Abstract
Chemoreceptors provide sensory specificity and sensitivity that enable motile bacteria to seek optimal positions for growth and metabolism in gradients of various physicochemical cues. Despite the abundance of chemoreceptors, little is known regarding the sensory specificity and the exact contribution of individual chemoreceptors to the lifestyle of bacteria. Azospirillum brasilense are motile bacteria that can fix atmospheric nitrogen under microaerophilic conditions. Here, we characterized a chemoreceptor in this organism, named AerC, which functions as a redox sensor that enables the cells to seek microaerophilic conditions that support optimum nitrogen fixation. AerC is a representative of a widespread class of soluble chemoreceptors that monitor changes in the redox status of the electron transport system via the FAD cofactor associated with its PAS domains. In A. brasilense, AerC clusters at the cell poles. Its cellular localization and contribution to the behavioral response correlate with its expression pattern and with changes in the overall cellular FAD content under nitrogen-fixing conditions. AerC-mediated energy taxis in A. brasilense prevails under conditions of nitrogen fixation, illustrating a strategy by which cells optimize chemosensing to signaling cues that directly affect current metabolic activities and thus revealing a mechanism by which chemotaxis is coordinated with dynamic changes in cell physiology.
Keywords: FAD, nitrogen fixation, signal transduction
Motile bacteria detect changes in environmental conditions and respond by navigating toward niches where conditions are optimal for growth. This widespread behavior is collectively known as chemotaxis (1). Various physicochemical cues are detected by dedicated chemoreceptors (methyl-accepted chemotaxis proteins or MCPs) that relay information to the flagellar motors via a signal transduction cascade (2). The model organism for chemotaxis, Escherichia coli, possesses five transmembrane chemoreceptors (Tar, Tsr, Tap, Trg, and Aer) and the sensory specificities for each of these have been determined (2). Most bacterial species have a greater number of chemoreceptors than E. coli (3); however, their sensory specificities and contribution to the lifestyle of bacteria are virtually unknown.
Azospirillum brasilense are motile bacteria that can fix atmospheric nitrogen under microaerophilic conditions. These bacteria respond tactically to various chemoeffectors that affect their metabolism and the resulting changes in energy levels function as signals (4). In oxygen gradients, A. brasilense cells quickly navigate to a specific zone where oxygen concentration is low enough (3–5 μM) to support their microaerobic lifestyle and to be compatible with nitrogen fixation (5). Chemotactic responses to changes in energy metabolism have been identified in several bacterial species and are collectively referred to as “energy taxis” (6). In E. coli, two of the five chemoreceptors have been implicated in mediating energy taxis: Aer, which senses the redox status via an FAD cofactor (7, 8) and Tsr, which likely monitors the proton motive force (8, 9). In A. brasilense, a transmembrane chemoreceptor Tlp1 modulates many energy taxis responses (10). The sensory domain of Tlp1 is located in the periplasm and lacks a recognizable motif for redox or energy sensing, and thus the energy-related parameter sensed by Tlp1 is unknown (10).
Here, we describe a chemoreceptor, named AerC (transducer for aerotaxis and related responses, cytoplasmic), which functions as an energy taxis transducer in A. brasilense. Strikingly, the cellular localization of AerC and its contribution to behavior correlate with metabolic changes under nitrogen-fixing conditions suggesting a mechanism by which chemotaxis is coordinated with dynamic changes in cell physiology. Given the widespread distribution of AerC-like chemoreceptors revealed by comparative genomics, these findings are directly relevant to many other bacterial species.
Results
Identification of AerC, a Cytoplasmic Chemoreceptor with Elevated Expression Under Nitrogen-Fixing Conditions.
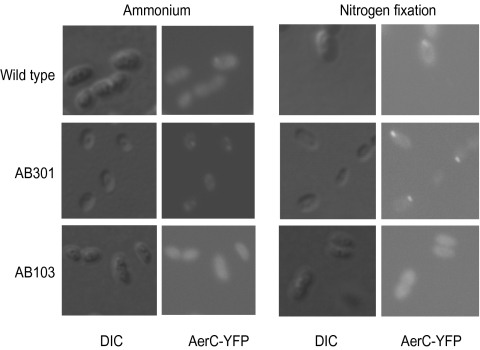
The available genome sequence of A. brasilense (http://genome.ornl.gov/microbial/abra/19sep08/) revealed 48 chemoreceptor genes. Five chemoreceptors were predicted to contain PAS domains that often serve as redox and oxygen sensors (11), including one encoded by a gene located between two nif operons, which was named aerC (Fig. 1). Although the chemoreceptor-encoding gene in this genomic region was identified previously (12), its function has not been elucidated. AerC consists of a C-terminal MCP signaling domain (3) and two N-terminal PAS domains, but lacks transmembrane regions (Fig. 1A). We compared the cellular levels of the AerC protein in the presence of ammonium and under nitrogen-fixing conditions. A polyclonal antibody targeted against the N-terminal PAS domain of the E. coli Aer (Aer2–166) protein cross-reacted with a protein of about 40 kDa (the predicted molecular weight of AerC) in the wild type, but not in a ΔaerC::Kan mutant strain (AB301), suggesting that it specifically recognizes AerC (Fig. 1B). Using this antibody, we found that AerC is present under both growth conditions but it is upregulated under conditions of nitrogen fixation (Fig. 1B). AerC was only found in the soluble fraction, an observation consistent with its predicted topology. A C-terminal translational fusion of the yellow fluorescent protein (Yfp) with AerC, expressed under the control of its native promoter from a broad host range low-copy plasmid, was used to determine the subcellular localization of this chemoreceptor (Fig. 2). When expressed in the ΔaerC::Kan mutant background (strain AB301), the plasmid could restore chemotaxis and aerotaxis to wild-type levels (Fig. S1). In plasmid-containing wild-type and AB301 cells grown in the presence of ammonium, fluorescence was seen within the cell cytoplasm and often in dim polar foci. Under conditions of nitrogen fixation, however, bright fluorescent foci were detected at either one or both cell poles in more than 90% of the cells examined (Fig. 2). No localization of the AerC–Yfp fusion in polar foci was detected when AerC–Yfp was expressed in strain AB103, which is deleted for the Che1 chemotaxis pathway (13) (Fig. 2). Bright fluorescence was observed throughout the cytoplasm indicating that the AerC–Yfp fusion was expressed. The expression levels of AerC–Yfp were similar in the wild-type and AB301 strains, but lower in the AB103 strain (Fig. S2), suggesting that AerC–Yfp is less stably expressed and/or at reduced levels in this strain.
Fig. 1.
AerC is a chemoreceptor homolog found in the nif/fix gene region of the genome of A. brasilense. (A) Organization of the genomic region around aerC (Upper) and protein domains found in AerC (Lower). The arrows indicate the direction of transcription. (B) Expression of AerC in A. brasilense strain Sp7 (wild type) and its ΔaerC mutant derivative (strain AB301) grown in presence of ammonium and under nitrogen-fixation conditions. An equivalent amount of protein extracted from whole cells was analyzed. AerC expression was detected by using an affinity-purified E. coli anti Aer2–166 antibody that cross-reacts with a protein of about 40 kDa in Sp7 but not AB301 (indicated by an arrow on the Right). The unidentified low molecular weight cross-reacting band present in all lanes was used as an internal control. The experiment was repeated three times and representative results are shown.
Fig. 2.
AerC–Yfp localization in A. brasilense. The wild-type and mutant derivative strains were grown in presence of ammonium (Left) and under nitrogen-fixation conditions (Right). The AB301 strain is a ΔaerC::Kan derivative of Sp7 and the AB103 strain is a ΔcheOp1::Cm derivative of Sp7 (13) (Table S2). In each panel, DIC images are shown on the Left and fluorescent images on the Right. Representative images are shown.
AerC Is a FAD-Binding Chemoreceptor.
In contrast to the best studied PAS-domain containing chemoreceptor, Aer of E. coli, and similar chemoreceptors characterized in other bacteria (14, 15), AerC is a soluble protein. The PAS domain of Aer contains FAD (16, 17); however, PAS domains are ubiquitous and contain various other cofactors (11). To determine which cofactor is associated with the PAS domains of AerC, we compared PAS domains from bacterial chemoreceptors identified in public databases. A total of 1,056 chemoreceptor sequences containing 1,649 PAS domains have been analyzed with respect to their domain architecture and PAS domain conservation (Fig. 3; Figs. S3 and S4). Three groups of PAS domains were identified: (i) a conserved class I exemplified by the PAS domain of the E. coli Aer chemoreceptor; (ii) a conserved class II exemplified by both PAS domains of the A. brasilense AerC chemoreceptor, and (iii) a diverse group of apparently unrelated PAS domains. All MCPs containing class I PAS domains were predicted to be membrane bound, whereas >98% of MCPs containing class II PAS domains were predicted to be cytoplasmic (Fig. S3).
Fig. 3.
Comparative sequence analysis of the PAS domains from microbial chemoreceptors. (A) A neighbor-joining tree generated from multiple sequence alignment of 1,649 PAS domains. Class I members are shown in brown (designated as PAS_FAD1) and exemplified by the PAS domain from the Aer protein of E. coli. Members of the bipartite subfamily of Aer sensors (34) are shown in light brown. Class II members are shown in blue (designated as PAS_FAD2) and exemplified by the two PAS domains from AerC protein of A. brasilense. Members of the general PAS group are shown in black. (B) Sequence alignment of PAS domains from NifL, Aer, and AerC proteins. Positions that are conserved (>95%) within PAS_FAD1 and PAS_FAD2 classes are highlighted in gray. Positions that are suggested to bind FAD in the NifL protein (16) are indicated by asterisks. A tryptophan residue that is involved in FAD binding in NifL and is conserved (>95%) in both classes of PAS domains is highlighted in yellow.
The pattern of sequence conservation indicates that class I and II PAS domains are closely related in structure and function (Fig. S4). To predict the cofactor bound to PAS domains in AerC, we analyzed the multiple sequence alignment of the PAS domain for the presence of conserved residues that are involved in cofactor binding in PAS domains containing known cofactors: FAD (18), FMN (19), heme (20), hydroxycinnamic acid (21), and citrate (22). Strikingly, only one such residue, a tryptophan corresponding to Trp87 of the FAD-containing PAS domain in the NifL protein of Azotobacter vinelandii (18), was conserved in both class I and class II PAS domains (Fig. 3). In NifL, Trp87 participates in multiple interactions with FAD, specifically with its adenine moiety and the ribityl chain (18). We conclude that this residue is the only irreplaceable position specifically involved in FAD binding and its presence in both PAS domains of AerC (Trp77 in PAS1 and Trp199 in PAS2) strongly suggests that they both contain FAD. This prediction was verified experimentally as shown below.
The PAS domain of E. coli Aer contains a noncovalently bound FAD that does not copurify well with the native protein, and an indirect assay was developed to confirm the association of FAD with the PAS domain (7, 16, 17, 23). Overexpression of a functional Aer protein (but not nonfunctional Aer alleles) from a plasmid is associated with an increase in the FAD content of cell membranes (7, 23). Overexpressing AerC in wild-type A. brasilense cells causes a significant increase in the FAD content of cells, which was not observed in wild-type cells carrying an empty vector (Table S1). Overexpressing AerC alleles that carried either W77F or W199F substitutions yielded a modest increase in the FAD content, whereas no significant increase was detected in cells expressing an AerCW77FW199F allele (Table S1). The cellular levels of the expressed mutant AerC proteins were comparable to the wild type and similar between all strains (Fig. S5). These results support the computational prediction that conserved tryptophan residues in the PAS1 and PAS2 domains of AerC bind FAD.
AerC Is an Energy Taxis Transducer.
We compared the motile behavior of strain AB301 with that of the wild-type strain in a set of behavioral assays performed in the presence of ammonium and under nitrogen-fixing conditions (Fig. S6). Strain AB301 was significantly impaired in chemotaxis to organic acids, glycerol, and certain sugars under all growth conditions (Fig. 4A and Fig. S6A). Interestingly, the ΔaerC mutant strain was null for chemotaxis to succinate when grown with combined nitrogen, but had only a slight defect under conditions of nitrogen fixation. Strain AB301 was also affected in chemotaxis under anaerobic conditions (Fig. S6B). The chemotactic defect could be complemented by expressing the wild-type aerC gene from its own promoter in trans from a plasmid (Fig. 4A and Fig. S1).
Fig. 4.
Role of tryptophan residues W77 and W199 of the PAS1 and PAS2 domains of AerC in chemoreceptor function. (A) Chemotaxis in the soft agar plate assay of the wild-type A. brasilense, the AB301 strain carrying an empty pRK415 vector (controls), or complemented with wild-type AerC and AerC alleles expressed from their own promoter on pRK415, after 24 h incubation at 28 °C. The soft agar plates contained malate as the carbon and energy source and ammonium chloride as the source of combined nitrogen. Longer incubation times did not change the results. (B) Aerotaxis in the capillary assay. An equivalent number of cells were inoculated in each capillary tube and the photograph was taken after a 5-min incubation period. The arrow indicates the direction of the air gradient from the air–liquid interface. (C) Effect of AerC on the propensity of cells to aggregate by cell-to-cell interaction and to form clumps. Pictures were taken directly from actively growing cultures in MMAB with 10 mM malate. Magnification, ×400. Representative pictures within a field of view are shown. The arrows point to clumps. (D) Subcellular localization of AerCW77F–Yfp, AerCW199F–Yfp, and AerCW77FW199F–Yfp in the AB301 strain under nitrogen-fixation conditions by fluorescence.
In oxygen gradients, A. brasilense cells rapidly (less than 5 min) form an aerotactic band at a preferred oxygen concentration (5). No difference in the position of the aerotactic band formed by the wild-type and the AB301 strains was detected when cells were grown in presence of ammonium (Fig. 4B and Fig. S6C); however, under conditions of nitrogen fixation, the AB301 strain formed the band farther away from the meniscus (Fig. 4B and Fig. S6C) and formation of the band was significantly delayed (∼15 min for the AB301 strain). Wild-type and AB301 cells were responding and adapting to the removal or addition of oxygen in a temporal gradient assay; however, AB301 cells had a much shorter adaptation time when grown under nitrogen-fixation conditions (Table 1).
Table 1.
Role of AerC in aerotaxis and redoxtaxis in A. brasilense
| Growth conditions/strains | Response time in seconds to adaptation* in a temporal assay for aerotaxis, ±SD | Threshold concentration for a repellent response in a miniplug assay, μM | |
| −Air† | +Air | ||
| Ammonium | |||
| Sp7 | 62.4 ± 5.1 | 58.1 ± 1.1 | 50 |
| AB301 | 76.7 ± 7.2 | 62.4 ± 2.0 | 50 |
| Nitrogen fixation | |||
| Sp7 | 71.6 ± 1.7 | 62.0 ± 2.3 | 50 |
| AB301 | 45.3 ± 3.3 | 37.2 ± 2.1 | 100 |
The response time of free-swimming cells until they return to a prestimulus swimming pattern was measured. The removal of air triggers a repellent response (increase in the reversal frequency of swimming cells) and the addition of air triggers an attractant response (decrease in the reversal frequency) under these conditions.
The “−” and “+” signs refer to the removal (repellent) or addition (attractant) of air, respectively, in the temporal assay.
The function of AerC as an energy taxis transducer was further confirmed by comparing the motile behavior of wild-type and AB301 cells in gradients of 1,4-benzoquinone, which elicits redoxtaxis (4). A difference in the sensitivity of the AB301 strain and the wild-type strain to the repellent effect of 1,4-benzoquinone was detected only when cells were grown under conditions of nitrogen fixation but not in the presence of ammonium (Table 1).
Observed behavioral defects in the AB301 strain could be complemented by the wild-type protein, but not by any of the mutated proteins that carry W77F or W199F substitutions, indicating that mutations abolish protein function (Fig. 4 A and B). Furthermore, these mutations caused the cells to clump, which could explain the null aerotactic phenotypes and the more severe chemotactic phenotypes observed when expressing these proteins (Fig. 4C). These phenotypes are also consistent with a Che1-dependent function of AerC because Che1 was implicated in controlling taxis as well as clumping (13). The subcellular localization of AerCW77F–Yfp, AerCW199F–Yfp, and AerCW77FW199F–Yfp was similar to that observed for the wild-type AerC protein, suggesting correct folding of these proteins (Fig. 4D).
Contribution of AerC to Locomotor Behavior Correlates with Changes in Metabolism.
Consistent with the expression pattern of AerC, behavioral assays showed that this chemoreceptor plays a major role under nitrogen-fixing conditions whereas its contribution in the presence of ammonium is minor. Given that FAD, which is likely a cofactor associated with the AerC PAS domains, is also a major metabolite intermediate, we hypothesized that changes in AerC expression and its contribution to locomotor behavior might correlate with changes in the energy metabolism of the cells. To test this hypothesis, we measured the total FAD content of cells grown in the presence of ammonium and under nitrogen-fixing conditions. The total FAD content of cells grown under nitrogen-fixing conditions (21.9 ± 1.16 fmol FAD/mg protein) was significantly higher than that of cells grown in the presence of ammonium (14 ± 2.8 fmol FAD /mg protein) (statistically different at the P < 0.05 level). Therefore, cells increase their total cellular FAD content under conditions of nitrogen fixation. This increase coincides with the elevated expression and functional contribution of AerC. Thus, the lack of AerC might affect function(s) that rely on the ability of cells to locate environments that support optimum energy metabolism. Given the delayed aerotactic band formation of the ΔaerC mutant cells in oxygen gradients, we hypothesized that the expression of nitrogen-fixation genes, which requires a specific oxygen concentration, should be affected. We compared the time course of expression of the promoter for the nitrogenase structural gene nifH as cells move along an oxygen gradient (Fig. 5A). The expression of nifH in the mutant strain was affected and appeared delayed relative to the wild-type strain (Fig. 5B). We confirmed that in wild-type cells the promoter for aerC was expressed as cells navigate the gradient (Fig. 5C). Changes in the total cellular FAD content as cells navigated the gradient paralleled the pattern of AerC expression (Fig. 5D). Therefore, cells navigating in the oxygen gradient coordinate changes in metabolism with changes in the concentration of the energy taxis transducer to locate the optimal position for metabolism.
Fig. 5.
Time course of aerotactic band movement in a spatial gradient of oxygen under conditions of nitrogen fixation. (A) An equivalent number of cells were inoculated at the bottom of tubes containing soft agar and a source of carbon and energy and incubated under conditions of nitrogen fixation. Photographs were taken at regular intervals (24, 48, and 72 h postinoculation). There was no band formed during the first 10 h of incubation, after which bands were seen moving up the gradients as shown. A representative photograph is shown. The arrows indicate the center of the aerotactic band in each photograph, which also corresponds to the position where samples were collected (directly from the bands) for analyses shown in panels B, C, and D. The star symbols represent statistically significant differences (P < 0.05). (B) Activity of a PnifH–gusA transcriptional fusion. (C) Activity of a PaerC–gusA transcriptional fusion. (D) Total cytosolic FAD content.
Discussion
The ability of motile cells to locate preferred niches for energy generation is essential for their survival. Given the ecological advantage provided by energy taxis (24), energy taxis transducers are expected to be widespread in bacteria; however, only a few have been identified thus far (7, 8, 10, 14, 15). Here we characterize an energy taxis chemoreceptor, AerC, in the nitrogen-fixing bacterium, A. brasilense. We show that AerC has elevated expression and localizes to bright polar foci at the cell poles under nitrogen-fixing conditions, but not in the presence of combined nitrogen. This chemoreceptor plays a role in many redox-mediated behaviors that comprise energy taxis, but it is most significant under nitrogen-fixing conditions. Similarly to the E. coli Aer chemoreceptor (7, 8, 16, 17), AerC contains FAD as a cofactor within its PAS domains; however, there is a striking difference between the two proteins. Whereas Aer has a single PAS domain and is membrane bound, AerC has dual PAS domains and is soluble. PAS domains of Aer and AerC exemplify two subfamilies that are dominant among PAS domain-containing chemoreceptors. Using protein sequence analysis and available structural information (18), we have identified conserved tryptophan residues in both PAS domains of AerC that are essential for FAD binding and sensory function. Two large conserved PAS domain subfamilies exemplified by Aer and AerC are now termed PAS_FAD1 and PAS_FAD2, respectively. This finding is important for the function prediction in thousands of PAS-domain containing sensors. The two classes of PAS domains differ by their predicted topology with PAS_FAD1 being separated from the signaling domain by at least one transmembrane region. In contrast, PAS_FAD2 never is separated from the signaling domain by transmembrane regions. Because both PAS domains contain FAD, but have different topologies, amino acid residues that are uniquely conserved in PAS_FAD1 and PAS_FAD2 classes (Fig. S4) likely represent differences in transmembrane versus cytoplasmic signaling.
In E. coli, the concentration of individual chemoreceptors within the chemoreceptor arrays is variable and affects signaling (25). Changes in the chemoreceptor content have been reported in Pseudomonas aeruginosa (26) and Rhodobacter sphaeroides (27); however, the effect of such changes on chemotaxis has not been established. Here we presented a case where changes in the chemoreceptor content dramatically affect motile behavior under very specific conditions: chemosensory input to AerC (changes in redox homeostasis) acts as a major cue to modulate behavior under nitrogen-fixing conditions.
Nitrogen fixation is an energy-demanding process, and at the same time nitrogenase must be protected from inhibition by oxygen (28). Nitrogen-fixing microorganisms developed several strategies to deal with this dilemma. Both Klebsiella pneumonia, an anaerobe, and A. vinelandii, an aerobe, monitor oxygen levels using the NifA transcriptional regulator that controls nitrogen fixation (29, 30). Many other prokaryotes also regulate the expression of nitrogen-fixation genes in response to oxygen concentration and energy availability. A. brasilense employs energy taxis to find a position in chemical gradients where low oxygen concentrations protect the nitrogenase while supporting energy generation (5, 31). AerC-mediated energy taxis prevails under conditions of nitrogen fixation, illustrating a strategy by which motile cells optimize chemosensing (increased expression of FAD-containing chemoreceptor) to signaling cues (redox conditions, monitored via FAD) that directly affect current metabolic activities (nitrogen fixation). Our findings further highlight the dynamic interplay between metabolism and sensing.
Materials and Methods
See SI Materials and Methods for details.
Media, Bacterial Strains, and Growth Conditions.
A. brasilense Sp7 (ATCC 29145), a wild-type strain for chemotaxis, was used throughout this study. Growth conditions are listed in SI Materials and Methods. Bacterial strains and plasmids are listed in Table S2.
Mutant Construction and Complementation.
The ΔaerC mutant was constructed by deletion of an internal 1,101-bp fragment in-frame and inserting a nonpolar kanamycin cassette (Tables S2 and S3). Correct allelic replacement was verified by PCR. One such mutant strain was used for further analysis (AB301). Complementation of the AB301 strain was performed by expressing wild-type aerC from its native promoter on a broad host range low-copy plasmid (pRK415). Single residue mutations in AerC_PAS1 and AerC_PAS2 were introduced at residue W77 (PAS1) and W199 (PAS2) by PCR using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Tryptophan codons (W77 in AerC_PAS1 and W199 in AerC_PAS2) were substituted with phenylalanine codons singly and in combination (Table S2). The mutations were confirmed by DNA sequencing. Plasmids were introduced into A. brasilense strains using triparental mating as described previously (13).
Generation of AerC–yfp Fusions and Fluorescence Microscopy.
C-terminal translational fusions of AerC with Yfp were generated essentially as described (32). For fluorescence microscopy, cultures were embedded in a 1% agarose pad on microscope slides and covered with a coverslip. Differential interference contrast (DIC) and fluorescence images were acquired using a Nikon 80i microscope. At least three independent experiments were performed, and at least five different fields of view from each experiment were analyzed.
Total Cellular FAD Content.
Total cellular FAD content was extracted from A. brasilense cells and quantitated by luminescence following a protocol developed by Watts et al. (23).
β-Glucuronidase Assay for Promoter Activity.
A 651-bp fragment at the 5′ region of aerC containing the putative promoter region and the start codon was cloned into the pFUS1 vector (33) to generate a transcriptional fusion with the promoterless gusA, yielding PaerC-gusA (pFUSPaerC; Table S2). The nifH promoter activity was assessed using a PnifH–gusA fusion from the pFAJ318 vector (Table S2). The β-glucuronidase activity was determined according to Reeve et al. (33).
Behavioral Assays.
Semisoft plates, capillary, miniplug, and temporal assays were performed essentially as previously described (4).
Computational Analysis.
Database searches and protein sequence analyses were carried out using standard bioinformatics techniques (see SI Materials and Methods for details).
Supplementary Material
Acknowledgments
The authors thank R. Goodchild and K. Watts for providing antibodies; K. Watts and M. S. Johnson for advice with the FAD measurement assays; and Q. Ma, B. L. Taylor, and W. Black for participating in the early stages of computational analysis. The authors also thank B. R. Crane for insightful discussion. This work was supported by National Science Foundation CAREER Award MCB-0622277 and MCB-0919819 (to G.A.), the National Institutes of Health Grant GM072285 (to I.B.Z.) and the DOE BioEnergy Science Center, which is supported by the Office of Biological and Environmental Research in the DOE Office of Science. This research used resources of the National Center for Computational Sciences at Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725.
Footnotes
The authors declare no conflict of interest.
This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910055107/DCSupplemental.
References
- 1.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandre G, Greer SE, Zhulin IB. Energy taxis is the dominant behavior in Azospirillum brasilense. J Bacteriol. 2000;182:6042–6048. doi: 10.1128/jb.182.21.6042-6048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhulin IB, Bespalov VA, Johnson MS, Taylor BL. Oxygen taxis and proton motive force in Azospirillum brasilense. J Bacteriol. 1996;178:5199–5204. doi: 10.1128/jb.178.17.5199-5204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor BL, Zhulin IB, Johnson MS. Aerotaxis and other energy-sensing behavior in bacteria. Annu Rev Microbiol. 1999;53:103–128. doi: 10.1146/annurev.micro.53.1.103. [DOI] [PubMed] [Google Scholar]
- 7.Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebbapragada A, et al. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JC, Johnson MS, Taylor BL. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol Microbiol. 2006;62:823–837. doi: 10.1111/j.1365-2958.2006.05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer-Phillips SE, Stephens BB, Alexandre G. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J Bacteriol. 2004;186:6595–6604. doi: 10.1128/JB.186.19.6595-6604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potrich DP, Bressel TA, Schrank IS, Passaglia LMP. Sequencing and promoter analysis of the nifENXorf3orf5fdxAnifQ operon from Azospirillum brasilense Sp7. Braz J Med Biol Res. 2001;34:1379–1395. doi: 10.1590/s0100-879x2001001100003. [DOI] [PubMed] [Google Scholar]
- 13.Bible AN, Stephens BB, Ortega DR, Xie Z, Alexandre G. Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J Bacteriol. 2008;190:6365–6375. doi: 10.1128/JB.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarand I, et al. Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ Microbiol. 2008;10:1320–1334. doi: 10.1111/j.1462-2920.2007.01546.x. [DOI] [PubMed] [Google Scholar]
- 15.Yao J, Allen C. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J Bacteriol. 2007;189:6415–6424. doi: 10.1128/JB.00398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikov SI, Barnes LA, Gitin Y, Parkinson JS. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5830–5835. doi: 10.1073/pnas.100118697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Repik A, et al. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 19.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Key J, Moffat K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry. 2005;44:4627–4635. doi: 10.1021/bi047942r. [DOI] [PubMed] [Google Scholar]
- 21.Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: A structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 23.Watts KJ, Johnson MS, Taylor BL. Minimal requirements for oxygen sensing by the aerotaxis receptor Aer. Mol Microbiol. 2006;59:1317–1326. doi: 10.1111/j.1365-2958.2005.05012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandre G, Greer-Phillips S, Zhulin IB. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev. 2004;28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 26.Güvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 27.Harrison DM, Skidmore J, Armitage JP, Maddock JR. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 28.Peters JW, Fisher K, Dean DR. Nitrogenase structure and function: A biochemical-genetic perspective. Annu Rev Microbiol. 1995;49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 29.Grabbe R, Klopprogge K, Schmitz RA. Fnr Is required for NifL-dependent oxygen control of nif gene expression in Klebsiella pneumoniae. J Bacteriol. 2001;183:1385–1393. doi: 10.1128/JB.183.4.1385-1393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. The NifL-NifA System: A multidomain transcriptional regulatory complex that integrates environmental signals. J Bacteriol. 2004;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchal K, Sun J, Keijers V, Haaker H, Vanderleyden J. A cytochrome cbb3 (cytochrome c) terminal oxidase in Azospirillum brasilense Sp7 supports microaerobic growth. J Bacteriol. 1998;180:5689–5696. doi: 10.1128/jb.180.21.5689-5696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallez R, Letesson JJ, Vandenhaute J, De Bolle X. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: Application to the Min system in Brucella abortus. Appl Environ Microbiol. 2007;73:1375–1379. doi: 10.1128/AEM.01873-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeve WG, Tiwari RP, Kale NB, Dilworth MJ, Glenn AR. ActP controls copper homeostasis in Rhizobium leguminosarum bv. viciae and Sinorhizobium meliloti preventing low pH-induced copper toxicity. Mol Microbiol. 2002;43:981–991. doi: 10.1046/j.1365-2958.2002.02791.x. [DOI] [PubMed] [Google Scholar]
- 34.Elliott KT, Zhulin IB, Stuckey JA, DiRita VJ. Conserved residues in the HAMP domain define a new family of proposed bipartite energy taxis receptors. J Bacteriol. 2009;191:375–387. doi: 10.1128/JB.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.