Abstract
Whereas exogenously acquired proteins are the major source of antigens feeding the MHC class II pathway in antigen-presenting cells, some endogenously expressed antigens also access that pathway but the rules governing such access are poorly understood. Here we address this using Epstein–Barr virus (EBV)-coded nuclear antigen EBNA1, a protein naturally expressed in EBV-infected B lymphoblastoid cell lines (LCLs) and a source of multiple CD4+ T cell epitopes. Using CD4+ T cell clones against three indicator epitopes, we find that two epitopes are weakly displayed on the LCL surface whereas the third is undetectable, a pattern of limited epitope presentation that is maintained even when nuclear expression of EBNA1 is induced to high supraphysiological levels. Inhibitor and siRNA studies show that, of the two epitopes weakly presented under these conditions, one involves macroautophagy, and the second involves antigen delivery to the MHC II pathway by another endogenous route. In contrast, when EBNA1 is expressed as a cytoplasmic protein, all three CD4 epitopes are processed and presented much more efficiently, and all involve macroautophagy. We conclude that EBNA1’s nuclear location limits its accessibility to the macroautophagy pathway and, in consequence, limits the level and range of EBNA1 CD4 epitopes naturally displayed on the infected cell surface.
Keywords: autophagy, antigen processing, T cell
Classically, antigen-presenting cells (APCs) initiate help for immune responses against viral infection by processing exogenously acquired viral proteins and displaying their derived epitopes as MHC II:peptide complexes to CD4+ helper T cells. However, viruses that actually infect APCs may also be subject to CD4+ T cell control through direct target cell recognition, that is, if endogenously expressed viral antigens access the MHC II pathway within the infected cell and are processed to CD4 epitopes. Whereas such access might be expected of endogenously expressed proteins that naturally traffic through endolysosomal compartments (1) or are released and reinternalized into endosomes (2), how proteins at other intracellular locations might enter the MHC II pathway is less well understood. One potential route is via macroautophagy (hereafter referred to as autophagy), a degradative process in which the cell sequesters portions of cytoplasm into double-membrane vesicles that then fuse with lysosomes. Thus autophagy appears necessary for the MHC II-restricted presentation of two endogenously expressed model antigens, mouse complement C5 (3) and neomycin phosphotransferase (4). Whereas autophagy usually targets cytoplasmic proteins, analysis of peptides eluted from surface MHC II molecules after inducing autophagy in human B cells suggests that nuclear proteins may also be susceptible to this process (5), and at least one nuclear protein is reportedly presented to CD4+ T cells in this way (6). However, the rules governing endogenously expressed protein, particularly nuclear protein, access into the MHC II pathway remain poorly understood.
Here we address this using a virus, Epstein–Barr virus (EBV), which naturally infects and growth transforms an MHC II-positive cell type, the human B cell, expressing a range of latent cycle proteins that are targets for both CD4+ and CD8+ T cell responses. One of the most closely studied of these proteins is the nuclear antigen EBNA1. This is generally a weak CD8 immunogen, because it contains a glycine/alanine repeat (GAr) that partially limits access to the MHC I processing pathway (7). By contrast EBNA1 elicits generally strong CD4 responses and contains several well-defined CD4 epitopes (8, 9). Interestingly, however, only CD4+ T cell clones specific for a subset of those epitopes are able to recognize EBV-transformed lymphoblastoid cell lines (LCLs) in vitro and then at generally low levels, whereas other epitopes are not detectably presented (10 –13). Here we investigate the reasons underlying the limited CD4 epitope display from endogenously expressed EBNA1.
Results
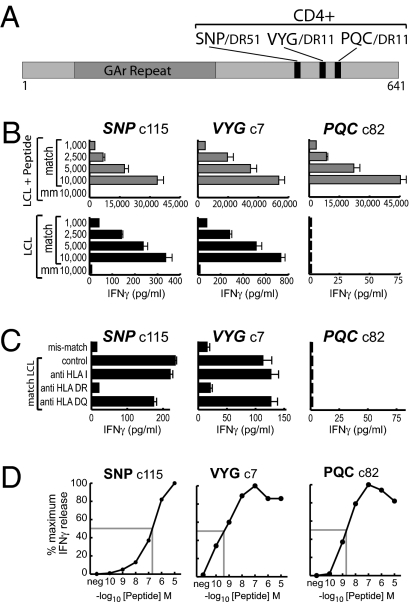
Using published procedures, we established CD4+ T cell clones against three EBNA1 epitopes, designated SNP restricted through HLA-DR51, VYG, and PQC, both restricted through HLA-DR11 (Fig. 1A, details in Table S1). All clones were then screened for LCL recognition in IFN-γ release assays. Because absolute levels of release are dependent upon the individual clone and upon numbers of T cells in the assay, all assays are internally controlled for T cell input and include HLA-matched and mismatched LCLs with and without brief preexposure to 5 μM epitope peptide. Fig. 1B shows representative results testing SNP-, VYG-, and PQC-specific clones on one matched and one mismatched target; data from a wider LCL panel, illustrating the specificity of recognition, are presented in Fig. S1. CD4+ T cells against all three epitopes showed strong recognition of HLA-matched, peptide-loaded targets. However, as seen in earlier work (13), assays on unmanipulated LCLs revealed clear interepitope differences. Clones against the PQC epitope consistently failed to recognize such targets, whereas clones against the SNP and VYG epitopes showed low level recognition (typically 1–2% of that seen with peptide loading) that titrated with T cell input and was clearly specific, being restricted to LCLs with the appropriate HLA-DR restricting allele (Fig. 1B) and being blocked by anti-DR but not by not by anti-DQ or anti-HLA class I antibodies (Fig. 1C). Note that these interepitope differences in LCL recognition could not be ascribed to differences in T cell clonal avidities. Thus in peptide titration assays (Fig. 1D) the PQC-specific clone, which failed to recognize unmanipulated LCL targets, was almost as avid as the VYG-specific and considerably more avid than the SNP-specific clone, both of which did recognize LCLs. Importantly, as shown in Fig. S2, there was no absolute block to PQC epitope generation by LCL cells because, when EBNA1 was provided to these cells as an exogenous antigen, all three epitopes were presented. Even here, however, PQC presentation was least efficient (possibly reflecting partial susceptibility of the PQC epitope sequence to destructive processing by lysosomal proteases; Fig. S2) and was the first to be lost when exogenous antigen supply to the MHC II pathway became limiting.
Fig. 1.
Characterization of EBNA1-specific CD4+ T cell clones. (A) Map showing the location and HLA restriction of the three EBNA1 CD4 epitopes (denoted by the first three amino acids of the epitope sequence). The GAr is also shown. (B) Results of an assay exposing T cell clones (1,000–10,000 T cells/well) to a matched and mismatched (mm) LCL either prepulsed with epitope peptide and washed well (shaded bars) or left unmanipulated (solid bars). IFN-γ release was measured after overnight coculture and is expressed in picograms per milliliter. Note the difference in horizontal scales for the Upper and Lower panels. (C) Results of a separate assay exposing T cell clones (1,000/well) to the same unmanipulated LCLs in the absence or presence of the indicated HLA blocking antibodies. A representative result from one of three experiments is shown. (D) Results of peptide titration assays using the above CD4+ T cell clones. Effector cells (100 per well) were incubated with the autologous LCL either unmanipulated (neg) or loaded with the indicated concentrations of epitope peptide and washed well, before use as targets. Results are expressed as a percentage of the maximum IFN-γ release seen in each assay. Functional avidities, measured as the peptide concentration required to achieve 50% maximum recognition (shaded lines), were 200 nM for the SNP-specific clone, 0.5 nM for the VYG-specific clone, and 2 nM for the PQC-specific clone.
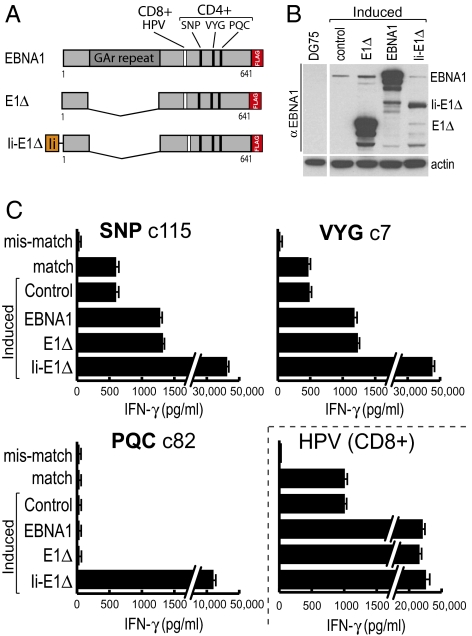
Turning to how endogenously expressed EBNA1 was accessing the MHC II presentation pathway in LCL cells, we first conducted a series of cross-feeding experiments with concentrated culture supernatants to show that access must be occurring by an entirely intracellular route and not, as has been observed with the other EBV-coded nuclear antigens EBNA2 and EBNA3C (14), through the release of antigenic species into culture medium and their uptake as exogenous antigen by neighboring cells (Fig. S3A). To explore this intracellular route in more detail, we then asked to what extent epitope display could be enhanced by increasing EBNA1 expression. To this end we used a recently developed episomal vector, pRTS1 (15), which is stably maintained in silent form in LCLs but from which antigen expression can be induced to high levels by doxocycline (dox). Fig. 2A shows the three EBNA1 constructs, all containing a carboxyl-terminal FLAG tag to aid detection, that were thus expressed: full-length EBNA1, GAr-deleted EBNA1 (E1Δ), and an invariant chain-E1Δ fusion (Ii-E1Δ) that is targeted to endosomes and the MHC II processing pathway (16). All three vector constructs, and an empty vector control, were stably introduced into LCLs, and then the cells were dox induced for 72 h and harvested, and protein extracts were subjected to SDS/PAGE and immunoblotted using an EBNA1-specific antibody. Full-length EBNA1 and E1Δ were expressed at levels >100-fold greater than was EBNA1 from the resident EBV genome (Fig. 2B); by contrast, steady-state levels of Ii-E1Δ were much lower, as expected of a protein targeted for endosomal degradation. These cells were all generated on an LCL background expressing the HLA-DR51 and DR11 restricting alleles for SNP, VYG, and PQC epitope presentation and also the HLA-B35 class I allele that presents an EBNA1-derived CD8 epitope HPV (Fig. 2A and Table S1) against which CD8 clones were also available. Fig. 2C shows the results when the above dox-induced targets (and standard LCLs matched and mismatched for the same restricting alleles) were assayed using T cell clones against the four epitopes. Cells induced to express the Ii-tagged antigen were recognized very strongly by the HPV-specific clone (in line with earlier work showing efficient MHC I processing of Ii-tagged constructs) (14, 17) and also by clones against all three CD4 epitopes including PQC; this clearly demonstrates that PQC can be processed from endogenously expressed EBNA1 providing the antigen is directly targeted to the endolysosomal system. However, high overexpression of the EBNA1 or E1Δ nuclear proteins led to only a 2- to 3-fold increase in SNP and VYG epitope display above the low baseline values seen on unmanipulated LCLs; furthermore there was still no detectable presentation of the PQC epitope, suggesting that antigen delivery into the MHC II pathway was still limited. By contrast, CD8 epitope display was greatly increased from the overexpressed nuclear proteins. Note that even with cells expressing such high levels, we could find no evidence for intercellular transfer of EBNA1 antigen (Fig. S3B), again strongly indicating that EBNA1 processing to the SNP and VYG epitopes must be occurring by an entirely intracellular route. Overall these data show that, in LCLs, the presentation of CD4 epitopes from endogenously expressed EBNA1 remains very limited even when the nuclear antigen is massively overexpressed.
Fig. 2.
EBNA1 overexpression only slightly increases SNP and VYG epitope display. (A) Map of the three EBNA1 constructs cloned into dox-regulated vectors. Each has a carboxy terminal FLAG tag; Ii represents amino acids 1–80 of the invariant chain. The locations of the three CD4 epitopes and an additional CD8 control epitope (epitope HPV) are shown. (B) LCLs stably transfected with an empty (control) or the indicated EBNA1 vectors were induced with dox for 72 h and lysed, and protein content was analyzed by SDS/PAGE and immunoblotting with an EBNA1-specifc mAb. Note that EBV-encoded EBNA1 is also detected in LCLs but not in the EBV-negative B cell line DG75. The blot was also probed with an anti-actin mAb as a loading control. (C) LCL cells, HLA matched for all T cell clones and either untransfected (match) or stably transfected with the indicated plasmids, were treated with dox for 72 h, washed, and used as targets in T cell assays (104 T cells/well); an HLA mismatched target LCL was also included. Results are shown as in Fig. 1; note the break in scale for some bars. A representative result from one of four experiments is shown. Similar results were obtained on a second HLA-matched LCL background.
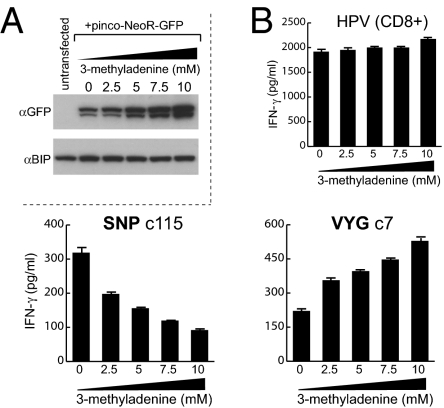
Given an earlier report that EBNA1 can be processed by autophagy (6), we asked whether baseline levels of SNP and VYG epitope display on the LCL surface were affected by a commonly used autophagy inhibitor, 3-methyladenine (3-MA) (18). To confirm the drug’s activity, we used LCL cells expressing NeoR, a model antigen turned over by autophagy (4), and showed that 72-h exposure to increasing 3-MA doses led to increasing accumulation of the NeoR protein (Fig. 3A). LCLs expressing physiological levels of EBNA1 from the EBV genome were therefore exposed to 3-MA as above, washed well, and then used as targets for T cell clones against the SNP and VYG CD4+ epitopes and, as controls, against the HPV CD8 epitope. Fig. 3B shows typical results, here using an LCL matched as before for all three HLA restricting alleles. Whereas display of the control CD8 epitope was largely unchanged, 3-MA treatment caused a dose-dependent fall in display of the SNP epitope but a concomitant increase in VYG-specific recognition. These differential effects of 3-MA on SNP and VYG epitope display were seen with a range of SNP- and VYG-specific clones and on several appropriately matched LCLs. Thus, of the two EBNA1 CD4 epitopes naturally presented on LCL cells, the processing of one (SNP) appears to be autophagy dependent whereas processing of the other (VYG), perhaps benefiting from reduced competition for limited antigen, is enhanced by autophagy inhibition. Although this latter pathway remains to be fully characterized, additional experiments showed that VYG presentation clearly requires the export of newly synthesized HLA II molecules, but unlike other candidate pathways reported in the literature (19), does not require the proteasome (Fig. S4)
Fig. 3.
Role of macroautophagy in EBNA1 CD4 epitope generation. (A) An LCL either untransfected or transfected with a plasmid expressing the autophagy substrate Neo-GFP, treated with the indicated concentrations of 3-MA for 72 h, and analyzed by SDS/PAGE and immunobloting with mAbs specific for GFP or BIP as a loading control. (B) An LCL incubated with 0–10 mM 3-MA for 72 h was used as targets for T cells (104 T cells/well) against the HPV (CD8), SNP, or VYG (CD4) epitopes. Results are shown as in Fig. 1. A representative result from one of three experiments is shown. In control experiments, 3-MA-treated and untreated cells were equally able to present limiting concentrations of exogenously loaded epitope peptides.
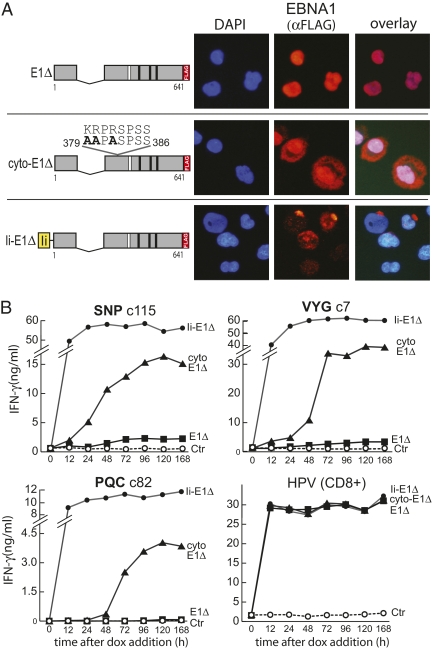
The link between autophagy and the presentation of some EBNA1 epitopes (shown here and reported by others) (6) raised the question why in LCL cells with high autophagic activity (20), EBNA1 epitope display is so limited. Because autophagosomes are predominantly cytoplasmic vesicles (21), we asked whether EBNA1’s exclusive nuclear location might limit its availability for processing by autophagy. We therefore mutated the nuclear localization sequence (nls) of EBNA1 (22) (using E1Δ as the template because of close proximity of the nls to the GAr), introduced this construct, cyto-E1Δ, into the dox-regulated vector, and generated stable transfectants on the same LCL background as with the other vectored antigens. Fig. 4A shows immunofluoresence staining of dox-induced LCL cells carrying the cyto-E1Δ, E1Δ, and Ii-E1Δ constructs, here staining with an anti-FLAG antibody to distinguish vector-coded antigen from the native virus-coded EBNA1. E1Δ was exclusively nuclear whereas cyto-E1Δ was distributed throughout the cytoplasm and nucleus, as seen previously for EBNA1 with this nls mutation (22); note that Ii-E1Δ localized to discrete cytoplasmic structures, confirming that Ii targeting overrides the EBNA1 nls signal. Protein levels were now assayed by immunoblotting up to 168 h after dox addition (Fig. S5). All three antigens were induced within 12 h and by 48–72 h both E1Δ and cyto-E1Δ had reached stable plateau levels well above that of native EBV-coded EBNA1; by comparison Ii-E1Δ levels were always much lower, consistent with this protein's targeting to endosomes. Using the same cells as targets in T cell assays (Fig. 4B), all three antigens were rapidly processed for CD8+ T cell recognition using the HPV indicator epitope, reaching maximal levels within 12 h. In the CD4+ T cell assays, as expected, endosomally targeted Ii-E1Δ was rapidly processed for recognition by SNP-, VYG-, and PQC-specific T cell clones whereas expression of nuclear-localized E1Δ led to just small increments of SNP and VYG epitope display that were apparent from 72 h onward; here again there was no detectable presentation of the PQC epitope, this time up to 168 h after dox induction. By contrast, the cyto-E1Δ antigen gave a very different pattern of results. Now all three CD4 epitopes were processed and presented to high levels that, over time, reached 25–50% of those observed with endosomally targeted Ii-E1Δ; interestingly, although all plateaued at high levels, there were some kinetic differences with SNP and VYG first being detected at 24–48 h and PQC at 48–72 h. Again these results reflected intracellular processing because, in experiments similar to those in Fig S3B, we never detected any intercellular antigen transfer.
Fig. 4.
Effect of antigen localization upon recognition by CD4+ T cells. (A) (Left) Maps of the same EBNA1 constructs from Fig. 2 and a new construct, cyto-E1Δ, in which three amino acids in the NLS have been altered to alanine. Locations of the CD8 (white bar) and CD4 (black bars) epitopes and the FLAG tag are shown. (Right) Immunofluorescence micrographs of LCLs stably transfected with the above constructs treated with dox for 72 h to induce expression. Nuclei are visualized using DAPI (blue) and EBNA1 was detected with anti-FLAG and alexa fluor-conjugated mAbs (red). (B) Results of T cell assays using an LCL HLA matched for all three CD4 T cell clones and the CD8 control. This LCL, stably transduced with one of the three vectors shown above or with the empty vector as a negative control (ctrl), was treated with dox for various lengths of time to induce EBNA1 expression before testing in T cell assays (104 T cells/well). Results are expressed as IFN-γ release in nanograms per milliliter; note the break in some vertical axes. A representative result from one of three experiments is shown.
Two further experiments were designed to check the significance of these findings with cytoplasmically targeted antigen. First, to avoid effects caused by antigen overload, we used a different vector system capable of expressing the E1Δ and cyto-E1Δ constructs at physiologic levels. LCL cells successfully transfected with this cyto-E1Δ vector showed a marked increment in SNP and VYG epitope display whereas transfectants with the E1Δ vector did not; yet both targets showed equally elevated levels of CD8 (HPV) epitope display (Fig. S6). Second, to check that the effects of nls mutation were indeed due to antigen relocalization, we introduced a heterologous nls into the cyto-E1Δ construct and expressed both this and cyto-E1Δ at physiologic levels in LCL cells using that same vector. The nls insertion restored nuclear localization and abrogated the increment in SNP and VYG epitope display that occurs with cytoplasmically targeted antigen (Fig. S7).
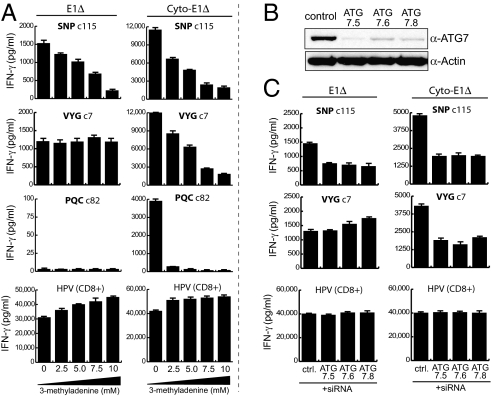
We then used the dox-inducible system to determine how processing of the different CD4 epitopes from nuclear and cytoplasmically targeted EBNA1 was affected by autophagy inhibition. As shown in Fig. 5A (Left), presentation of the SNP epitope from dox-induced nuclear E1Δ was inhibited by 3-MA whereas that of the VYG epitope was not; this result recapitulated the result seen with native EBNA1 in LCL cells (Fig. 3) and was also reproduced with full-length EBNA1 expressed from the dox vector (Fig. S8). However, the findings with cytoplasmically targeted antigen were again quite different. Not only were all three epitopes (SNP, VYG, and PQC) efficiently generated (as in Fig. 4) but also now each one's presentation was blocked by 3-MA (Fig. 5A, Right). As a further check, we took a second approach to inhibiting autophagy, using siRNA to transiently knock down the ATG7 protein essential for autophagosome formation (Fig. 5B) and timing dox-induced antigen expression to coincide with decreased autophagic activity. The resultant T cell assays (Fig. 5C) confirmed that only the SNP epitope was autophagy dependent when processed from nuclear antigen, whereas the much more efficient processing of cytoplasmic antigen to both SNP and VYG epitopes required autophagy.
Fig. 5.
Effect of cellular location on mechanism of EBNA1 processing. (A) Results of T cell assays using an LCL stably transduced with pRTS1-E1Δ (Left) or pRTS1-cyto-E1Δ (Right) and HLA matched for all three CD4+ and the CD8+ T cell clone. Antigen expression was induced with dox for 72 h in the presence of the indicated concentrations of 3-MA, and the cells were washed and used as targets in T cell assays (104 T cells/well). (B) Immunoblot of LCLs 72 h after electroporation with either control or one of three different ATG7-specific siRNA duplexes. The blot was probed with antibodies specific for ATG7 or actin as a loading control. (C) Results of T cell assays using the same approach described in A, except LCLs were electroporated with either control (ctrl) or ATG7-targeting siRNAs as in B. Antigen expression was induced 24 h (E1Δ) or 48 h (cyto-E1Δ) after siRNA electroporation and cells were harvested at 72 h for targets in T cell assays (104 T cells/well). Representative results are shown from one of three experiments. Note that 3-MA or ATG7 knockdown had no effect on MHC II processing capacity per se because, when these same cells were exposed to EBNA1 protein (as in Fig. S2A), levels of SNP and VYG epitope display from exogenously added antigen were not reduced by either treatment.
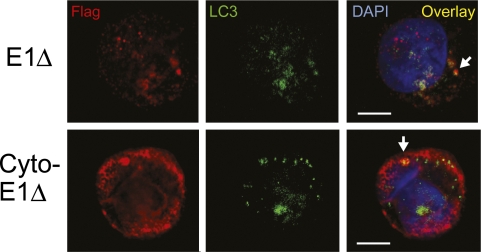
Finally, we used confocal microscopy to ask where EBNA1 was intersecting the autophagic pathway in LCLs, prompted by an earlier report of EBNA1 detection in cytoplasmic vesicles described as autophagosomes on the basis of their staining with monodansylcadaverine (MDC) (6). Because MDC is no longer considered a marker of bona fide autophagosomes (23), here we dual stained the above dox-induced cells for LC3, the only protein known to be specifically associated with autophagosomes (21), and for the FLAG-tagged EBNA1 antigens. As shown in Fig. 6, when ongoing endolysosomal proteolysis was halted by chloroquine, we were able to detect FLAG-tagged antigen colocalizing with LC3-positive autophagosomes in some cells in E1Δ -expressing cultures, whereas more dual-stained structures could be seen in cells induced to express cytoplasmically targeted antigen.
Fig. 6.
Confocal microscopy of nuclear- and cytoplasmic-localized EBNA1 in LCLs. An LCL, stably transfected with pRTS1 vectors expressing E1∆ or cyto-E1∆, was treated with dox for 72 h to induce protein expression. Cells were then treated with 50 mM chloroquine for a further 12 h and stained for EBNA1 (red) using an anti-FLAG mAb and for autophagosomes (green) using an anti-LC3 mAb, followed in both cases by appropriate Alexa-fluor-labeled secondary mAbs. Nuclei (blue) were visualized using DAPI. Note that, under the mild permeabilization conditions necessary to preserve cytoplasmic vesicular structures, detection of FLAG-tagged EBNA1 protein in the nucleus is reduced. Colocalization of EBNA1 and LC3 in the cytoplasm is shown as yellow in the overlay. (Scale bars, 5 μm.) Representative cells from one of three experiments are shown.
Discussion
Viruses naturally infecting MHC II-positive cells are potentially open to recognition by CD4+ T cells specific for endogenously expressed viral antigens, providing those antigens gain access to the MHC II processing pathway. To probe the rules governing such access, we focused on EBNA1, one of several EBV proteins constitutively expressed in virus-transformed LCL cells. Most healthy EBV-infected individuals possess CD4+ T cell memory to EBNA1 epitopes (8, 9), presumably as a result of a conventionally cross-primed response, and these cells, when cloned in vitro, provide sensitive probes of epitope display (10 –13). Focusing on three such epitopes, we found that two (SNP and VYG), were detectably displayed on the surface of unmanipulated LCL cells but at very low levels, whereas a third (PQC) was never detectable; indeed, this held true even when EBNA1 expression was increased by >100-fold from a dox-regulated vector. Such findings reinforce a theme seen in earlier work where EBNA1 epitope display on the LCL surface was generally poor, with several epitopes never detected, compared to epitopes from other endogenously expressed EBV proteins, EBNA2, -3A, and -3C (10, 12, 13).
We find that this is associated with a fundamental difference in the way these antigens access the MHC II pathway in LCL cells. Whereas EBNA2, -3A, and -3C, or antigenic fragments thereof, are shed into LCL culture supernatant and then acquired and processed as exogenous antigen by coresident cells (14), such intercellular antigen transfer is never detectable for EBNA1, whether expressed at physiologic levels or hyperexpressed as native or even as cytoplasmically targeted protein. We infer that, in LCL cells, endogenously expressed EBNA1 must access the MHC II presentation pathway by an intracellular route. For the native nuclear protein, however, such access is extremely limited, a limitation that cannot be overcome simply by increasing antigen expression. This contrasts with the MHC-I-restricted proteasomal processing of EBNA1 in the same cells, where CD8 epitope display was easily detectable from the EBV genome-coded antigen and was greatly enhanced by the dox-induced increase in antigen expression.
Interestingly, a recent study (6) reported that the endogenous presentation of some CD4 epitopes from EBNA1 required autophagy, a process that is known to be highly active in LCL cells (20). To ask if this was true of the SNP and VYG epitopes, we took the same approach and used 3-MA to inhibit autophagy, first confirming the drug’s inhibitory effect in LCLs expressing an indicator antigen, NeoR, a known substrate of autophagy. We found that 3-MA treatment of LCL cells reduced display of SNP, consistent with a role for autophagy in EBNA1 processing to that epitope, but had no such effect on the other epitope, VYG. The same held true when antigen supply was increased by dox-induced overexpression of EBNA1 or E1Δ. There we used two independent means of inhibiting autophagy, by 3-MA and by siRNA-mediated knockdown of the ATG7 protein, and both reduced display of the SNP but not of the VYG epitope. We infer that there are at least two intracellular routes whereby nuclear EBNA1 can be processed for CD4+ T cell recognition and these may lead to different epitope displays. One route involves antigen delivery to the endolysosomal system via autophagosomes as previously reported (6) whereas the other, yet to be characterized, is independent of autophagy. Interestingly, inhibition of autophagy actually led to an increase in VYG epitope presentation by this other route, an effect that was most marked when antigen supply was limiting (cf. Figs. 3 and 5), as one might expect if normally there were competition between the two pathways.
A key observation came when EBNA1 was targeted to the cytoplasm by nls mutation. Now all three CD4 epitopes, including PQC, were very efficiently presented. Furthermore this clearly involved autophagy, because the high levels of SNP, VYG, and PQC epitope display now observed were all markedly reduced both by 3-MA and siRNA targeting of ATG7. We acknowledge the need for caution when interpreting experiments that involve overexpression of a mutant protein within antigen-presenting cells. However, the greater susceptibility of cytoplasmic EBNA1 to autophagic processing did not seem to be a result of overexpression per se, as the effect was also observed with the same nls-mutated protein expressed at physiologic levels, or an artifact of the nls mutation, as redirection of that protein back to the nucleus with a heterologous nls reduced epitope display down to levels seen with nuclear EBNA1 itself. Our results therefore strongly suggest that cytoplasmic location increases susceptibility to autophagic processing, an idea already hinted at by other studies. Thus most examples of intracellular antigen delivery into the MHC II pathway involve cytoplasmic proteins (1, 3, 4, 19). Furthermore, although the processing route was not investigated, the tumor antigen CDC27 is better presented to CD4+ T cells when located in the cytoplasm rather than the nucleus (24). In addition, two polyglutamine-expanded proteins involved in the pathogenesis of neurologic disease, ataxin-5 (25) and androgen receptor (26), resist degradation by autophagy in the nucleus but not the cytoplasm. Such findings are consistent with autophagy being predominantly a cytoplasmic process, with autophagosomes mainly forming in the cell periphery far away from the nucleus (21). Indeed, in the present work, when endolysosomal processing was halted by chloroquine to reveal intermediates in the antigen delivery pathway (6), colocalization of EBNA1 staining with LC3, a marker of autophagosomes, was frequently detected by confocal microscopy in cells expressing cytoplasmically targeted EBNA1. However, as others have also observed (6), such colocalization with cytoplasmic autophagosomes could also be detected, albeit less frequently, in cells expressing EBNA1 as a native nuclear protein. This emphasizes a point, clear from work both in this viral system (ref. 6 and the present study) and on nonviral model antigens (27), that nuclear or nuclear-targeted antigens are not completely protected from delivery into the MHC II processing pathway via autophagy. How such antigens are captured into autophagosomes, whether within the nucleus itself, or by cytoplasmic structures originating from the nuclear envelope as recently seen in herpes simplex virus-infected cells (28), or by sequestration of some newly synthesized protein into a cytoplasmic store, remains to be determined.
The central message of this work coming from the study of EBNA1 is 3-fold. First, although the endogenously expressed protein can be processed for CD4+ T cell recognition in its native nuclear form, access to the MHC II pathway is very limited; as a result, only some epitopes are presented and then at very low levels. Second, that processing may occur by one of (at least) two pathways; for some epitopes, this involves autophagy, and for other epitopes, it does not. Third, cytoplasmic targeting of EBNA1 markedly sensitizes the protein to autophagy and leads to a much more abundant and wide-ranging epitope display. We therefore conclude that the nuclear location of native EBNA1 markedly limits its access to autophagy and, as a result, its visibility to the CD4+ T cell response.
Materials and Methods
T cell clones were generated and their HLA restriction, functional avidity, and ability to recognize LCLs were determined as described (13). T cell assays were performed in triplicate by coculturing LCL cells (5 × 104/well) overnight with T cells (104/well unless otherwise stated) in a 96-well V-bottom plate; IFN-γ in the supernatant was then measured by ELISA (13), and the mean ± SD are shown. 3-MA (Sigma-Aldrich) was dissolved in cell medium immediately before use; siRNA targeting the ATG7 gene was purchased from Dharmacon and control siRNA was from Sigma-Aldrich. More details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Elena Osdinatova (University of Birmingham) for expert assistance with confocal microscopy. This work was supported by the Medical Research Council, Cancer Research UK, and the Croucher Foundation (Hong Kong).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909448107/DCSupplemental.
References
- 1.Hegde NR, et al. Endogenous human cytomegalovirus gB is presented efficiently by MHC class II molecules to CD4+ CTL. J Exp Med. 2005;202:1109–1119. doi: 10.1084/jem.20050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eager KB, et al. Murine cell lines stably expressing the influenza virus hemagglutinin gene introduced by a recombinant retrovirus vector are constitutive targets for MHC class I- and class II-restricted T lymphocytes. J Immunol. 1989;143:2328–2335. [PubMed] [Google Scholar]
- 3.Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol. 1997;27:1506–1514. doi: 10.1002/eji.1830270629. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn F, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 5.Dengjel J, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paludan C, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 7.Lee SP, et al. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J Exp Med. 2004;199:1409–1420. doi: 10.1084/jem.20040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Münz C, et al. Human CD4(+) T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J Exp Med. 2000;191:1649–1660. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leen A, et al. Differential immunogenicity of Epstein-Barr virus latent-cycle proteins for human CD4(+) T-helper 1 responses. J Virol. 2001;75:8649–8659. doi: 10.1128/JVI.75.18.8649-8659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna R, et al. Isolation of cytotoxic T lymphocytes from healthy seropositive individuals specific for peptide epitopes from Epstein-Barr virus nuclear antigen 1: Implications for viral persistence and tumor surveillance. Virology. 1995;214:633–637. doi: 10.1006/viro.1995.0076. [DOI] [PubMed] [Google Scholar]
- 11.Paludan C, et al. Epstein-Barr nuclear antigen 1-specific CD4(+) Th1 cells kill Burkitt’s lymphoma cells. J Immunol. 2002;169:1593–1603. doi: 10.4049/jimmunol.169.3.1593. [DOI] [PubMed] [Google Scholar]
- 12.Mautner J, et al. Epstein-Barr virus nuclear antigen 1 evades direct immune recognition by CD4+ T helper cells. Eur J Immunol. 2004;34:2500–2509. doi: 10.1002/eji.200324794. [DOI] [PubMed] [Google Scholar]
- 13.Long HM, et al. CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines. J Virol. 2005;79:4896–4907. doi: 10.1128/JVI.79.8.4896-4907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor GS, et al. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol. 2006;177:3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 15.Bornkamm GW, et al. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 2005;33:e137. doi: 10.1093/nar/gni137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaux P, et al. Identification of MAGE-3 epitopes presented by HLA-DR molecules to CD4(+) T lymphocytes. J Exp Med. 1999;189:767–778. doi: 10.1084/jem.189.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonini C, Lee SP, Riddell SR, Greenberg PD. Targeting antigen in mature dendritic cells for simultaneous stimulation of CD4+ and CD8+ T cells. J Immunol. 2001;166:5250–5257. doi: 10.4049/jimmunol.166.8.5250. [DOI] [PubMed] [Google Scholar]
- 18.Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lich JD, Elliott JF, Blum JS. Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med. 2000;191:1513–1524. doi: 10.1084/jem.191.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27:2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 21.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura R, et al. Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (Importin alpha5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J Virol. 2006;80:1979–1991. doi: 10.1128/JVI.80.4.1979-1991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: From autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 24.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encoding MHC class II-restricted antigens: Mutated CDC27 as a tumor antigen. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 25.Iwata A, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montie HL, et al. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18:1937–1950. doi: 10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedel A, et al. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur J Immunol. 2008;38:2090–2095. doi: 10.1002/eji.200737900. [DOI] [PubMed] [Google Scholar]
- 28.English L, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.