol. 2002;11:2241–2249. doi: 10.1046/j.1365-294x.2002.01605.x. [DOI] [PubMed] [Google Scholar]
- 34.Fraser DJ, et al. Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Mol Ecol. 2007;16:3866–3889. doi: 10.1111/j.1365-294X.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 35.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- 37.Spong G, Hellborg L. A near-extinction event in lynx: Do microsatellite data tell the tale? Conserv Ecol. 2002;6:15. [Google Scholar]
- 38.Gonzalez EG, Beerli P, Zardoya R. Genetic structuring and migration patterns of Atlantic bigeye tuna, Thunnus obesus (Lowe, 1839) BMC Evol Biol. 2008;8:252. doi: 10.1186/1471-2148-8-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latch E, Dharmarajan G, Glaubitz JC, Rhodes OEJ. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet. 2006;7:295–302. [Google Scholar]
- 40.Alvarado Bremer JR, et al. Comparative phylogeography of Atlantic bluefin tuna and swordfish: The combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Mol Phylogenet Evol. 2005;36:169–187. doi: 10.1016/j.ympev.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 42.ICCAT. Report of the 2008 Atlantic Bluefin Tuna Stock Assessment Session, Madrid, Spain, June 23–July 4, 2008. Madrid: ICCAT; 2008. pp. 1–161. [Google Scholar]
- 43.Guinand B, Scribner KT. Evaluation of methodology for detection of genetic bottlenecks: Inferences from temporally replicated lake trout populations. C R Soc Biol. 2003;326(Suppl 1):S61–S67. doi: 10.1016/s1631-0691(03)00039-8. [DOI] [PubMed] [Google Scholar]
- 44.Schlötterer C, Kauer M, Dieringer D. Allele excess at neutrally evolving microsatellites and the implications for tests of neutrality. Proc R Soc London Ser B. 2004;271:869–874. doi: 10.1098/rspb.2003.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
- 46.Hauser L, et al. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus) Proc Natl Acad Sci USA. 2002;99:11742–11747. doi: 10.1073/pnas.172242899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutchinson WF, van Oosterhout C, Rogers SI, Carvalho GR. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua) Proc Biol Sci. 2003;270:2125–2132. doi: 10.1098/rspb.2003.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruzzante DE, Taggart CT, Doyle RW, Cook D. Stability in the historical pattern of genetic structure of Newfoundland cod (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv Genet. 2001;2:257–269. [Google Scholar]
- 49.Shrimpton JM, Heath DD. Census vs. effective population size in chinook salmon: Large- and small-scale environmental perturbation effects. Mol Ecol. 2003;12:2571–2583. doi: 10.1046/j.1365-294x.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- 50.Consuegra S, Verspoor E, Knox D, de Leaniz CG. Asymmetric gene flow and the evolutionary maintenance of genetic diversity in small, peripheral Atlantic salmon populations. Conserv Genet. 2005;6:823–842. [Google Scholar]
- 51.Waples RS. Conservation genetics of Pacific salmon. 2. Effective population-size and the rate of loss of genetic-variability. J Hered. 1990;81:267–276. [Google Scholar]
- 52.Goudet J. FSTAT version 1.2. A computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 53.Excoffier L. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 54.Raymond M. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 55.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 56.Rambaut A, Drummond AJ. TRACER. Version 1.3. 2004. Available at: http://beast.bio.ed.ac.uk/Tracer (accessed on September 5, 2007)
- 57.Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.