Abstract
Normal tissue homeostasis is maintained through asymmetric cell divisions that produce daughter cells with differing self-renewal and differentiation potentials. Certain tumor cell subfractions can self-renew and repopulate the heterogeneous tumor bulk, suggestive of asymmetric cell division, but an equally plausible explanation is that daughter cells of a symmetric division subsequently adopt differing cell fates. Cosegregation of template DNA during mitosis is one mechanism by which cellular components are segregated asymmetrically during cell division in fibroblast, muscle, mammary, intestinal, and neural cells. Asymmetric cell division of template DNA in tumor cells has remained elusive, however. Through pulse-chase experiments with halogenated thymidine analogs, we determined that a small population of cells within human lung cancer cell lines and primary tumor cell cultures asymmetrically divided their template DNA, which could be visualized in single cells and in real time. Template DNA cosegregation was enhanced by cell–cell contact. Its frequency was density-dependent and modulated by environmental changes, including serum deprivation and hypoxia. In addition, we found that isolated CD133+ lung cancer cells were capable of tumor cell repopulation. Strikingly, during cell division, CD133 cosegregated with the template DNA, whereas the differentiation markers prosurfactant protein-C and pan-cytokeratins were passed to the opposing daughter cell, demonstrating that segregation of template DNA correlates with lung cancer cell fate. Our results demonstrate that human lung tumor cell fate decisions may be regulated during the cell division process. The characterization and modulation of asymmetric cell division in lung cancer can provide insight into tumor initiation, growth, and maintenance.
Keywords: cancer stem cell, CD133, immortal DNA, template DNA
Asymmetric segregation of template DNA strands exclusively to one daughter cell during cell division was first reported by Karl Lark in 1966 (1). One explanation for this phenomenon was provided by John Cairns, who proposed the “immortal DNA strand hypothesis,” which states that stem cells asymmetrically divide the “immortal” template DNA to the daughter stem cell and segregate the newly synthesized DNA to the differentiating cell to prevent potentially carcinogenic DNA mutations in the long-lived stem cells (2). The immortal DNA strand hypothesis is controversial in terms of why some cells retain their template DNA and whether or not the phenomenon is restricted to stem cells (3, 4). Asymmetric division of template DNA has been shown in fibroblasts and epithelial, neural, and muscle satellite cells (1, 5–10). Other studies have demonstrated that hematopoietic stem cells (HSCs), epidermal basal cells, hair follicle bulge cells, and intestinal epithelial stem cells segregate their chromosomes randomly (11–15). Perhaps asymmetric division of template DNA is cell type–specific, or perhaps only a very limited number of stem cells retain their template DNA strands in some tissues, which may go undetected by the methods or mathematical models used. Most studies to date have been performed retrospectively at the population level, by virtue of label retention. More studies at the single-cell level are needed to unequivocally determine whether asymmetric separation of older and newer DNA strands occurs during mitosis.
Recent data suggest that certain primary solid tumors and cancer cell lines harbor a subpopulation of cells that when injected into immunocompromised mice are able to self-renew and regenerate phenotypically heterogeneous tumors (16–20), or when cultured in vitro are able to repopulate the entire unsorted tumor cell population (21, 22). These data imply that tumor cell repopulation occurs via asymmetric cell divisions within the isolated subpopulations; however, an equally plausible explanation is that the isolated cells divide symmetrically, but the progeny adopt different cell fates independent of the division, possibly through interactions between the daughter cells and their microenvironments. To the best of our knowledge, there is no direct evidence indicating that any malignant cells are capable of undergoing asymmetric cell division. Consequently, we set out to determine whether human lung cancer primary cell and cell lines compose a subpopulation of cells that asymmetrically divide their template DNA strands exclusively to only one daughter cell during cell division and, furthermore, whether this process could be modulated and linked to cell fate.
Results
Lung Cancer Cells Can Asymmetrically Divide Their Template DNA During Cell Division.
Pulse-chase experiments involving the halogenated thymidine analog 5-bromo-2-deoxyuridine (BrdU) have been used to label DNA and track label-retaining cells that are growing slowly or asymmetrically dividing (9, 12–15). Using a strategy similar to that reported for single mitotic cells (Fig. 1A) (5–8), we set out to determine whether non–small cell lung cancer (NSCLC) cells have the ability to asymmetrically divide their BrdU-labeled template DNA exclusively to one daughter cell. We grew NSCLC cell lines A549 and H441 for several passages in the presence of 1 μM BrdU, a concentration at least 200-fold below the limit at which cells take up detectable BrdU during DNA repair (23). A long pulse allowed for selection of cells that had undergone at least one symmetric division, because cells that exclusively asymmetrically divide or are senescent are “passaged out” due to expansion of symmetrically dividing cells (6, 7), ensuring that both sets of DNA strands were labeled in the vast majority of cells. To address this experimentally, we stained cells at the end of the pulse with an anti-BrdU antibody and then examined them by flow cytometry and immunofluorescence. As expected, 99.98% ± 0.01% of the cells were BrdU+ (SI Fig. S1 A and B), verifying that the BrdU pulse was sufficient.
Fig. 1.
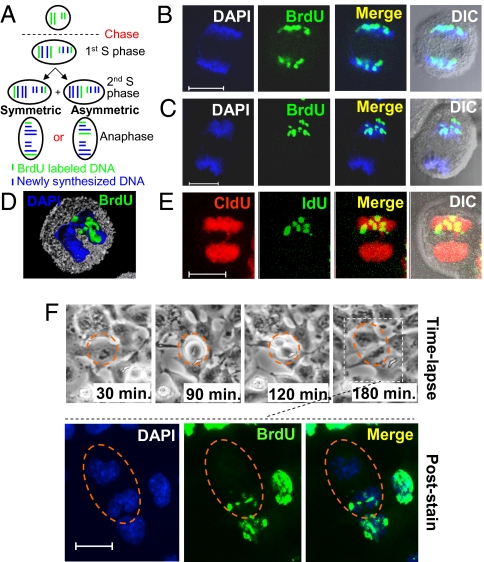
DNA is partitioned asymmetrically during mitosis in lung cancer cells. (A) Schematic representation of symmetric and asymmetric division of template DNA. Cells are grown in BrdU (green). During anaphase of the second cell division after BrdU is removed (the chase), the BrdU- labeled template chromosomes are segregated either randomly (symmetric) or exclusively to one side of the metaphase plate (asymmetric). (B–D) Representative anaphase A549 cells that partition their BrdU-labeled template DNA (green) either randomly to both top and bottom daughter cells (B) or exclusively to the top daughter cell (C and D). (E) An A549 cell asymmetrically partitions IdU-labeled template DNA (green) to the top daughter cell and randomly segregates the newly synthesized CldU-labeled DNA (red) to both daughter cells. (F) Time frames from live time-lapse imaging show a dividing label-retaining cell, outlined by a dotted circle. The two daughter cells, outlined by a dotted oval, were fixed and stained for BrdU. The BrdU-labeled template DNA was segregated exclusively to the upper daughter cell. DNA was stained with DAPI (blue). (Scale bar: 10 μm in B, C, E, and F.)
On removal of BrdU, we examined the BrdU-retaining pattern of the template DNA in individual lung cancer cells during cell division. All of the 1,200 anaphase A549 cells examined demonstrated a positive BrdU label segregated to both daughter cells during the first cell division (Fig S1C). According to the model, asymmetric division of template DNA should be observed after the first cell division of the chase. Most BrdU+ anaphase cells exhibited random segregation of the BrdU signal during the second cell division, and some cells in anaphase also demonstrated exclusive segregation of the BrdU-labeled template DNA to one side of the cell and no BrdU label on the opposing side (Fig. 1B and C). Differential interference contrast confirmed that the cells were in anaphase, rather than two cells in close proximity. An in-depth examination of three-dimensional images from confocal “Z-stacks” revealed that only one set of the sister chromatids exclusively inherited the BrdU-labeled template DNA (Fig. 1D and Movie S1). To verify that both sets of sister chromatids had equal accessibility to staining and that the cells underwent DNA synthesis during the chase period, cells were grown in the DNA analog iododeoxyuridine (IdU) for 2 weeks and then chased for two cell divisions. A second DNA analog, chlorodeoxyuridine (CldU) was added after the first cell division, similar to what was done in previous studies (5, 6). Confirming our previous results, we observed asymmetric segregation of heavily IdU-labeled template DNA exclusively to one set of chromosomes and CldU-labeled newly synthesized DNA to both sets of chromosomes during anaphase (Fig. 1E). Interestingly, during the first division of the chase, the BrdU stain was even, whereas during the second division, the labeled BrdU pattern was patchy, similar to that seen in asymmetrically dividing neural stem cells (6). Surprisingly, we did not observe exclusive asymmetric segregation of heavily BrdU-labeled template DNA beyond three cell divisions after the chase, suggesting that successive asymmetric divisions of template DNA were rare because of intervening symmetric divisions of DNA.
To verify that asymmetric chromosomal segregation is preserved through cytokinesis, we next used real-time imaging to track asymmetric DNA partitioning in the daughter cells. During the second division of the chase, cells were monitored by time-lapse imaging, then fixed and stained for BrdU retention. We visualized label-retaining cells (LRCs) that exclusively segregate their template DNA to only one daughter cell to give rise to BrdU+ and BrdU− cells (Fig. 1F and Movie S2). Thus, the anaphase images of asymmetric division reflect exclusive template DNA segregation after cytokinesis.
To validate the data, we quantified the frequency of asymmetric division of BrdU-labeled template DNA in seven human NSCLC cell lines and two immortalized normal human bronchial epithelial (HBET) cell lines. At least 250 anaphases per sample were scored in each of three independent experiments. In 6 of 7 NSCLC samples, the frequency of asymmetric division of template DNA ranged from 5.6 ± 1.8% to 0.7 ± 0.6% (Fig. 2A). No asymmetric divisions were detected in H1650 cells. The frequency of template DNA asymmetric division was significantly higher in three NSCLC cell lines compared with immortalized normal HBET cells (P < 0.01) and was similar in three NSCLC cell lines and HBET cells. Overall, asymmetric segregation of template DNA in NSCLC cell lines was within the range of established murine myoblast cultures (5%), and murine muscle satellite cells (0.5–6.8%) in vitro (5, 8).
Fig. 2.
Template DNA is divided asymmetrically in several human lung cancer cell lines, immortalized bronchial epithelial cells, and primary lung tumors. (A) The frequency of asymmetric division of BrdU-labeled template DNA is shown by bar graphs for two normal immortalized HBET cell lines and six NSCLC cell lines. Data are reported as the mean ± SD from three independent experiments. At least 250 anaphases were scored in each experiment. *P < 0.01, one-way ANOVA between individual NSCLC cell lines and combined HBET cell lines. (B) Primary human lung cancer cells from two samples were BrdU-pulsed, chased, and treated with nocodazole. Shown is an example of asymmetric partitioning of BrdU-labeled DNA in a single late-telophase cell from sample 2. The number of asymmetric cell divisions is shown. (C) Primary human lung cancer cells from three samples were BrdU-pulsed, and colony assays were performed during the BrdU chase. Single cells were cocultured with unlabeled A549 cells. Colonies were visualized with CTR (red) and stained for BrdU (green). A representative colony demonstrating an asymmetric division of template DNA from sample 3 shows two BrdU+ cells (asterisks) and one BrdU− cell (white arrow). The total number of colonies with both BrdU+ and BrdU− cells is shown, 220 cells were scored and 17 cells were BrdU−. NOS, not otherwise specified; AC, adenocarcinoma; PE, Pleural effusion. (Scale bars, 5 μm in B; 20 μm in C.)
We extended our observations to five primary NSCLC tumors through assessment of anaphase cells (two samples) or by colony assays (three samples). We verified that dissociated human tumor cells and pleural fluids were derived from the tumor by immunofluorescence examination of cell type–specific markers (Fig. S2), and these cells were grown in BrdU for up to four passages. For two samples, 100 ng/mL of nocodazole was added to the cultures before the second division of the chase, to arrest the cells in mitosis. Cells were washed to allow them to complete mitosis and enter cytokinesis, and then fixed and stained for BrdU label. Remarkably, 4 of 22 (18%) anaphase cells in tumor and 33 of 264 (12.5%) anaphase cells in tumor 2 were asymmetric for BrdU-labeled DNA (Fig. 2B). These data were verified in three additional primary NSCLC tumors by colony assays. BrdU-pulsed cells were labeled with Cell Tracker Red CMTPX (CTR), which allows tracking of the progeny. The cells were then mixed with a coculture of A549 cells and plated onto gasket slides at a dilution of one CTR-labeled cell per well. Single CTR-labeled tumor cells were verified by immunofluorescence and monitored until they expanded into three- or four-cell colonies (i.e., two cell divisions), at which point they were fixed and stained for BrdU label. Colonies containing both BrdU+ and BrdU− cells represented an asymmetric cell division. Two samples each had 5/20 colonies and the third sample had 5/25 colonies containing mixed BrdU+ and BrdU− cells (Fig. 2C). This extension of our findings in cancer cell lines to primary lung cancer cells suggests that asymmetric cell division of template DNA is a common feature of lung cancer (albeit one that occurs at a low frequency). The higher frequency observed in primary samples compared with malignant and normal cell lines could reflect selection for symmetric divisions during long-term culture or selective early growth of asymmetrically dividing primary tumor cells ex vivo.
The legitimacy of the premise that certain cells undergo asymmetric division of their template DNA has been challenged recently (3, 4). To address this issue, we vigorously verified that the observations were robust. We determined that the probability of observing asymmetric chromosomal segregation in an individual diploid anaphase cell by chance is exceedingly small (P = 2.8 × 10−14; SI Methods). The modal chromosome numbers of A549 and H441 cells are 66 and 52, respectively (http://www.atcc.org); thus, the probability of observing an exclusive asymmetric partitioning of labeled DNA in these cell lines by chance is considerably lower. We next determined that all chromosomes had incorporated BrdU by immunofluorescence examination of BrdU-stained metaphase cells at the end of the BrdU pulse (Fig. S3A), and verified that entire single chromosomes could be readily detected by examining BrdU-chased cells in metaphase (Fig. S3B). Finally, we extended the BrdU chase in A549 cells to an average of 15.2 cell doublings (2 weeks), stained for BrdU, and performed flow cytometry analysis. We calculated that 0.2% of A549 cells would be expected to be positive if the sensitivity of the assay were as low as one chromosome (SI Methods). Strikingly, 1.4 ± 0.3% of the cells were BrdU+. The staining pattern within the nuclei showed small single or double punctuate foci, suggesting that each positive signal represents a single positive chromosome (Fig. S3C). The higher percentage of BrdU+ cells observed likely includes the small percentage of cells that undergo asymmetric division of template DNA; therefore, the observed asymmetric segregation of BrdU-labeled template DNA probably was not due to chance or assay sensitivity.
Environmental Modulation Influences the Frequency of Asymmetric Division of Template DNA in Lung Cancer.
Mouse embryonic fibroblasts and muscle satellite cells asymmetrically divide their template DNA more frequently when seeded at a higher density, consistent with increased expansive self-renewal via symmetric cell divisions at low cell densities (8, 24). To determine whether asymmetric division of template DNA in human NSCLC cells is cell density dependent, we chased BrdU-labeled cells at various cell concentrations, so that collections were performed at specified cell confluences. At least 250 BrdU-stained anaphases were scored in three independent experiments. The frequency of template DNA asymmetric division at 90% confluence was 6.6 ± 0.7% for the A549 cell line and 8.2 ± 1.3% for the H441 cell line, both significantly higher than the asymmetric division frequencies at 45% confluence (3.8 ± 0.6% for A549 and 3.0 ± 1.1% for H441) and 20% confluence (3.2 ± 0.4% for A549 and 2.4 ± 1.4% for H441) (P < 0.01; Fig. 3A). The frequencies were normalized to the mitotic index to account for any changes in cell growth rates. As expected, higher cell densities were associated with a higher frequency of asymmetric cell division when the mitotic index was taken into account (P < 0.0001; Fig. S4). These data demonstrate that symmetric division of template DNA is more prevalent when there is a significant opportunity for repopulation.
Fig. 3.
Asymmetric segregation of template DNA is modulated by environmental changes. (A) Bar graph showing a decreased frequency of asymmetric division of template DNA at lower cell densities, as well as a decreased frequency of asymmetric template DNA division under hypoxic conditions and serum deprivation. A549 and H441 cells are indicated by black and gray bars, respectively. Data are reported as mean ± SD from three independent experiments. *P < 0.0001, one-way ANOVA. Cell density is reported as million/cm2, compared with 95% confluence. At least 250 anaphases were scored in each experiment. (B) Representative image of four-cell colonies derived from single A549 cells that were grown in BrdU, labeled with CTR (red), and cocultured with unlabeled A549 cells. Colonies contained all BrdU+ (green) cells (Upper), indicating symmetric divisions of the template DNA, or a mixture of BrdU+ and BrdU− cells (Lower), indicating an asymmetric division. DNA was stained with DAPI (blue). The total number of colonies, colonies with all BrdU+ cells, and colonies with mixed BrdU+ and BrdU− cells are shown. Numbers in parentheses represent the percentage of colonies with asymmetric divisions. The single cell–derived four-cell colonies comprise mixed BrdU+ and BrdU− cells only when cocultured with neighboring A549 cells (no membrane barrier). When the colonies were separated from the cocultured cells by a membrane barrier, no asymmetric divisions were observed. (Scale bar: 20 μm.)
To address the possibility that asymmetric division of template DNA in lung cancer requires cell–cell contact, we plated single BrdU-labeled and CTR-stained cells to the bottom chambers of transwell microwell plates. Wells contained a coculture cell layer with cells from the same cell line either in the top chamber (separated by a membrane) or in the bottom chamber (without a membrane barrier). Single cells were verified by immunofluorescence, tracked until they reached four-cell colonies, and then stained and analyzed for BrdU retention. When cocultured without a membrane barrier, 4.2% of A549 and 4.7% of H441 single cell-derived colonies contained a mixture of BrdU+ and BrdU− cells (Fig. 3B), similar to the frequency of asymmetric template DNA segregation observed in anaphase cells (Fig. 2A). In 333 two-cell colonies, every cell was BrdU+. Strikingly, with a membrane barrier, asymmetric segregation of labeled template DNA was abolished (Fig. 3B). These data indicate that cell–cell contact may be required for asymmetric template DNA segregation in cultured lung cancer cells.
Hypoxia induces neuroblastoma, glioma, and breast cancer cells to lose their differentiated cell gene expression patterns and acquire stem cell–like phenotypes (25–27). Likewise, the use of serum-free media designed for in vitro expansion of human embryonic stem cells results in maintenance of a cancer stem cell phenotype in prostate cancer (28). To determine whether these environments influence the frequency of asymmetric division of template DNA, we conducted chase experiments in ambient O2 with serum, 1% O2 with serum, or ambient O2 without serum. Cell densities were kept constant at the time of cell collection. After normalization to the mitotic index, the frequency of template DNA asymmetric divisions was decreased by at least four-fold in 1% O2 and at least three-fold in serum-free media (P < 0.0001; Fig. S4). The differences were not significant before normalization (Fig. 3A), reflecting that the decreased rate of cell division by hypoxia and serum deprivation affects the rate of asymmetric cell division as well. Thus, in agreement with data from other model systems, both hypoxia and serum deprivation might favor expansive self-renewal by increasing symmetric divisions in lung cancer.
Correlation Between Asymmetric Division of Template DNA and Cell Fate.
NSCLC tumor repopulating cells are known to be enriched within the side population (SP) fraction (29). To test whether the SP fraction within A549 lung cancer cells preferentially retains the template DNA strand, we chased BrdU-labeled cells for four cell divisions and analyzed the sorted SP and non-SP fractions by flow cytometry for BrdU retention. Interestingly, the mean fluorescence intensity (MFI) of the BrdU signal was significantly higher in SP cells compared with non-SP cells (718 ± 84 vs. 218 ± 17; P < 0.001) (Fig. S5 A–C). The higher BrdU retention in the SP fraction cannot be explained by slower cycling cells, because sorted SP cells cycled notably faster than non-SP cells, as evidenced by the fact that SP cells had a significantly lower Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) intensity (P < 0.001) (Fig. S5D), a cellular stain that decreased by half during each cell division (6), and a significantly higher mitotic index (0.53 ± 0.1% vs. 0.34 ± 0.05%; P = 0.04; Fig. S5E). These results were confirmed by performing colony assays on single sorted CTR-stained cells. The frequency of mixed BrdU+ and BrdU− colonies from the SP sorted fraction was 13.4% ± 5.6%, significantly higher than the frequency of mixed colonies from non-SP cells of 4.2% ± 0.8% (P = 0.05; Fig. S5F). Overall, these data demonstrate that the SP fraction in human NSCLC cells is enriched for cells that asymmetrically divide their template DNA, although depletion of SP cells does not reduce asymmetric cell divisions in the isolated non-SP fraction.
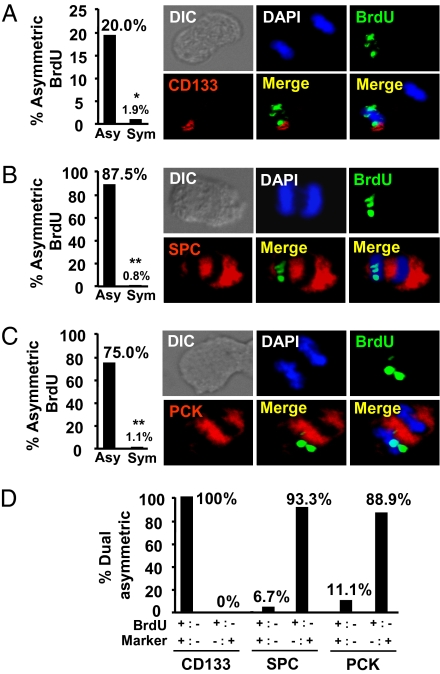
The surface marker protein CD133 is expressed by a subpopulation of NSCLC cells with enhanced self-renewal and repopulation abilities both in vitro and in vivo (30, 31), suggesting that they may be capable of asymmetric cell division. To determine whether CD133+ cells preferentially asymmetrically divide their template DNA, we examined BrdU-pulsed and -chased anaphase cells for both BrdU segregation and CD133 expression. The segregation pattern of CD133 observed during mitosis was reminiscent of punctuate staining within lipid rafts in human HSCs (Fig. S6) (32). We compared the frequency of asymmetric division of BrdU-labeled template DNA between cells that segregated CD133 either asymmetrically or symmetrically. Remarkably, 20% of cells that asymmetrically partitioned CD133 during mitosis also asymmetrically segregated their template DNA, compared with just 1.9% of cells that symmetrically segregated CD133 (P < 0.001; Fig. 4A). Furthermore, the majority of anaphases with dual asymmetric partitioning exhibited CD133 expression only in the daughters inheriting the BrdU-labeled template DNA (Fig. 4D). The localized pattern of asymmetric CD133 partitioning during mitosis was similar to that seen in polarized CD34+ HSCs (32). We extended these findings to primary tumor cells. We treated BrdU-pulsed primary lung cancer cells with nocodazole to induce mitotic arrest during the second cell division after the chase, then costained them for CD133 and BrdU. CD133 segregated with BrdU in 6 of 8 cells (75%) that demonstrated dual asymmetric segregation (Fig. S7A).
Fig. 4.
Differing cell fates correlate with asymmetric segregation of template DNA. (A) Cells that asymmetrically partition CD133 are 10-fold more likely to asymmetrically divide the template DNA A total of 715 cells were scored. A representative image of a dual-asymmetric anaphase cell with CD133 (red) cosegregating with the BrdU-labeled template DNA (green) is shown. (B and C) Cells that asymmetrically partition the differentiation markers pro-SP-C (B) and PCK (C) are more likely to asymmetrically divide the template DNA. A total of 4,244 pro-SP-C cells and 2,116 PCK cells were scored. Respective images are shown of pro-SP-C (B) and PCK (C) (both red) segregating toward the opposite side of the metaphase plate compared with the BrdU-labeled template DNA. (D) Bar graphs showing the frequency at which the asymmetrically segregated proteins were either segregated with or against the template DNA.
A key step in validating that the CD133+ population preferentially asymmetrically divides is determining whether CD133+ cells can repopulate the entire tumor cell population. We sorted cells into CD133+ and CD133− populations in triplicate and cultured them for 7 days. After each sample was restained for CD133, flow cytometric analysis revealed that the cultures derived from the unsorted cell sample contained a CD133+ compartment totaling 3.3 ± 1.2%, whereas the CD133− fraction represented 96.7 ± 1.2% of the total cells (Fig. S8A). Similarly, in the cultures derived from the CD133+ sorted cells, the CD133− fraction composed 97.9 ± 1.9% of the CD133+ cells, representing a 100 ± 1.1% repopulation of the original CD133− cell population. In contrast, in cultures derived from CD133− cells, the CD133+ fraction totaled 0.3 ± 0.03%, translating to an only 8.1 ± 3.4% repopulation of the original CD133+ cell population (Fig. S8 A and B). These results cannot be attributed to differences in proliferative capacity, because the sorted populations had a similar number of population doublings during the 7-day period (7.8 doublings for CD133+ cells and 7.0 doublings for CD133− cells). Thus, the CD133+ tumor cell subpopulation preferentially asymmetrically divides with the template DNA and also has the ability to repopulate the entire original population.
The asymmetric cell division of cell components should correlate with the difference in cell fate of the daughter cells if the event has a biological purpose. Upon differentiation, CD133+ adenocarcinoma NSCLC cells acquire expression of cytokeratins and prosurfactant apoprotein-C (pro-SP-C) and become nontumorigenic (30). We found that CD133+ NSCLC tumorspheres harbored only a small percentage of cells expressing pan-cytokeratin (PCK) and pro-SP-C, but when grown as a monolayer in the presence of serum, the majority of cells expressed PCK and pro-SP-C (Fig. S9 A and B). We examined anaphase cells from the A549 and H441 cell lines for the expression of PCK and pro-SP-C to determine whether the template DNA segregates to the opposite side of the metaphase plate to these markers. We further analyzed anaphase cells that demonstrated asymmetric staining of BrdU and expression of pro-SP-C or PCK. Cells that asymmetrically segregated the pro-SP-C and PCK markers were 116- and 70-fold more likely to asymmetrically divide their template DNA, respectively (P < 0.001; Fig. 4 B and C). Strikingly, of 15 cells that segregated both BrdU and pro-SP-C, 14 cells segregated the pro-SP-C marker to the opposite daughter receiving the BrdU-labeled template DNA. Likewise, of nine cells that demonstrated dual asymmetric cell division of the template DNA and PCK, eight cells had PCK partitioned to the opposite daughter receiving the template DNA. These data were confirmed in primary tumor samples in which 6 of 8 anaphases displaying dual asymmetric divisions of BrdU with pro-SP-C and 8 of 10 anaphases with dual asymmetric divisions of BrdU with PCK segregated the marker to the opposing daughter cells from the template DNA (Fig. S7 B and C). The CD133+ NSCLC cells underwent asymmetric cell divisions in which the template DNA segregated to the daughter that remained CD133+. In contrast, the other daughter acquired the newly synthesized DNA and adopted a more differentiated phenotype, characterized by expression of pro-SP-C and PCK. Thus, our data indicate that segregation of the template DNA strands in lung cancer is coincident with cell fate decisions during cell division.
Discussion
In this study, we found that a subfraction of cancer cells within several cell lines and primary tumor samples asymmetrically divide their template DNA exclusively to one daughter cell. We also found that the frequency of asymmetric division of template DNA can be modulated by environmental changes, suggesting that the phenomenon is of biological importance. Moreover, we provide evidence that the template DNA strands cosegregate with CD133, a surface protein that marks a tumor cell subpopulation that can repopulate the entire lung tumor cell population both in vitro and in vivo (30, 31). The pro-SP-C and PCK differentiation markers are inherited by the daughter cells receiving only newly synthesized DNA, suggesting that tumor cell repopulation occurs via asymmetric cell division. In our experiments, the staining pattern of the BrdU label was patchy. According to the immortal DNA strand model, all of the chromosomes should be labeled. Based on similar previous studies (5, 6), our extensive examination of potential artifactual explanations, and correlation with cell fate proteins, an unknown phenomenon related to stem cell self-renewal and template DNA cosegregation may explain these observations.
Previous studies have suggested that asymmetric division of template DNA occurs along multiple successive divisions (5, 6, 8). But in the presence of excessive tissue damage, the stem cell pool will expand via symmetric divisions, and a “new” immortal DNA strand will arise. Our data demonstrate that during in vitro growth of cancer cells, asymmetric cell divisions of template DNA often are interrupted by intervening symmetric divisions. Perhaps a propensity towards excessive self-renewal via symmetric divisions among CSCs contributes to the exponential expansion of tumors. Indeed, defects in asymmetric division genes have been shown to lead to neoplastic proliferation and cancer formation in Drosophila melanogaster and Caenorhabditis elegans (33). In our study, the requirement of cell–cell contact for asymmetric division of template DNA suggests that polarity may be maintained through interactions with neighboring cells, but that symmetric divisions abound in the absence of that signaling. Our data also show that additional conditions known to favor normal stem cell expansion also increase symmetric divisions in tumor cells. An in-depth examination of intercellular and environmental response signaling pathways is needed to decipher exactly how neighboring cells and the environment influence cancer cell fate decisions. The examination of genes known to disrupt asymmetric cell division in lower organisms in human cancers could elucidate the pathways controlling cancer maintenance and provide new therapeutic targets.
It was originally hypothesized that template DNA cosegregation may provide a mechanism through which normal stem cells prevent potentially tumorigenic replication-related mutations (2). Asymmetric DNA partitioning in cancer also may represent a deregulated remnant used by cancer cells to protect themselves from lethal DNA mutations. Because oxidative DNA damage would affect both DNA strands, asymmetric segregation of the template strands in cancer cells would only protect the genome from replication errors. Normal and cancer stem cells are capable of repairing DNA damage more efficiently than differentiated progeny, and DNA repair defects in stem cells are associated with several cancer syndromes (34, 35). Thus, it is feasible that enhanced DNA repair mechanisms acquired by CSCs protect the template strand from lethal oxidative DNA damage. An alternative explanation for the retention of template DNA is that cells harboring the template DNA strands would not be subject to telomere shortening (6, 36), because telomere shortening occurs during successive rounds of DNA replication of the newly synthesized DNA. But because nearly all human cancers maintain telomere length via telomerase or recombination-based alternative lengthening of elomeres (ALT) mechanisms (37), telomere maintenance is unlikely to be the mechanism driving asymmetric division of template DNA in cancer. A possible alternative explanation is that the template DNA strands retain an epigenetic profile different from the newly synthesized DNA, contributing to differing cell fate potentials during asymmetric cell division (3, 6, 38). Evidence supporting this possibility is provided in the fission yeast, in which epigenetic gene control between complementary sister chromatids is necessary for cellular differentiation (39).
The findings presented herein may restimulate efforts to unmask how cells distinguish cDNA strands during DNA replication and cell division. A molecular mechanism driving asymmetric chromosomal segregation has not yet been elucidated in any cell type or organism. Several hypotheses have been generated, including the recognition of DNA methylation sites, chromatin patterns, histones, ribosomal synthesis, and microtubule/centrosomal assemblies (4, 40). The next challenge is to determine how dysregulation of these putative mechanisms may participate in template DNA recognition, as well as self-renewal, in cancer cells.
Materials and Methods
BrdU, CldU, and IdU Administration, and Chase.
BrdU (or IdU, where indicated) was added to the culture medium at a concentration of 1 μM for 2 weeks to ensure labeling of all cells. Primary tumor cells were grown in 1 μM BrdU for up to four passages. During the pulse, the medium was supplemented with fresh BrdU every 72 h, and cell growth was maintained in log phase.
For the single mitotic cell assays, cells were cultured for two cell divisions in the absence of BrdU (the chase), and then collected by mitotic shake-off. For the IdU-CldU pulse-chase experiments, 1 μM CldU was added after the first cell division. Unless specifed otherwise, mitotic shake-offs were performed when the cells were at 70%–75% confluence.
More detailed descriptions of the methods are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Karen Yarrick [Laboratory of Human Carcinogenesis (LHC)] for bibliographic assistance, Elisa Spillare and Mary McMenamin (LHC) for technical assistance, Susan Garfield [National Cancer Institute (NCI) Confocal Core Facility] for assistance with confocal microscopy, Barbara J. Taylor and Subhadra Banerjee (NCI FACS Core Facility) for cell sorting, and Alan Berger (Johns Hopkins University) for calculating the probability of DNA cosegregation. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, and a NIH Clinical Center Bench-to-Bedside Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.M.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909390107/DCSupplemental.
References
- 1.Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154:1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 3.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Rando TA. The immortal strand hypothesis: Segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpowicz P, et al. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. and erratum (2005) 170:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- 8.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 9.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 11.Kuroki T, Murakami Y. Random segregation of DNA strands in epidermal basal cells. Jpn J Cancer Res. 1989;80:637–642. doi: 10.1111/j.1349-7006.1989.tb01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 13.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 15.Waghmare SK, et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev. 2008;18:48–53. doi: 10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Clarke MF, et al. Cancer stem cells: Perspectives on current status and future directions. AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 18.Charafe-Jauffret E, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multilineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirschmann-Jax C, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrawala L, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 23.Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425:479–494. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Rambhatla L, et al. Cellular senescence: Ex vivo p53-dependent asymmetric cell kinetics. J Biomed Biotechnol. 2001;1:28–37. doi: 10.1155/S1110724301000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jögi A, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci USA. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helczynska K, et al. Hypoxia promotes a dedifferentiated phenotype in ductal breast carcinoma in situ. Cancer Res. 2003;63:1441–1444. [PubMed] [Google Scholar]
- 27.Kang SK, Park JB, Cha SH. Multipotent, dedifferentiated cancer stem-like cells from brain gliomas. Stem Cells Dev. 2006;15:423–435. doi: 10.1089/scd.2006.15.423. [DOI] [PubMed] [Google Scholar]
- 28.Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 30.Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 31.Bertolini G, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giebel B, et al. Segregation of lipid raft markers including CD133 in polarized human hematopoietic stem and progenitor cells. Blood. 2004;104:2332–2338. doi: 10.1182/blood-2004-02-0511. [DOI] [PubMed] [Google Scholar]
- 33.Gönczy P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 34.Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35:7557–7565. doi: 10.1093/nar/gkm1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyler CE, Rich JN. Survival of the fittest: Cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 37.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 38.Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- 40.Fichelson P, et al. Live imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol. 2009;11:685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.