Abstract
Bacillus subtilis forms biofilms whose constituent cells are held together by an extracellular matrix. Previous studies have shown that the protein TasA and an exopolysaccharide are the main components of the matrix. Given the importance of TasA in biofilm formation, we characterized the physicochemical properties of this protein. We report that purified TasA forms fibers of variable length and 10–15 nm in width. Biochemical analyses, in combination with the use of specific dyes and microscopic analyses, indicate that TasA forms amyloid fibers. Consistent with this hypothesis, TasA fibers required harsh treatments (e.g., formic acid) to be depolymerized. When added to a culture of a tasA mutant, purified TasA restored wild-type biofilm morphology, indicating that the purified protein retained biological activity. We propose that TasA forms amyloid fibers that bind cells together in the biofilm.
Keywords: cell-to-cell interactions, extracellular matrix, TasA
Bacteria form complex, structured communities called biofilms in response to diverse environmental cues (1). The metabolic pathways activated by these cues are remarkably different depending on the species studied. However, they all lead to the formation of an extracellular matrix that holds the cells together (2). Although the specific composition of the extracellular matrix also varies from species to species, polysaccharides, proteins, and nucleic acids are important components of the matrix that help maintain the structural integrity of biofilms, mediating both cell-to-cell and cell-to-surface interactions (3 –8).
An important group of biofilm matrix-associated proteins are those that polymerize into fibers variously known as pili or fimbriae (9). In the specific case of Escherichia coli, the so-called curli pili have been shown to be an integral part of this bacterium’s biofilm matrix (10). Curli subunits assemble into fibers which display amyloid properties. From bacteria to humans, amyloid fibers share the ability to fold into a characteristic cross-β-strand structure, where the β-sheets are oriented perpendicular to the axis of the fibers (11, 12). This structure endows amyloid fibers with resistance to environmental attacks such as proteolysis as well as the ability to bind certain dyes, a property that has been commonly exploited to detect amyloids in vitro and in vivo (11, 13). For a long time, amyloid fibers have been associated with cellular malfunctioning in human degenerative diseases such as Alzheimer’s disease (14). However, “functional amyloids” that participate in normal biological processes in bacteria, fungi, insects, invertebrates and humans have begun to be appreciated (15). Although not directly shown to be part of the extracellular matrix of biofilms, amyloid fibers have been identified in numerous microorganisms. These include the harpins of Xanthomonas campestris and Pseudomonas syringae (16), fimbriae recently described from Mycobacterium tuberculosis (17), chaplins from Streptomyces coelicolor (18, 19) and hydrophobins from fungi (20, 21). Although recent studies suggest that amyloid adhesins are abundant in natural biofilms (22), curli fimbriae remain the sole example of an amyloid protein that has been shown to be an important functional component of a biofilm matrix.
The endospore forming Gram-positive bacterium Bacillus subtilis makes matrix-encased biofilms on the surface of agar plates as well as robust floating biofilms (pellicles) at the air–liquid interface of standing cultures (23). These biofilms, far from being mere aggregates of cells, exhibit an ordered spatiotemporal distribution of subpopulations of cells involved in tasks as diverse as sporulation, motility, and matrix formation (24). This organization is largely dependent upon the presence of the extracellular matrix, which is composed mainly of an exopolysaccharide and the protein TasA (25). TasA was first reported to be a protein associated with the spore and was proposed to have antimicrobial activity (26, 27). It was later shown that TasA is indeed the major protein component of the extracellular matrix of B. subtilis biofilms (25). Here, we show that TasA forms amyloid fibers and that the formation of these fibers is essential for the integrity of the extracellular matrix and thus, the biofilm. To accomplish this, we developed a protocol to purify TasA to homogeneity directly from B. subtilis. The purified TasA protein was functional in that it was able to restore wild-type biofilm forming ability to a mutant lacking tasA . Using biochemical, microscopic and dye-binding analyses, we show that fibers of purified TasA display amyloid properties and appear to mediate cell-to-cell interactions within the biofilms.
Results
Electron Microscopy Reveals TasA Fibers in B. subtilis Biofilms.
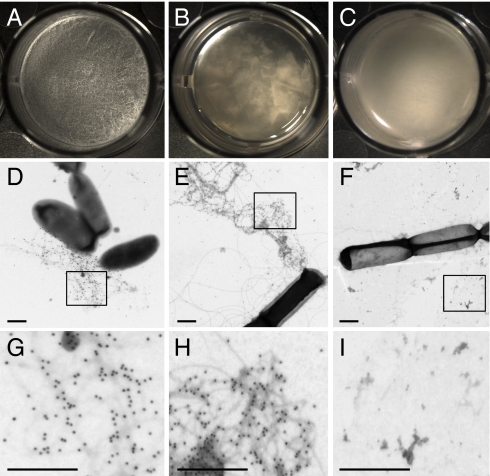
B. subtilis forms pellicles at the liquid–air interface of standing cultures. Fig. 1A shows a typical pellicle formed by the wild-type strain B. subtilis NCIB3610 after 48 h of incubation in MSgg medium. The two main components of the extracellular matrix that hold these pellicles together are an exopolysaccharide (EPS), synthesized by the products of the eps operon and TasA (25). Consequently, mutants defective in EPS (Fig. 1B) or TasA production (Fig. 1C) are both impaired in biofilm formation and form flat, fragile pellicles. We analyzed the localization of TasA in B. subtilis biofilms by using gold-labeled anti-TasA antibodies and visualizing the wild type and matrix mutants by transmission electron microscopy (TEM). In samples from both the wild type and the eps mutant, gold-labeled fibers 10–15 nm in width, and of variable length, emanating from the cells were easily observed (Fig. 1 D, E, G, and H for close-up). In marked contrast, the tasA mutant exhibited no such fibers or immunogold signal (Fig.1 F and I for close-up), strongly suggesting that the observed fibers were composed of TasA.
Fig. 1.
The presence of fibers in pellicles is related to the presence of TasA. Pellicle formation in MSgg at 30 °C as observed under a stereomicroscope at a magnification of 32× of (A) wild type, (B) eps mutant, and (C) tasA mutant. (D–F) Electron micrographs of negatively stained, immunogold labeled samples from pellicles after 24 h of incubation. (D) Wild type, (E) eps mutant, and (F) tasA mutant. (Scale bars, 500 nm.) The rectangles shown in D, E, and F represent the areas shown at higher magnification in G, H, and I, respectively.
Biofilms of B. subtilis Bind the Amyloid-Specific Dye Congo Red.
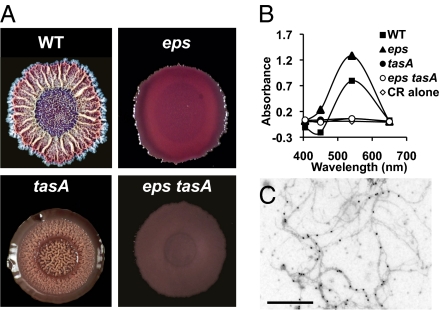
The method developed to detect the presence of amyloid curli fibers of E. coli biofilms (10) was adapted for the growth of B. subtilis biofilms to determine whether TasA, like curli, binds Congo Red and Coomassie Brilliant Blue dyes. Fig. 2A shows colonies formed by the wild-type strain as well as those formed by eps, tasA, and eps tasA mutants. The wild-type and eps colonies both stained red, consistent with the presence of an amyloid fiber. In contrast, the two strains unable to make TasA did not stain red, suggesting that TasA fibers bind Congo Red.
Fig. 2.
Dye binding properties of colonies and cell extracts. (A) Colonies of B. subtilis wild-type and matrix mutants grown for 72 h on Congo Red indicator plates. (B) Absorbance of a 10 μM solution of Congo Red (CR) with 1 M NaCl extracts of wild-type and matrix mutants. (C) Immunogold-labeled electron micrographs of TasA fibers from 1 M NaCl extracts of wild-type B. subtilis. (Scale bar, 500 nm.)
Previous studies report that Congo Red may exhibit nonspecific binding when the samples being analyzed are too complex (28). We reduced the complexity of our sample by extracting the cell-surface-associated protein fraction with 1M NaCl and used this fraction to determine if indeed Congo Red binding was the due to an extracellular factor. As shown in Fig. 2B, extracts from the wild type and the eps mutant showed a peak of increased absorbance of the Congo Red solution with a maximum at 541 nm. In contrast, extracts from tasA and eps tasA mutants did not show an increase in the absorbance of the Congo Red solution. Interestingly, visualization of the crude extract from the wild-type strain using TEM showed the presence of fibers that specifically reacted with gold-labeled anti-TasA antibody (Fig. 2C). Fibers were absent in extracts of single tasA and double eps tasA mutants. These results all suggest that TasA forms amyloid fibers.
Purified TasA Forms Amyloid Fibers.
In an effort to characterize TasA further, we wanted to purify TasA to homogeneity directly from B. subtilis. To simplify this process, we used a strain that overproduced TasA and that lacked EPS so that TasA would not be associated with EPS in the matrix. We accomplished this by using a strain defective in the SinR repressor, the regulator that represses the expression of both the eps and tasA operons (29). The strain was also deleted for the eps operon, which encodes the protein machinery required for the production of EPS. Using this sinR eps strain, we followed the kinetics of TasA production over time in shaken liquid cultures using immunoblotting (Fig. S1A). TasA was first detectable after 15 h of growth when the population entered stationary phase, and its abundance increased until levels peaked between 24 and 36 h. Under these conditions, TasA was mostly cell-associated at 20 h, with only a small amount of protein detected in the medium (Fig. S1B Left).
TasA was next extracted from the cells by treatment with a buffer containing 1M NaCl (Fig. S1B Right). To purify TasA further, this crude salt-extracted fraction was chromatographed through a gel-filtration column and fractions were analyzed by immunoblotting for the presence of TasA (Fig. S2A). Consistent with TasA forming a large polymer, most of the protein eluted in the void volume. This fraction was analyzed by SDS/PAGE and immunoblot with an anti-TasA antibody, showing that the preparation was homogeneous (Fig. S2A). The purified protein band was excised from the gel and subjected to electron spray ionization mass spectroscopy and N-terminal amino acid sequencing. The results obtained confirmed that we had purified mature TasA. When analyzed by immunogold and negative staining-TEM, this pure preparation of TasA appears mostly as fibers of variable length and 10–15 nm wide that are able to bind the anti-TasA antibody (Fig. S2B).
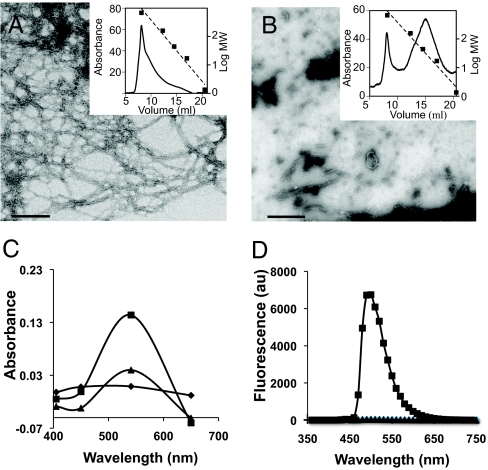
Amyloid proteins display a characteristic β-strand secondary structure irrespective of their primary sequence. It is this structure that gives amyloid proteins their dye-binding properties. Freshly purified TasA appeared as long fibers that eluted in the void volume of an analytical gel-filtration column with an exclusion size of 600 kilodaltons (Fig. 3A). Consistent with what has been described for other amyloid fibers (10), the addition of this purified TasA to a solution of Congo Red induced a noticeable increase in absorbance (Fig. 3C, squares). Another assay for amyloid fibers is their ability to increase the fluorescence of Thioflavin T with a peak emission at 482 mn. Adding purified TasA to a Thioflavin T solution indeed induced an increase in fluorescence (Fig. 3D, squares).
Fig. 3.
Amyloid properties of purified TasA. Negative staining electron micrographs of (A) freshly prepared TasA polymerized in long fibers of large MW and (B) depolymerization of fibrils into smaller oligomers with broad distribution of lower MW. Insets show chromatography traces of the samples run over a sizing column and standards of known molecular weights (dotted lines). (C) Differential spectra of the Congo Red absorbance after addition of polymerized (■) or unpolymerized TasA (▲). (D) Fluorescence of 20 μM Thioflavin solution alone or mixed with polymerized (■) or unpolymerized TasA (▲) after excitation at 450 nm. (Scale bars, 100 nm.)
Amyloid fibers are extremely stable structures. They are very difficult to disrupt using mild denaturation methods such as heat. They require harsher treatments, such as exposure to strong acids, to break them into their constituent monomers. Consistent with this, we observed that treatment with 10% formic acid depolymerized the TasA fibers. When analyzed by negative staining-TEM after formic acid treatment, a heterogeneous mixture of oligomers and monomers was observed instead of fibers (Fig. 3B). Accordingly, this soluble material lost the ability to bind both Congo Red and Thioflavin T (Fig. 3 C and D, triangles), which suggested the absence of amyloid fibers.
In addition to the dye binding assays, we performed CD spectroscopy on purified TasA fibers as well as dissociated, formic acid-treated TasA to characterize their secondary structure directly (Fig S3). TasA fibers (Fig. S3 circles) showed a decrease in ellipticity in the range of 220–210 nm and an increase at around 190 nm, suggesting the presence of β-sheets (30, 31). It does appear that the fibers possess some α-helix and random coil characteristics in addition to the β-sheets that confer the dye-binding properties of TasA. It is not unprecedented for amyloid fibrils to contain secondary structures other than β-sheets (31). A 1 min treatment with formic acid was enough to induce a dramatic difference in the spectra (Fig. S3, squares), suggesting a rearrangement during the dissociation of fibers into their constituent components.
Polymerization of TasA In Vitro.
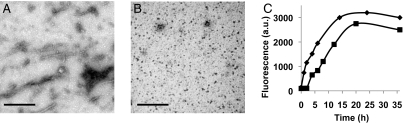
Different amyloids have been shown to repolymerize in vitro from soluble monomers into fibers (32). This property has allowed the identification of diverse aggregation states that display distinct biochemical properties and kinetics of polymerization (13). To investigate the repolymerization characteristics of TasA, purified TasA was treated for either 1 min or 5 min with 10% formic acid. This led to a partial and complete dissociation of the fibers, respectively, as judged by TEM (Figs. 4 A and B). After rapid evaporation of the formic acid and further resuspension in buffer (SI Material and Methods) the samples were incubated at room temperature to allow repolymerization. The kinetics of repolymerization were monitored using the Thioflavin T fluorescence assay (Fig. 4C). The repolymerization curve after the 5 min treatment fit nicely with the typical nucleation-dependent polymerization model, consisting of three clear phases: a lag phase of about 2 h, a fiber growth phase of about 20 h, and then a stationary phase (Fig. 4C squares) (13, 33). In contrast, the sample treated for 1 min repolymerized without a lag phase (Fig. 4C, diamonds, and Fig. S4B). This difference probably results from the fact that the sample treated for 1 min contains a population of short fibers that can serve to nucleate repolymerization (Fig. 4A). To test if nuclei were still present after a 5 min formic acid treatment that were undetectable by TEM, we increased the time of formic acid treatment to 10 min (Fig. S4 A and B). Under these conditions, the lag phase was longer than observed after 5 min treatment, suggesting that even after 5 min, there were still some TasA nuclei remaining.
Fig. 4.
Repolymerization of TasA in vitro. Electron micrographs of TasA immediately after 1-min (A) or 5-min (B) treatment with formic acid. Note that small fibers are seen in A but not in B. (Scale bars, 200 nm.) (C) Kinetics of TasA repolymerization in vitro. Purified TasA treated for 1 min (◆) or 5 min (▪) with 10% formic acid. After evaporation of formic acid, the repolymerization was followed by the Thioflavin T assay.
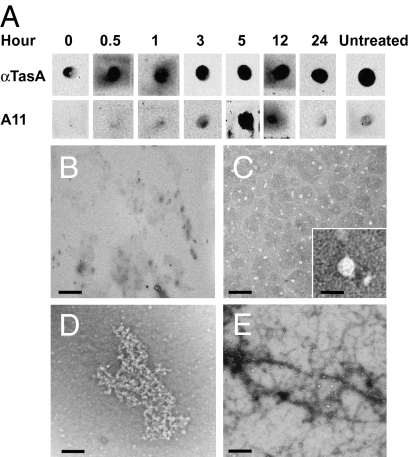
The polymerization of other amyloid proteins has been shown to involve the formation of transient aggregates with different morphologies (34). The commercially available antibody A11 detects these aggregates and is used in the diagnosis of human neurodegenerative disorders. An A11 antibody recognizes intermediates of diverse amyloid aggregates with similar morphologies (33, 35). We used the A11 antibody to determine whether TasA formed intermediate structures related to these diagnostic amyloid aggregates over a period of 24 h after complete dissociation of the TasA fibers with formic acid (Fig. 5A). At all stages, purified TasA protein positively reacted with anti-TasA antibody. At early time points, when TasA fibers were completely denatured and no aggregates were detected by TEM (Fig. 5B) A11 did not recognize TasA. However, at time points when particles of regular shape (Fig. 5C; see Inset for higher magnification of individual particle) and larger aggregates (Fig. 5D) were observed, the antibody A11 reacted positively with purified TasA. Finally, almost no A11 reaction was observed after 24 h of incubation, when the fibers had completely reformed and aggregates were no longer visible (Fig. 5E). Thus, the A11 antibody, used diagnostically to identify transient aggregates from different amyloid proteins, specifically recognizes TasA particles and aggregates observed transiently during the repolymerization process.
Fig. 5.
TasA particles and aggregates react with A11 antibody. (A) Detection of transient intermediates during polymerization of TasA. Samples were blotted onto nitrocellulose membranes and probed with anti-TasA (1:20,000) or A11 (1:10,000) antibodies. Samples were removed during the time course for TEM analysis. (B) No structures were observed immediately after treatment with formic acid. (C) After 3 h, when some A11 reactivity was observed in the dot-blot, small homogenous particles (5–12 nm wide) were observed. Detail of one particle (12 nm) is shown in the Inset at higher magnification. (D) After 5 h, larger amorphous aggregates were observed that coincided with the strongest reactivity with A11. (E) At 24 h, when A11 reactivity had dropped, fibers morphologically similar to those in untreated samples were observed. (Scale bars, 100 nm; Inset, 20 nm.)
TasA Amyloid-Fibers Restore the Formation of Pellicles.
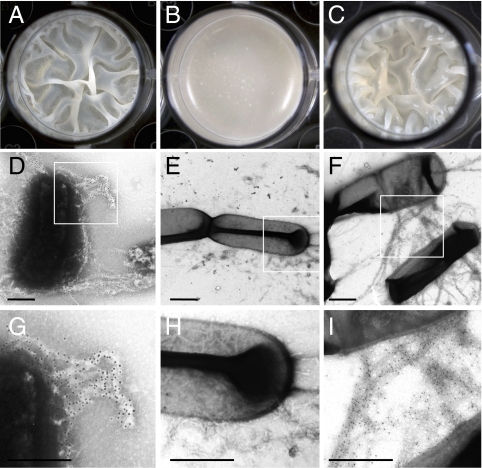
We wanted to determine whether purified TasA fibers had biological activity. To test this we added purified TasA to a tasA-defective mutant during the process of pellicle formation. As stated before, wild-type pellicles are extremely wrinkly whereas those formed by the tasA mutant are flat and fragile (Fig. 1). Strikingly, the addition of purified TasA to a tasA mutant resulted in complete restoration of the wrinkly pellicle phenotype. Extracellular complementation with purified TasA yielded pellicles that were undistinguishable from those formed by the wild-type strain (Fig. 6 A–C). When analyzed by TEM, the added TasA appeared as ordered fibers connecting tasA mutant cells, very similar to what was observed in the wild-type strain (Fig. 6 D–I). These results indicate that TasA amyloid fibers play a key structural role in giving mature pellicles their complex architecture.
Fig. 6.
Addition of purified TasA restores pellicle formation to a tasA mutant. Cells were grown in MOLP broth and incubated for 48 h without agitation at 30 °C: (A) wild-type, (B) tasA mutant, and (C) tasA mutant after addition of purified (40 μg) polymerized TasA. Immunogold-labeled electron micrographs of TasA fibers between cells of wild-type (D and G) and tasA mutant after addition of purified TasA (F and I) and absence of fibers in a tasA mutant alone (E and H). (Scale bars, 500 nm.) The rectangles shown in D, E, and F, represent the areas shown at higher magnification in G, H, and I, respectively.
Discussion
In this study, we have found that the major protein component of the B. subtilis biofilm matrix, TasA, forms amyloid fibers. Amyloid fibers have been extensively studied in human neurodegenerative diseases including Alzheimer’s and Parkinson’s or spongiform encephalopathies induced by prions (15, 36, 37). Besides having a role in pathogenicity, amyloids may also be an important factor in the physiology of the diverse organisms. For example, amyloids contribute to the attachment of bacteria to surfaces and the raising of aerial structures (18, 19), and they have a role in pathogenic processes such as adhesion to host cells (7, 10, 38) and induction of toxicity to host cells (16). However, in biofilm formation, only curli amyloid fibers of E. coli have been studied in detail (10). TasA is an amyloid protein shown to be specifically involved in the formation of a biofilm matrix by a Gram-positive organism. Now that two model systems that use amyloids to help structure their biofilms have been identified, it will be possible begin to identify commonalities. Although it is difficult to identify amyloid proteins from their primary structure, it is possible that this phenomenon may be more common in bacterial biofilms than is presently recognized. In fact, amyloid fibers have been proposed to be present in natural bacterial biofilms under different environmental conditions (22). We propose that amyloid formation may be a common mechanism to attain architectural complexity in biofilms.
Given the existence of amyloid fibers in biofilms of both Gram-positive and Gram-negative bacteria, it is possible that these structures might play a widespread and important function in supporting the structure of the biofilm. For instance, these fibers may promote cell-to-cell interaction as well as cell-to-surface adhesion in conjunction with other components of the extracellular matrix, such as exopolysaccharides (19). In support of this hypothesis, previous studies have shown that TasA and EPS are both required for pellicle formation, and only a mutant lacking both of these components is completely defective in biofilm formation (25). It is possible that the TasA amyloid fibers interact in some specific way with the matrix EPS whereby the fibers serve as a rigid scaffold upon which is overlaid the more flexible and amorphous EPS. Such kind of interactions have been proposed to occur between fibers of chaplins and cellulose in S. coelicolor, giving rise to the fimbriae that mediate the attachment to surfaces and probably the covalent interaction of these fibers to the cell wall (19). Interestingly, tasA and eps mutants can be mixed in culture and they will complement each other extracellularly and form phenotypically wild-type biofilms (25). These data, coupled with our finding that pellicle formation in a tasA mutant can be restored by adding purified TasA, indicate that TasA fibers indeed are an important matrix component which provide the structural “backbone” of the biofilm.
In B. subtilis, the TasA protein was originally described as having antibiotic activity (27). Quite interestingly, the small aggregates that are precursors to amyloid fibers have been reported to be toxic (35, 39). There is a generic structural similarity not only between amyloid fibrils formed from diverse proteins, but also their intermediate structures. This is supported by the ability of the A11 antibody to recognize fiber-forming intermediate aggregates of proteins as diverse as Aβ, synuclein, islet amyloid polypeptide, insulin, prions, lysozyme, and curli (33, 35, 37). We have shown that TasA also forms these small aggregate intermediates that are recognized by the A11 antibody (Fig. 5). It is tempting to speculate that these small aggregates may be used as a defense mechanism by B. subtilis. In this way, the TasA filaments would serve a dual purpose. First, they would provide structure to the extracellular matrix of the biofilm and hold the cells together. Second, once it is outside the cell at high concentrations in amyloid form, small aggregates may be released to help defend the cells within the biofilm from potential competitors. Clearly this speculative idea requires extensive further testing.
The widespread existence of amyloidogenesis suggests it is an evolutionarily conserved process in which functional fibers are formed from transient intermediates that may exhibit toxicity. Studies in other systems have reinforced the perception of amyloid fibers as functional products that result from detoxifying their intermediate aggregates (33, 35, 39, 40). TasA forms amyloid fibers that we have shown are essential in the formation of robust biofilms. Given the ease of genetic manipulation of this system, it should quickly become an excellent model for the study and design of drugs to target not only the formation of amyloids but also biofilms.
Material and Methods
Bacterial Strains and Culture Conditions.
The strains and medium used in this study are described in SI Material and Methods.
Protein Expression, Purification, and Molecular Weight Estimation.
Detail procedures are described in SI Material and Methods.
Congo Red and Thioflavin T Assays.
Protein solutions were analyzed for they ability to bind amyloid stains Congo Red and Thioflavin T solutions as previously described (10, 41, 42) (a brief summary is found in SI Material and Methods).
In Vitro Polymerization Assay.
Freshly purified TasA was depolymerized by short (1 min) or long (5 min) treatment with 10% formic acid solution. After the treatment, formic acid was eliminated by evaporation in a speed-vac at 30 °C. The remaining product containing TasA was resuspended in 50mM NaCl, 20 mM Tris, (pH 7) buffer and incubated at room temperature. Samples were taken at different time intervals and mixed with 20 μM Thioflavin T for fluorescence analyses, as described above.
Transmission electron microscopy, immunolabeling, immunoblot, and dot-blot analyses were done according to standard procedures (see SI Material and Methods for detailed description).
Supplementary Material
Acknowledgments
We thank M. Ericsson, L. Trakimas, and E. Benecchi for help and guidance in the electron microscope; E. Sattely for help in the use of FPLC. K; Arnett for help in the CD spectroscopy analysis; A. Earl and E. Shank for critical reading of this manuscript; and D. López and H. Vlamakis and other members of Losick and Kolter laboratories for helpful discussion and suggestions. This work was funded by National Institutes of Health grants to R.K. (GM58213) and R.L. (GM18546). D.R. is the recipient of a MEC/Fulbright post-doctoral fellowship from Secretaría General de Estado de Universidades e Investigación del Ministerio de Educación y Ciencia (Spain).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910560107/DCSupplemental.
References
- 1.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Itoh Y, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemm P, Schembri MA. Bacterial adhesins: Function and structure. Int J Med Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 5.Lasa I, Penadés JR. Bap: A family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Lemon KP, Earl AM, Vlamakis HC, Aguilar C, Kolter R. Biofilm development with an emphasis on Bacillus subtilis . Curr Top Microbiol Immunol. 2008;322:1–16. doi: 10.1007/978-3-540-75418-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157:H7 on stainless steel: Effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005;71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 9.Epstein EA, Chapman MR. Polymerizing the fibre between bacteria and host cells: the biogenesis of functional amyloid fibres. Cell Microbiol. 2008;10:1413–1420. doi: 10.1111/j.1462-5822.2008.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 12.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 13.Naiki H, Gejyo F. Kinetic analysis of amyloid fibril formation. Methods Enzymol. 1999;309:305–318. doi: 10.1016/s0076-6879(99)09022-9. [DOI] [PubMed] [Google Scholar]
- 14.Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wösten HA. Amyloids—a functional coat for microorganisms. Nat Rev Microbiol. 2005;3:333–341. doi: 10.1038/nrmicro1127. [DOI] [PubMed] [Google Scholar]
- 15.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Oh J, et al. Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem. 2007;282:13601–13609. doi: 10.1074/jbc.M602576200. [DOI] [PubMed] [Google Scholar]
- 17.Alteri CJ, et al. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci USA. 2007;104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claessen D, et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 20.Wösten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 21.Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Larsen P, et al. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 23.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 26.Serrano M, et al. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J Bacteriol. 1999;181:3632–3643. doi: 10.1128/jb.181.12.3632-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stöver AG, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khurana R, Uversky VN, Nielsen L, Fink AL. Is Congo red an amyloid-specific dye? J Biol Chem. 2001;276:22715–22721. doi: 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- 29.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis . Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 30.Tjernberg L, Hosia W, Bark N, Thyberg J, Johansson J. Charge attraction and beta propensity are necessary for amyloid fibril formation from tetrapeptides. J Biol Chem. 2002;277:43243–43246. doi: 10.1074/jbc.M205570200. [DOI] [PubMed] [Google Scholar]
- 31.Tompa P. Structural disorder in amyloid fibrils: its implication in dynamic interactions of proteins. FEBS J. 2009;276:5406–5415. doi: 10.1111/j.1742-4658.2009.07250.x. [DOI] [PubMed] [Google Scholar]
- 32.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 35.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 36.Nordstedt C, et al. The Alzheimer A beta peptide develops protease resistance in association with its polymerization into fibrils. J Biol Chem. 1994;269:30773–30776. [PubMed] [Google Scholar]
- 37.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collinson SK, et al. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valincius G, et al. Soluble amyloid beta-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: implications for cell toxicity. Biophys J. 2008;95:4845–4861. doi: 10.1529/biophysj.108.130997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douglas PM, et al. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klunk WE, Jacob RF, Mason RP. Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 1999;309:285–305. doi: 10.1016/s0076-6879(99)09021-7. [DOI] [PubMed] [Google Scholar]
- 42.LeVine H., 3rd Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.