Abstract
Using a multidimensional genomic data set on glioblastoma from The Cancer Genome Atlas, we identified hsa-miR-26a as a cooperating component of a frequently occurring amplicon that also contains CDK4 and CENTG1, two oncogenes that regulate the RB1 and PI3 kinase/AKT pathways, respectively. By integrating DNA copy number, mRNA, microRNA, and DNA methylation data, we identified functionally relevant targets of miR-26a in glioblastoma, including PTEN, RB1, and MAP3K2/MEKK2. We demonstrate that miR-26a alone can transform cells and it promotes glioblastoma cell growth in vitro and in the mouse brain by decreasing PTEN, RB1, and MAP3K2/MEKK2 protein expression, thereby increasing AKT activation, promoting proliferation, and decreasing c-JUN N-terminal kinase-dependent apoptosis. Overexpression of miR-26a in PTEN-competent and PTEN-deficient glioblastoma cells promoted tumor growth in vivo, and it further increased growth in cells overexpressing CDK4 or CENTG1. Importantly, glioblastoma patients harboring this amplification displayed markedly decreased survival. Thus, hsa-miR-26a, CDK4, and CENTG1 comprise a functionally integrated oncomir/oncogene DNA cluster that promotes aggressiveness in human cancers by cooperatively targeting the RB1, PI3K/AKT, and JNK pathways.
Keywords: microRNA, CDK4, miR-26a, PTEN, RB1
Glioblastoma multiforme (GBM) is one of the most malignant of all brain tumors, with a median survival of ∼14 months (1, 2). The recently published Cancer Genome Atlas (TCGA) data set (3) includes high-resolution information on DNA copy number, mRNA and microRNA expression, DNA methylation, single-nucleotide polymorphisms, and somatic mutations in GBM. One of the most common copy-number alterations in GBM is amplification at chromosome 12q13.3–14.1, which is also amplified in melanoma (4), breast (5), and lung cancers (6). The CDK4 gene has been postulated as the target of this amplification. CDK4 promotes proliferation by inhibiting the RB1 tumor suppressor and by sequestering p27Kip1 and p21Cip1, thereby promoting E2F- and Cdk2-dependent cell cycle progression (7). However, CDK4 overexpression alone does not induce spontaneous tumorigenesis in transgenic animal models, suggesting that CDK4 cooperates with other genetic changes during tumorigenesis (7). Frequently coamplified with CDK4 is CENTG1. CENTG1 encodes PIKE, which activates AKT and PI3 kinase (8), thus promoting cell growth and transformation (9).
Recent studies have implicated microRNAs in carcinogenesis (10, 11), and some microRNAs are the targets of genomic amplifications or deletions in a range of cancers. In GBM, however, the role of microRNAs in GBM tumorigenesis is only beginning to be unraveled. Using data from the TCGA GBM project, we show here that hsa-miR-26a is frequently coamplified with CDK4 and CENTG1. miR-26a decreases expression of the tumor suppressors PTEN and RB1 and inhibits expression of MAP3K2/MEKK2, an activator of c-Jun N-terminal kinase (JNK) (12). JNK has also been implicated in tumor suppression (13). In this manner, miR-26a transforms NIH 3T3 cells and cooperates with CDK4 and CENTG1 to increase GBM growth. Amplification of this oncomir/oncogene cluster correlates with shortened survival in GBM patients.
Results
miR-26a Is a Frequent Target of the 12q13.3–14.1 Amplicon.
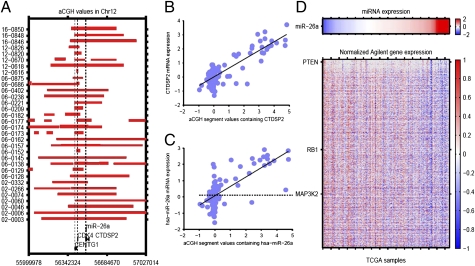
Analysis of TCGA data from GBM revealed numerous genomic alterations (3). To identify microRNAs that are likely to play a key role in GBM, we searched for microRNAs whose expression is driven by copy number. Using Pearson’s correlation across 176 patients, we identified miR-26a with a correlation of 0.72 between copy number and microRNA expression (Table S1). hsa-miR-26a was located within an amplicon at 12q13.3–14.1. This amplicon was present in 32 samples and often contained CDK4 and CENTG1 (Fig. 1A). hsa-miR-26a was included in 21 of the amplicons.
Fig. 1.
hsa-miR-26a is a component of the 12q13.3–14.1 amplicon. (A) Schematic map of 12q13.3–14.1 amplifications in the 32 GBM specimens (y axis), including the genomic locations of CENTG1, CDK4, hsa-miR-26a, and CTDSP2. The thickness of each horizontal line indicates the degree of amplification. (B) Scatter plot of DNA copy number versus mRNA expression for CTDSP2. The Pearson’s correlation coefficient is 0.83. (C) Scatter plot of DNA copy number versus microRNA expression values for hsa-miR-26a. Each circle represents the expression value from a single GBM specimen. The horizontal dotted line represents the median miR-26a expression value of the samples in which the DNA segment was neither amplified nor deleted (−0.2 < segment value < 0.2). The solid line shows a linear fit of data. Pearson’s correlation coefficient is 0.72. (D) Correlations between miR-26a and mRNA expression profiles identified using the method described in the text. Gray areas represent data not used for correlation computations to filter extraneous effects such as copy-number alterations and DNA methylation.
The hsa-miR-26a sequence is embedded within the CTDSP2 gene, which encodes carboxyl-terminal domain RNA polymerase II polypeptide A small phosphatase 2. Pearson’s correlation coefficient between CTDSP2 mRNA expression and gene copy number was 0.83 in GBMs harboring the 12q13.3–14.1 amplicon, indicating that CTDSP2 expression is copy-number driven (Fig. 1B). Importantly, many miR-26a expression values in tumors with 12q13.3–14.1 amplification exceeded the median miR-26a expression value for the entire sample population (Fig. 1C).
The hsa-miR-26a sequence is also encoded by another gene, CTDSPL, which is located on chromosome 3. However, we saw no correlation between CTDSPL and miR-26a expression (Fig. S1a). Thus, amplification of miR-26a on 12q is the primary determinant of miR-26a overexpression in GBM.
Identification of microRNA Targets by Integrative Genomics.
Several microRNA target prediction algorithms exist [e.g., miRanda (ref. 14), miRBase (ref. 15), and TargetScan (ref. 16)] that are based upon the predicted binding between microRNA and the 3′-UTR sequence of the target messenger RNAs. However, the biologically relevant targets of each microRNA may vary from one tissue to the next, depending on the expression of the target mRNAs and the cellular context. Such considerations are especially relevant in cancer, where genetic and epigenetic modifications can radically alter RNA expression patterns and protein function. Consequently, we developed a strategy for identifying microRNA targets that takes into account tissue-specific DNA, mRNA, microRNA, and epigenetic modifications (Fig. S1b).
We first used clinical annotations to exclude tumor samples with excessive necrosis or nontumor tissue. Copy number and DNA methylation data from the remaining samples were then used to eliminate loci that were amplified, deleted, or aberrantly methylated, thereby filtering out changes in mRNA expression due to copy-number alterations or promoter silencing. Although this filtering procedure may result in a decrease in sensitivity, it allows for a more specific identification of microRNA-driven changes in mRNA expression. We selected for further analysis those mRNAs whose expression was inversely correlated (r < −0.15) with miR-26a expression. Genes that are predicted targets of transcription factors that regulate expression of the miR-26a host gene (CTDSP2) were also filtered. Complementary binding sites between the miR-26a sequence and the 3′-UTR sequences of these candidate mRNAs were identified, and the unfolding free energy of the microRNA as well as the energy of binding for potential mRNA targets [computed by RNAup (refs. 17 and 18)] was used to rank the targets. Using this approach, 961 putative miR-26a target transcripts were identified in GBM (Table S2). The mRNA expression of these targets was anti-correlated with miR-26a expression (Fig. 1D). Additional details of this strategy are described in SI Methods.
We reasoned that miR-26a might promote GBM growth by decreasing the expression of a tumor suppressor, and we identified the PTEN and RB1 tumor suppressors among the list of candidate target genes. We also identified MAP3K2/MEKK2, an activator of the JNK pathway, which has been implicated in apoptosis and tumor suppression (13, 19). Thus, PTEN, RB1, and MAP3K2/MEKK2 were chosen for further study. Reporter assays in which the PTEN, RB1, or MAP3K2 3′-UTRs (containing predicted binding sites for miR-26a) were fused to GFP or luciferase confirmed that the 3′-UTRs of these mRNAs confer regulation by miR-26a (Fig. S2 a and b).
miR-26a Regulates PTEN Expression and AKT Activation in GBM.
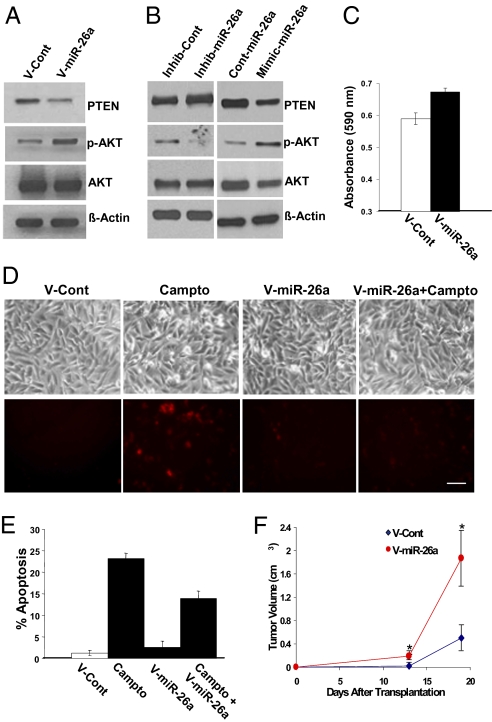
To examine the effect of miR-26a in GBM, we generated a lentivirus containing a 300-bp sequence encoding the miR-26a-2 pri-miRNA. Real-time PCR confirmed overexpression of miR-26a in LN229 GBM cells transduced with the virus (Fig. S2c). As predicted, miR-26a overexpression in LN229 GBM cells decreased PTEN protein expression and increased AKT activation (Fig. 2A).
Fig. 2.
miR-26a regulates PTEN and AKT. (A) PTEN expression and AKT phosphorylation in LN229 GBM cells transduced with a miR-26a lentivirus or a control lentivirus. (B) PTEN expression and AKT phosphorylation in LN229 cells after exposure to the miR-26a inhibitor, mimic, or control oligonucleotides (200 nM). (C) MTT growth assay for LN229 cells transduced with a miR-26a virus or a control virus. Mean ± SEM is shown (P < 0.003, t test). (D) Phase and fluorescence micrographs illustrating effect of mir-26a on DNA damage-induced apoptosis in LN229 cells. Cells were transduced with a miR-26a virus or a control virus and exposed overnight to the DNA-damaging agent, camptothecin (Campto, 50 μM) or vehicle alone before staining for Annexin V (red) to identify apoptotic cells. (Scale bar, ≈100 μm.) (E) Quantitation of effect of miR-26a on DNA damage-induced apoptosis in LN229 cells. Mean ± SEM is shown (P ≤ 0.005, t test). (F) Effect of miR-26a on LN229 cells transduced with a miR-26a virus or a control virus and transplanted s.c. into nude mice. Tumor volume was measured over 3 weeks. Mean ± SEM is shown (*P ≤ 0.05, t test).
To investigate this phenomenon further, we inhibited endogenous miR-26a function using an oligonucleotide microRNA inhibitor (antagomir), and we also used a miR-26a oligonucleotide mimic (a double-stranded oligonucleotide chemically modified to enhance programming of the RISC complex with the active miR-26a microRNA strand). A control oligonucleotide was used as a control. The miR-26a mimic decreased PTEN protein levels and increased AKT phosphorylation, although the miR-26a inhibitor had the opposite effect (Fig. 2B).
Overexpression of miR-26a increased LN229 GBM cell growth when compared to control cells, consistent with the ability of miR-26a to activate AKT (Fig. 2C). The miR-26a mimic (200 nM) also increased GBM cell growth (Fig. S2d). We also observed a significant decrease in apoptosis occurring after DNA damage in LN229 cells overexpressing miR-26a (Fig. 2 D and E). When transplanted subcutaneously (s.c.) into mice, LN229 GBM cells overexpressing miR-26a formed larger tumors than control cells (Fig. 2F). Thus, miR-26a decreases PTEN expression, activates AKT, and promotes tumor growth.
miR-26a Inhibits RB1 Expression in GBM.
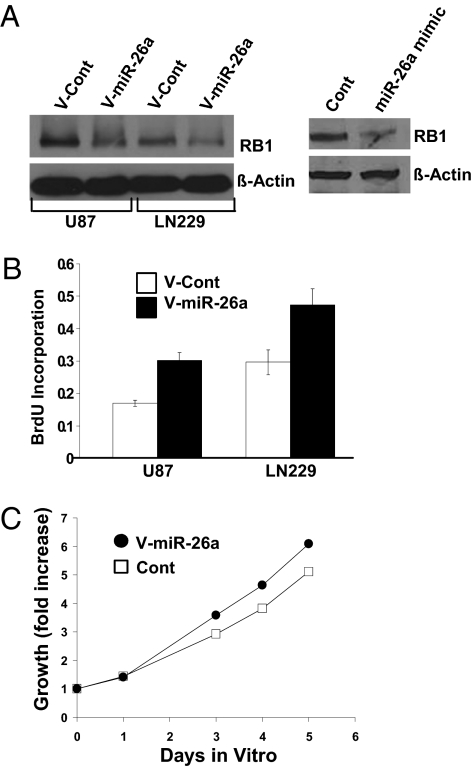
The GBM-specific microRNA target identification strategy used here also identified RB1 as a probable target of miR-26a in GBM. To investigate the effect of miR-26a on RB1 separately from its effects on PTEN, we used human U87 GBM cells that lack functional PTEN (20). The miR-26a mimic or lentiviral-mediated overexpression of miR-26a in U87 or LN229 GBM cells decreased RB1 expression (Fig. 3A) and increased DNA synthesis (Fig. 3B). The miR-26a mimic (100 nM) also increased DNA synthesis in LN428 GBM cells (Fig. S2e). In addition, miR-26a overexpression or the miR-26a mimic increased U87 GBM cell growth despite the absence of PTEN (Fig. 3C). The proliferative effect of miR-26a was antagonized by overexpression of PTEN or RB1 (Fig. S2f).
Fig. 3.
miR-26a decreases RB1 expression and promotes proliferation. (A) (Left) RB1 protein expression in LN229 and U87 GBM cells transduced with a miR-26a lentivirus or a control lentivirus. (Right) RB1 protein expression in U87 cells after exposure to miR-26a mimic or a control oligonucleotide (200 nM). (B) BrdU proliferation assay for LN229 (PTEN-wild type) and U87 (PTEN-deficient) GBM cells transduced with a miR-26a lentivirus or a control lentivirus. Mean ± SEM is shown (P < 0.02, t test). (C) MTT growth assay for U87 GBM cells transduced with a miR-26a lentivirus or a control virus. MTT measurements were normalized to data obtained at the time of plating. Mean of eight replicates is shown (P = 0.0001, t test).
miR-26a Inhibits MAP3K2/MEKK2 Expression and JNK-Dependent Apoptosis.
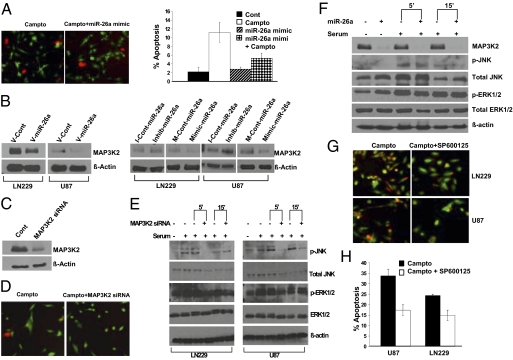
miR-26a inhibited DNA damage-induced apoptosis in PTEN-deficient U87 GBM cells (Fig. 4A), indicating the presence of a PTEN-independent mechanism for apoptosis regulation by miR-26a. Our microRNA target identification strategy identified MAP3K2 as a probable target of miR-26a in GBM. The MAP3K2 gene encodes MEKK2, a mitogen-activated protein kinase kinase. MEKK2 is involved in JNK and ERK5 activation, and JNK activation can promote apoptosis in GBM cells (12, 19).
Fig. 4.
miR-26a decreases MAP3K2/MEKK2 expression and inhibits JNK-dependent apoptosis. (A) (Left) Live cell (green)/dead cell (red) assay for U87 GBM cells after exposure to camptothecin (50 μM) overnight. U87 cells were treated with the miR-26a mimic or a control oligonucleotide (200 nM) before exposure to camptothecin. (Right) Quantification of the data shown to the Left. Mean ± SEM is shown (P = 0.05, t test). (B) (Left) MAP3K2/MEKK2 protein expression in LN229 and U87 GBM cells transduced with a miR-26a virus (V-miR-26a) or a control virus (V-Cont). (Right) Effect of miR-26a mimic or inhibitor (200 nM) on MAP3K2/MEKK2 protein expression in U87 and LN229 cells. (C) Effect of MAP3K2 siRNA on MAP3K2/MEKK2 protein expression in 293T cells. (D) Live cell (green)/dead cell (red) assay for U87 cells transfected with MAP3K2/MEKK2 siRNA or a control oligonucleotide before exposure to camptothecin. (E) Effect of MAP3K2 siRNA on serum-stimulated JNK phosphorylation and ERK1/2 phosphorylation in LN229 and U87 cells. Protein isolates were collected at 0, 5, and 15 min after the addition of serum. Of note, serum alone induced a decrease in total JNK expression over time. (F) Effect of miR-26a mimic (200 nM) on MAP3K2/MEKK2 expression, serum-stimulated JNK phosphorylation, and ERK1/2 phosphorylation in LN229 GBM cells. (G) Live cell (green)/dead cell (red) assay for U87 and LN229 cells treated with the specific JNK inhibitor SP600125 (50 μM) or an inactive control compound before exposure to camptothecin overnight. (H) Quantification of data shown in G. Mean ± SEM is shown (P = 0.0041 and P = 0.013 for U87 and LN229 cells, respectively, t test).
As predicted, miR-26a and the miR-26a mimic decreased MAP3K2/MEKK2 protein expression in U87 and LN229 GBM cells, whereas the miR-26a inhibitor increased it (Fig. 4B). The anti-apoptotic effect of miR-26a could be mimicked by siRNA-mediated knockdown of MAP3K2/MEKK2 in U87 cells (Fig. 4 C and D). MAP3K2/MEKK2 knockdown decreased serum-dependent JNK phosphorylation in LN229 and U87 cells (Fig. 4E). The miR-26a mimic also inhibited JNK activation in U87 cells (Fig. 4F). Finally, a JNK inhibitor (SP600125) decreased DNA damage-induced apoptosis in GBM cells (Fig. 4 G and H). Thus, miR-26a decreases JNK-dependent apoptosis in GBM via MAP3K2/MEKK2 inhibition.
Collaborative Effect of miR-26a, CDK4, and CENTG1 in GBM.
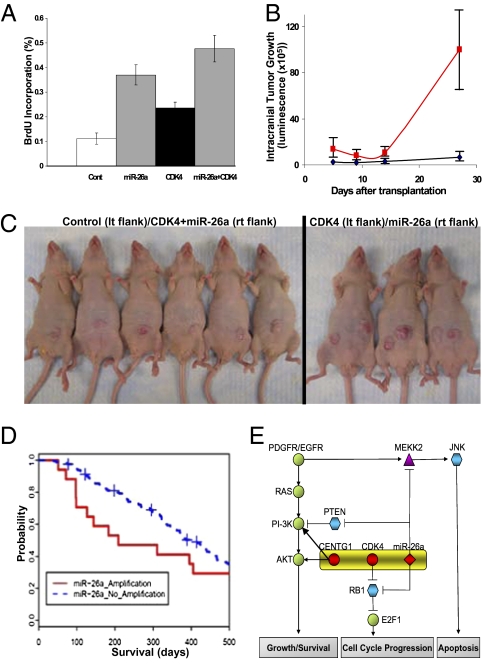
Inspection of the 12q13.3–14.1 amplicon in 32 GBMs indicated that hsa-miR-26a was amplified in 66%, CENTG1 was amplified in 81%, and CDK4 was amplified in 100% (Fig.1A). hsa-miR-26a was coamplified with CDK4 and CENTG1 in 56% of the amplicons and with CDK4 alone in an additional 9% of the amplicons. CDK4 and CENTG1 were coamplified without hsa-miR-26a in 25% of the amplicons, and CDK4 was amplified alone in 9% of the amplicons. We observed a significant correlation between mRNA expression for CDK4, CENTG1, and miR-26a in tumors harboring 12q13.3–14.1 amplifications (Fig. S3a), indicating coordinated copy-number driven expression of all three genes. We therefore overexpressed these genes in various combinations in 293T or U87 cells and measured DNA synthesis. Overexpression of miR-26a, CDK4, or CENTG1 alone increased 293T cell proliferation, but coexpression of miR-26a and CDK4 or CENTG1 produced further increases (Fig. S3b). In PTEN-deficient U87 GBM cells, a similar pattern was observed, except that no additional effect of miR-26a was observed after RB1 was inactivated by CDK4 overexpression (Fig. S3c). This is consistent with the idea that miR-26a promotes proliferation via RB1 suppression in cells lacking PTEN and promotes proliferation via AKT activation in PTEN-intact cells overexpressing CDK4. Accordingly, either mir-26a or CDK4 increased proliferation in PTEN-intact NIH 3T3 cells, and coexpression of CDK4 and miR-26a further increased proliferation (Fig. 5A, P < 0.0036, t test).
Fig. 5.
miR-26a promotes tumorigenesis and regulates survivorship. NIH 3T3 cells transfected with a CDK4 vector or a control vector were exposed to the miR-26a mimic or a control oligonucleotide (200 nM) and BrdU proliferation assays were performed. Mean ± SEM (five replicates) is shown. miR-26a promoted the proliferation of control cells (P < 0.0014, t test) and cells overexpressing CDK4 (P < 0.0036, t test). (B) Effect of miR-26a overexpression on the growth of U87 GBM cells stably expressing luciferase after intracranial transplantation in nude mice. Tumor growth was imaged noninvasively using luminescence. Mean ± SEM is shown (P < 0.0089, t test). (C) NIH 3T3 cells stably expressing CDK4 or a control vector were transduced with either a miR-26a lentivirus or a control virus and transplanted s.c. into the left or right flank of nude mice. After 1 month, control cells (Left, left flank) failed to produce tumors, whereas cells overexpressing both CDK4 and miR-26a (Left, right flank) uniformly produced tumors. Cells overexpressing CDK4 alone (Right, left flank) or miR-26a alone (Right, right flank) also produced tumors in 100% of the animals. (D) Kaplan–Meier survival analysis for 178 GBM patients harboring amplifications of hsa-miR-26a. Patients were divided into two groups on the basis of the presence or the absence of high amplifications of hsa-miR-26a (segment value >2.0). Survival was calculated from the time of first diagnosis. The P value of the log-rank test was 0.0233. Fifty-eight patients with survival >500 days are included in the P-value computation (Table S3), but are not shown here. (E) Diagram illustrating the collaborative effect of genes in the 12q13.3–14.1 amplicon (hsa-miR-26a, CDK4, and CENTG1) on the PI3K/AKT, RB1, and JNK pathways in GBM.
To confirm a PTEN-independent effect of miR-26a on GBM growth in vivo, we transplanted PTEN-deficient U87 GBM cells transduced with either a miR-26a lentivirus or a control virus into the brains of nude mice. After 4 weeks, tumors formed by U87 cells overexpressing miR-26a were significantly larger than control tumors (Fig. 5B and Fig. S3d, P < 0.009, t test).
To examine the role of miR-26a in cell transformation, we stably overexpressed miR-26a, CDK4, or control vectors in nontransformed NIH 3T3 fibroblasts and transplanted the cells s.c. into nude mice. After 4 weeks, none of the mice transplanted with control NIH 3T3 cells developed tumors. In contrast, 100% of the mice transplanted with cells overexpressing miR-26a, CDK4, or miR-26a and CDK4 together developed tumors (Fig. 5C). Taken together, these data indicate that miR-26a alone promotes transformation and growth and that coamplification of miR-26a, CDK4, and CENTG1 collaboratively promotes tumor growth.
Using clinical annotations from the TCGA data set, we compared the survival of GBM patients with tumors that either contained or lacked the 12q13.3–14.1 amplicon (Fig. 5D). Both hsa-miR-26a and CDK4 amplifications were associated with a significantly shorter median survival (P = 0.0233 and P = 0.0175, respectively, log-rank test; see SI Methods and Table S3). The median survival for patients with tumors harboring the miR-26a amplicon was 209 days, whereas the median survival for those lacking it was 383 days. Strikingly, no long-term survivors (>2 years) were observed among patients with amplifications at 12q13.3–14.1.
On the basis of these observations, we propose a model in which hsa-miR-26a, CDK4, and CENTG1 comprise a functionally integrated oncomir–oncogene DNA cluster (Fig. 5E). Frequent coamplification of these genes leads to combined activation of the PI3K/AKT pathway and inactivation of the RB1 and JNK pathways. The coordinated biological effect of this amplification thus potentiates GBM growth and markedly decreases survival in patients harboring this genomic alteration.
Discussion
Numerous microRNAs have been implicated in cancer, including the let-7 family of microRNAs (21, 22), the microRNA 200 family (23), the microRNA 34 family (24), microRNA 21 (25), microRNA 221 and 222 (26), and others. In GBM, microRNA 21 acts as an anti-apoptotic factor (27). More recently, microRNA 7 has been shown to regulate the EGFR and AKT pathways in GBM (28). In addition, microRNAs 221 and 222 have been shown to regulate p27Kip1 levels in GBM (29).
Here we provide evidence that the amplicon at 12q13.3–14.1 contains an oncogenic microRNA, miR-26a. We do not believe that CTDSP2 itself is the biological target of this amplification, as small oligonucleotides mimicking the miR-26a sequence and the pri-miR-26a sequence alone promoted tumor growth. A majority of amplicons at 12q13.3–14.1 contained miR-26a, suggesting that miR-26a amplification provides a significant growth advantage. Our data confirm that overexpression of this microRNA with CDK4 enhances growth. Interestingly, CDK4 alone does not produce tumors when overexpressed in astrocytes or keratinocytes in vivo. However, it does increase tumor formation in animal models of tumor promotion (7) and in NIH 3T3 cells in the current study. We find that miR-26a alone can transform NIH 3T3 cells and increase GBM growth in the absence of PTEN or CDK4 overexpression.
Another opportunity for synergism among genes at 12q13.3–14.1 involves coamplification of miR-26a and CENTG1. CENTG1 encodes a family of GTPases (PIKE) that bind PI3K and AKT and stimulate their activity (8, 30). Our data indicate that miR-26a and CENTG1 collaboratively promote growth in GBM. Taken together, these observations reveal the presence of a functionally integrated oncomir/oncogene cluster that is frequently amplified in GBM. The net effect of this amplification is the coordinated antagonism of the JNK and RB1 pathways and the activation of the PI3K/AKT pathway. These latter two pathways are among the most frequently altered in GBM. Importantly, amplification of this oncomir/oncogene cluster is associated with a particularly poor prognosis among GBM patients.
Methods
TCGA Data Analysis.
Array CGH, microRNA, mRNA, DNA methylation, and clinical data for 178 GBM patients were downloaded from The Cancer Genome Atlas project data portal (http://cancergenome.nih.gov/dataportal) in March, 2009. Data use certification was obtained for the Controlled-Access data. Details on the data processing and platforms are available in the SI Methods and the publication describing the GBM data analysis (3).
Lentiviruses and Stable Cell Lines.
The hsa-miR-26a gene with ≈150 bp of flanking sequence was cloned from human genomic DNA by PCR and confirmed by DNA sequencing. The forward primer was 5′-ACTAGTCAGAGCAAGACTCGGCAGGGTGTCTG-3′, and the reverse primer was 5′-GTCGACCACCAGGCTTCCAATGGATCAGTGGTC-3′. The PCR product was transferred into the pLenti6-IRES-EGFP vector and packaged in 293FT cells. LN229, U87, and LN428 GBM cells were transduced with the appropriate lentiviruses, and stable cell lines were selected using blasticidin where appropriate. NIH 3T3 cells were transiently transfected with a CDK4 vector or a control vector, and stable transfectants were selected in G418 (500 μM). For some experiments, NIH 3T3 cells were transduced with the mir-26a or control lentiviruses.
EGFP and Luciferase Reporter Assays.
Fusion proteins were generated in which EGFP or firefly luciferase was fused to the PTEN, RB1, or MAP3K2 3′-UTR containing putative miR-26a binding sites. Expression of the EGFP or luciferase fusion proteins in 293T cells was then determined in the presence of the miR-26a mimic or a control oligonucleotide (100 nM). See SI Methods for details.
RNAi Studies.
miRIDIAN microRNA oligonucleotide mimic and inhibitor for mir-26a, as well as the corresponding control oligonucleotides, were purchased from Dharmacon. MAP3K2 siRNA and a matched oligonucleotide control were purchased from Invitrogen. miRIDIAN miR-26a mimic or inhibitor was added to the medium without the use of additional transfection reagents at a concentration of 100–200 nM for 48 h before performing experiments. For MAP3K2 siRNA experiments, oligofectamine was used to transiently transfect the cells overnight before performing experiments.
Real-Time PCR.
Total RNA was extracted from LN229 or U87 GBM cell lines. cDNA was prepared using 1 μg of total RNA from each sample, using a miR-26a-specific primer (Applied Biosystems). Six nanograms of cDNA were then used for real-time PCR analysis in a final reaction volume of 20 μL. Samples were analyzed in triplicate and statistical analysis was performed using the t test.
Western Blots.
Total protein was extracted and separated by gel electrophoresis. Protein was then transferred to nitrocellulose membranes and probed overnight using the appropriate primary antibodies. The antibodies used were AKT, phosphorylated AKT (ser473), or RB1 (Cell Signaling); PTEN and MAP3K2/MEKK2 (Abcam); and total SAPK/JNK, phospho-SAPK/JNK (Thr183/Tyr185), p44/42 MAP Kinase (ERK), and phospho-p44/42 MAPK (Thr202/Tyr204, p-ERK) (Cell Signaling). After incubation in the appropriate secondary antibodies, immunoreactive bands were visualized using chemiluminescence.
Growth and Proliferation Assays.
Cell growth was assayed in vitro using MTT and CCK-8 colorimetric assays, and cell proliferation was assayed using BrdU incorporation, using standard methods (SI Methods).
Tumor Growth Assays.
Animal studies were conducted under the auspices of an Institutional Review Board approved protocol. LN229 GBM cells, U87 GBM cells expressing luciferase, or NIH 3T3 cells expressing CDK4 or a control vector were transduced with either a miR-26a lentivirus or a control virus. Totals of 1 × 105 cells (U87, intracranial) or 5 × 106 cells (LN229 or NIH 3T3, s.c.) were then injected into nude mice (n = 3–7 animals per condition). s.c. tumor growth was measured serially, and volume was calculated using the formula for a spheroid. Significance was determined using the t test. Intracranial tumor growth was imaged noninvasively using luminescence (SI Methods).
Supplementary Material
Acknowledgments
While this manuscript was in revision, a separate report confirming the effect of miR-26a on PTEN in GBM was published (31). Data were obtained from The Cancer Genome Atlas pilot project (downloadable from http://tcga.cancer.gov/dataportal). This work was supported by a National Institutes of Health Director’s New Innovator Award (to M.D.J.) and National Institutes of Health grants 1U24 CA126554 (The Cancer Genome Atlas) and R01 GM082798 (to P.J.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909896107/DCSupplemental.
References
- 1.Wiedemeyer R, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLendon R, et al. Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muthusamy V, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2008;45:447–454. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- 5.An HX, Beckmann MW, Reifenberger G, Bender HG, Niederacher D. Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am J Pathol. 1999;154:113–118. doi: 10.1016/S0002-9440(10)65257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikman H, et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes Chromosomes Cancer. 2005;42:193–199. doi: 10.1002/gcc.20122. [DOI] [PubMed] [Google Scholar]
- 7.Miliani de Marval PL, Macias E, Conti CJ, Rodriguez-Puebla ML. Enhanced malignant tumorigenesis in Cdk4 transgenic mice. Oncogene. 2004;23:1863–1873. doi: 10.1038/sj.onc.1207309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CB, Ye K. PIKE GTPase are phosphoinositide-3-kinase enhancers, suppressing programmed cell death. J Cell Mol Med. 2007;11:39–53. doi: 10.1111/j.1582-4934.2007.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Hu Y, Hao C, Rempel SA, Ye K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene. 2007;26:4918–4927. doi: 10.1038/sj.onc.1210290. [DOI] [PubMed] [Google Scholar]
- 10.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 11.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Su B, Cheng J, Yang J, Guo Z. MEKK2 is required for T-cell receptor signals in JNK activation and interleukin-2 gene expression. J Biol Chem. 2001;276:14784–14790. doi: 10.1074/jbc.M010134200. [DOI] [PubMed] [Google Scholar]
- 13.Lee JH, et al. JNK pathway mediates apoptotic cell death induced by tumor suppressor LKB1 in Drosophila. Cell Death Differ. 2006;13:1110–1122. doi: 10.1038/sj.cdd.4401790. [DOI] [PubMed] [Google Scholar]
- 14.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Mückstein U, et al. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 18.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, et al. Glioma pathogenesis-related protein 1 exerts tumor suppressor activities through proapoptotic reactive oxygen species-c-Jun-NH2 kinase signaling. Cancer Res. 2008;68:434–443. doi: 10.1158/0008-5472.CAN-07-2931. [DOI] [PubMed] [Google Scholar]
- 20.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SM, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 23.Nam EJ, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 24.Rokhlin OW, et al. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–1296. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 26.Visone R, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 27.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 28.Kefas B, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 29.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 30.Ahn JY, Hu Y, Kroll TG, Allard P, Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc Natl Acad Sci USA. 2004;101:6993–6998. doi: 10.1073/pnas.0400921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.