Abstract
There are many striking examples of phenotypic convergence in nature, in some cases associated with changes in the same genes. But even mutations in the same gene may have different biochemical properties and thus different evolutionary consequences. Here we dissect the molecular mechanism of convergent evolution in three lizard species with blanched coloration on the gypsum dunes of White Sands, New Mexico. These White Sands forms have rapidly evolved cryptic coloration in the last few thousand years, presumably to avoid predation. We use cell-based assays to demonstrate that independent mutations in the same gene underlie the convergent blanched phenotypes in two of the three species. Although the same gene contributes to light phenotypes in these White Sands populations, the specific molecular mechanisms leading to reduced melanin production are different. In one case, mutations affect receptor signaling and in the other, the ability of the receptor to integrate into the melanocyte membrane. These functional differences have important ramifications at the organismal level. Derived alleles in the two species show opposite dominance patterns, which in turn affect their visibility to selection and the spatial distribution of alleles across habitats. Our results demonstrate that even when the same gene is responsible for phenotypic convergence, differences in molecular mechanism can have dramatic consequences on trait expression and ultimately the adaptive trajectory.
Keywords: adaptation, genetics, lizard, Mc1r, speciation
Convergent evolution of similar phenotypes in similar environments has long been taken as evidence of adaptation driven by natural selection (1, 2). An outstanding question, however, is whether such convergence results from similar or different mechanisms. There are multiple levels at which mechanistic causes of convergence can occur. Are changes in the same developmental or physiological pathways implicated in phenotypic convergence? If so, are changes at the same specific gene(s) responsible for convergent phenotypes? And, finally, if the same genes are employed, are the underlying molecular and functional mechanisms linking genotype to phenotype conserved? Whereas several studies have shown that the same pathway or the same gene is responsible for convergence (3), few studies have tested the functional effects of evolutionarily independent mutations within the same gene to determine whether they are equivalent. Different mutations in the same gene can act through unique functional mechanisms even if they result in similar phenotypic effects—alleles may differ in their dominance and net selection coefficients (e.g., due to differences in pleiotropy and/or epistatic interactions), which can affect the probability and rate of fixation of alleles in the wild. Thus, understanding the functional basis of adaptation has far-reaching implications, as it sheds light on the predictability of mechanisms generating adaptive change.
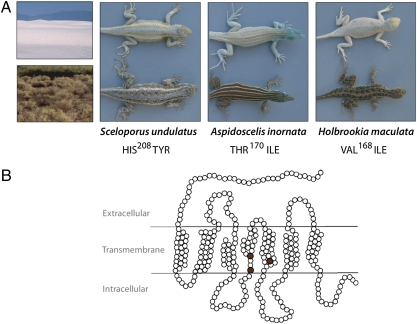
Here we test whether independent mutations in the melanocortin-1 receptor (Mc1r) gene have similar functional consequences in three lizard species with convergent adaptations to a common novel environment. The White Sands formation in the Chihuahuan Desert is a stark landscape of white gypsum dunes. Formed less than 6000 years ago (4), these sand dunes represent a dramatic new selective environment for small diurnal animals subject to visual predation. Three species of lizards [Eastern Fence Lizard (Sceloporus undulatus), Little Striped Whiptail (Aspidoscelis inornata), and Lesser Earless Lizard (Holbrookia maculata)] have each evolved blanched, substrate-matched phenotypes at White Sands (Fig. 1A). The convergent phenotypes have evolved rapidly under strong divergent selection from a brown, ancestral phenotype found on the dark adobe soils in the surrounding desert (5).
Fig. 1.
Mutations associated with blanched coloration in White Sands lizards. (A) Blanched morphs from white sands on top and dark morphs in ancestral dark soil habitat on bottom. (B) Amino acid schematic of the melanocortin-1 receptor (Mc1r); replacements statistically associated with coloration in the focal taxa are shown in red.
Previously, we identified an association between adaptive pigmentation and mutations for all three taxa in Mc1r, a gene that plays a critical role in the production of vertebrate melanin pigments (6). In each species, a single derived amino acid replacement (different in each species) was statistically associated with the blanched coloration of White Sands lizards (Fig. 1B). Here we use functional assays to connect the mechanism of disruption of the Mc1-receptor to patterns of allelic dominance and the distribution of alleles in natural habitats.
Results and Discussion
Using a large sample of lizards from white sand and dark soil habitats, we confirm that the statistical association between a single Mc1r mutation in each species and color was highly significant for each species (S. undulatus: n = 114, P < 10−9; A. inornata: n = 100, P < 10−15; H. maculata: n = 88, P < 10−24; Table S1). The genotype-phenotype association was not always perfect (some ancestral alleles were found in white sand populations of both S. undulatus and A. inornata), suggesting that additional genes also contribute to color variation. All three mutations occur in transmembrane (TM) regions, which are important for maintaining the structural integrity of the receptor and play a known role in both ligand binding and signaling (7). In S. undulatus the replacement from histidine (wild-type) to tyrosine (derived) at amino acid residue 208 (His208Tyr) is a change from a positively charged to an aromatic, uncharged amino acid. Histidine is conserved at this residue across vertebrates, suggesting it is important for Mc1r function. In A. inornata the Thr170Ile replacement is a polarity-changing replacement at a position that is also implicated in mutationally induced changes of Mc1-receptor function in humans (8). Unlike the other two mutations, in H. maculata the Val168Ile replacement is perfectly associated with color variation, but is a conservative change between physiochemically similar amino acids, both of which are common in other species (e.g., human Mc1r, Ile168) and Mc1r paralogs (e.g., human Mc4r, Ile173).
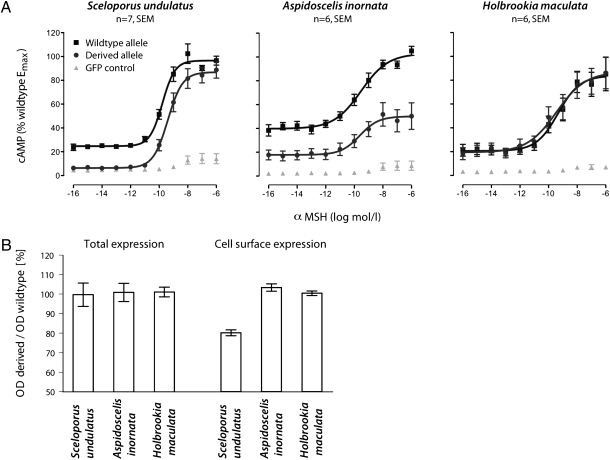
To investigate the functional consequences of these Mc1r mutations, we expressed wild-type and derived Mc1r alleles from all three lizard species and measured receptor signaling. Specifically, we performed heterologous cell-based assays of signal transduction efficiency for all six Mc1r alleles in mammalian COS-7 cells by measuring agonist-induced intracellular cyclic adenosine monophosphate (cAMP) levels (Fig. 2A; Table S2). All alleles responded to the natural agonist α-MSH [which is highly conserved across vertebrates (9)] with an increase in intracellular cAMP levels. However, for S. undulatus and A. inornata, the derived alleles exhibited reduced basal and agonist-induced cAMP formation relative to their respective wild-type alleles. The 208Tyr allele from blanched S. undulatus showed a 24% reduction in agonist-induced cAMP formation and a 68% decrease in basal cAMP formation (paired t test, P < 0.05) compared to the wild-type allele. Similarly, the 170Ile allele from blanched A. inornata showed a 62% reduction in agonist-induced cAMP formation (paired t test, P < 0.05) and a 62% decrease in basal cAMP formation (paired t test, P < 0.05). Reductions in receptor signaling of similar magnitude have pronounced effects on pigmentation and are associated with light-colored hair in mice and humans (10, 11). Thus, this assay demonstrates repeated use of the same gene in adaptive evolution in phylogenetically distant species. Both S. undulatus and A. inornata [which share a most recent common ancestor ≈175 million years ago (12)] evolved blanched phenotypes via partial loss-of-function Mc1r mutations.
Fig. 2.
Functional assays for Mc1r in White Sands lizards. (A) Cell-based assays identifying partial loss-of-function Mc1r alleles for blanched S. undulatus and A. inornata. Derived and wild-type alleles in a mammalian expression vector were tested for basal and agonist-induced cAMP accumulation in response to increasing concentrations of α-MSH. Green fluorescent protein (GFP) plasmid-transfected cells served as controls. (B) ELISAs identifying reduced cell-surface expression for the derived S. undulatus Mc1r allele. Specific optical density (OD) readings (OD value of HA-tagged construct minus OD value of control-transfected cells) are given for each species as a percentage of the wild-type variant for total and cell-surface expression.
Although H. maculata showed the strongest statistical association between Mc1r mutation (Val168Ile) and color, our functional assays showed no measurable differences in agonist-induced cAMP levels. Thus, the Val168Ile mutation may be (i) simply a spurious statistical association, (ii) in linkage disequilibrium with a functionally relevant noncoding mutation, or (iii) affecting Mc1r function in a way that we did not measure (e.g., mRNA stability). In any case, it is clear that H. maculata has evolved coloration via a different mechanism than the other two species and underscores the importance of functional tests in evolutionary studies (13).
The reduction in basal and agonist-induced cAMP levels in derived S. undulatus and A. inornata Mc1r alleles suggested two possible functional mechanisms: lower cell-surface expression levels and/or a reduced coupling efficiency of Mc1r. To discriminate between these two possibilities, we quantified cell-surface expression levels of wild-type and derived alleles. Specifically, we measured Mc1r protein-expression levels in all cellular compartments and at the plasma membrane using a total cellular ELISA and a cell-surface ELISA, respectively. Total receptor protein expression did not differ between the wild-type and derived alleles in transiently transfected cells in either species; however, cell-surface expression level of the S. undulatus 208Tyr allele was reduced by 20% (paired t tests, P < 0.0001; Fig. 2B; Table S2). This reduction in cell-surface expression suggests a partial intracellular retention of this protein similar to that observed for human Mc1r alleles associated with red hair (14). In other words, the reduced activity of the derived S. undulatus receptor is caused by a reduction in its ability to integrate into the melanocyte membrane efficiently (i.e., trafficking deficiency). In contrast, the dysfunction of the A. inornata derived allele cannot be attributed to a difference in the receptor’s cell-surface expression. Instead, the reduced activity of the 170Ile allele is likely due to a change in its ability to transduce signals (e.g., impaired G-protein-coupling efficacy), as is the mechanism for well-characterized Mc1r mutations in humans (e.g., ref. 11). Therefore, the independent Mc1r replacements produce similar phenotypic results via entirely different functional mechanisms.
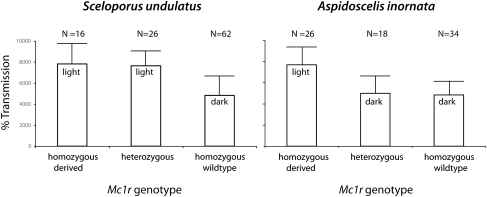
These differences in the molecular underpinnings of Mc1r disruption lead to important organismal-level consequences. We evaluated the effect of Mc1r genotype on dorsal coloration for samples of both species (Table S3) and found significant differences in color among genotypic classes [S. undulatus: F(2,49) = 42.15, P < 10−5; A. inornata: F(2,36) = 15.00, P < 10−5]. In both species, individuals homozygous for the derived Mc1r allele were significantly lighter in color than individuals homozygous for the wild-type allele. However, for S. undulatus, heterozygotes were statistically indistinguishable in color from individuals homozygous for the derived allele, whereas for A. inornata, heterozygotes were statistically indistinguishable in color from individuals homozygous for the wild-type allele (Fig. 3). Thus, we can determine dominance patterns for Mc1r alleles in the focal taxa, even though controlled breeding studies cannot be conducted for these species. The derived Mc1r allele appears to be dominant in S. undulatus but recessive in A. inornata. Therefore, mutations in the same gene associated with similar phenotypes differ in dominance.
Fig. 3.
Dominance relationships of Mc1r alleles. Dorsal coloration (mean and standard deviation for area under the spectral curve) for Mc1r genotypes showing the derived allele is dominant in S. undulatus and recessive in A. inornata. n, number of alleles sampled; “light” and “dark” refer to statistically distinguishable groups.
This difference in genetic dominance is consistent with what is known about Mc1r function. Mutations causing a reduction in membrane integration efficiency (as demonstrated in S. undulatus) are expected to be dominant. Because G-protein-coupled receptors (GPCRs) dimerize (15), if wild-type and mutant receptors are coexpressed (in a heterozygote), the mutant receptor can retain the wild-type receptor in the cellular interior and prevent its integration in the membrane. This mechanism is one explanation for the many examples of dominant reduced-function mutations in Mc1r paralogs (16) and other GPCRs (17). In contrast, mutations causing a disruption of receptor signaling (as observed for A. inornata) are expected to be recessive, and recessive Mc1r signaling mutations are found to cause lighter coloration in other taxa (18, 19).
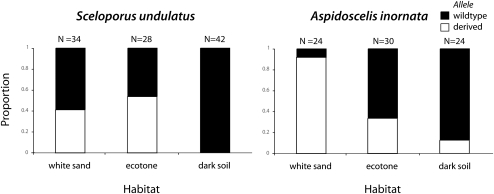
The observed differences in dominance also affect the spatial patterns of allelic variation in the wild. We measured Mc1r allele frequencies in three habitats: white sand (center of the gypsum dunes), dark soil (surrounding desert grasslands), and the ecotone (edge of the dune field with more variable, intermediate substrate). The distribution of Mc1r alleles across habitats differed between the taxa (Fig. 4). For example, nearly all light-colored A. inornata in white sand habitat were homozygous for the derived (recessive) Mc1r allele, whereas many light-colored S. undulatus in white sand habitat (in which the derived Mc1r allele is dominant) were heterozygous. The difference in the distribution of Mc1r alleles across habitat types can be largely explained by allelic differences in dominance; that is, selection will efficiently remove the visible dominant allele in the mismatched habitat. Moreover, because the distribution of Mc1r allelic variation is consistent with expectations based on the distribution of phenotypes, this provides strong evidence that color phenotype is, in fact, the direct target of selection (rather than a pleiotropic effect of another selection target).
Fig. 4.
Spatial distribution of Mc1r alleles in the wild for S. undulatus (derived Mc1r allele dominant) and A. inornata (derived Mc1r allele recessive). Proportion of wild-type (black) and derived (white) alleles across dark soil, ecotone, and white sand habitat. n, number of alleles sampled.
Here we report a natural system of multiple species under an identical selection regime using the same gene but different molecular mechanisms to generate convergent phenotypes. We suspect that convergence through different mutations with different functional effects may be common for partial loss-of-function (as opposed to gain-of-function) mutations because there are likely to be many ways to “break” a pathway or disrupt gene function. Importantly, we show that differences in functional mechanisms of mutations can affect color phenotype, allelic dominance, and the geographic distribution of alleles in nature. These differences in the genetic architecture of locally adapted populations can influence rates of adaptation and gene flow, which are also critical parameters in predicting the likelihood and rate of parapatric speciation (20). Further, in addition to changes in concealing coloration, white sand and dark soil populations exhibit population differences in color patches used for social signaling (21). The observed Mc1r mutations and their consequences for receptor function are therefore associated with convergent phenotypes important for both adaptation and speciation in recently diverged lizards in the wild.
Materials and Methods
Determining the Functional Effects of Mc1r Mutations.
Functional assays.
To determine the functional consequences of the amino acid replacements observed in Mc1r for White Sands lizards, we inserted full-length reptile Mc1r cDNA from dark soil individuals with wild-type Mc1r alleles (Sceloporus undulatus AY586117.1, Aspidoscelis inornata AY586036.1, Holbrookia maculata AY586075.1) into the mammalian expression vector pcDps and epitope-tagged with an N-terminal hemagglutinin (HA) and a C-terminal Flag epitope by PCR mutagenesis. We then introduced Mc1r mutations associated with the derived allele in White Sands individuals (S. undulatus AY586146.1, A. inornata AY586051.1, H. maculata AY586091.1) into the tagged wild-type Mc1r constructs using a PCR-based site-directed mutagenesis and restriction fragment replacement strategy. We confirmed the accuracy of all PCR-derived sequences by restriction analysis and sequencing.
Cell culture and transfection.
We cultivated COS-7 cells in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified 7% CO2 incubator. For cell transfection, we used Lipofectamine 2000 (Invitrogen) following the manufacturer’s instruction.
ALPHAScreen cAMP assay.
Because MC1R mediates its signal via Gs/adenylyl cyclase activation, we measured agonist-induced intracellular cAMP levels. The cAMP content of cell extracts was determined by a nonradioactive cAMP assay based on the ALPHAScreen technology (Perkin-Elmer, ref 22). Thus, we split cells into 50-mL cell-culture flasks (1 × 106 cells/flask) and transfected them with 5 μg of plasmid DNA. One day after transfection, we seeded cells into 48-well plates (5 × 104 cells/well). One day later, we performed cAMP accumulation assays. Cells were washed once and incubated in serum-free DMEM containing 1 mM 3-isobutyl-1-methylxanthine (Sigma) in the absence or in increasing amounts of agonist (α-MSH; Sigma) for 1 h at 37°C. We terminated the reactions by aspirating media and lysed cells in 50 μL lysis buffer (see ALPHAScreen manual) containing 1 mM 3-isobutyl-1-methylxanthine. From each well, we transferred 5 μL lysate into a 384-well plate and added acceptor/donor beads according to the manufacturer’s protocol. We analyzed the data from the ALPHAScreen cAMP assay using the GraphPad Prism program (version 5.01 for Windows).
ELISAs.
To ligand-independently estimate the amounts of receptor proteins expressed at the cell surface and in all cellular compartments, we used an indirect cell-surface ELISA and a total cellular ELISA, respectively (23). In brief, to estimate cell-surface expression of receptors carrying an amino-terminal HA tag, transfected COS-7 cells were seeded into 48-well plates 24 h after transfection. Cells were fixed with 4% paraformaldehyde without disrupting the cell membrane 72 h after transfection, and incubated with a peroxidase-coupled monoclonal anti-HA antibody (3F10; Roche). We then detected bound anti-HA antibody by adding H2O2 and o-phenylenediamine (2.5 mM each in 0.1 M phosphate-citrate buffer, pH 5.0) as substrate and chromogen, respectively. After 15 min at room temperature, we terminated the enzyme reaction by adding 1 M HCl containing 0.05 M Na2SO3, and measured color development bichromatically at 492 and 620 nm using an ELISA reader (Sunrise; Tecan). For detection of total cellular expression, we harvested COS-7 cells 3 days after transfection (4 μg of plasmid DNA/60-mm dish), added 150 μL solubilization buffer [10 nM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM DTT, 1 mM EDTA, 1% deoxycholate, 1% Nonidet P-40, 0.2 mM phenylmethylsulfonylfluoride, and 10 μg/mL aprotinin] and incubated them at 4°C for 12 h. We removed cell debris by centrifugation and used the supernatant for ELISA. Microtiter plates were coated (at 4°C for 16 h) with a monoclonal antibody directed against the carboxy-terminal Flag tag (Sigma). After blocking (with 10% FBS in PBS), we incubated cell lysates at 37°C for 2 h. We washed plates three times with PBS containing 0.05% Triton X-100 (PBS-T). Thereafter, a peroxidase-coupled anti-HA antibody (3F10; Roche) was added, and plates were incubated at 21°C for 2 h. Then we washed the plates with PBS-T three times with color reaction and took measurements as described above.
Understanding the Phenotypic Effects of Mc1r Mutations in Nature.
Sampling.
To understand the relationship between Mc1r genotype, color phenotype, and their spatial distribution in nature, we collected additional genotypic and phenotypic data for the two species with evidence for functional disruption of the blanched Mc1r allele (S. undulatus and A. inornata). We sampled three habitat categories: (i) “white sand” localities are in the heart of the White Sands formation and have white gypsum substrate; (ii) “dark soil” localities are allopatric to white sands and have brown adobe substrate typical of the Chihuahuan Desert region; and (iii) “ecotone” localities are parapatric to white sands and are areas of transition from dark soil to white sands substrate. For each species, 12–21 adult individuals per habitat type were sampled: n = 39 for A. inornata and n = 52 for S. undulatus (sample numbers and collecting localities are listed in Table S3).
Color quantification.
For each sample, we took spectrophotometric readings to measure dorsal color variation (following ref. 5). Briefly, dorsal coloration was characterized by averaging spectrophotometric readings from three points along the dorsal midline using an Ocean Optics USB 2000 spectrophotometer with a dual deuterium/tungsten halogen light source. A custom-made probe holder was used to orient the probe at 45° and 1 cm away from the dorsal body surface. We took each spectral reading in reference to a white standard and calculated percent transmission at 0.3 nm intervals. We used readings from 400 to 700 nm, the visual spectrum, for analysis. Previous analyses have shown that color differences among lizards in this system are explained primarily by differences in brightness (intensity of light transmission), as opposed to hue or chroma (5). Therefore, we focus on a direct estimate of brightness from the spectrophotometric data: area under the spectral curve (AUC).
Mc1r genotyping.
To obtain Mc1r genotypes, we extracted whole genomic DNA from frozen tissue with Qiagen DNeasy extraction kits. The entire coding region of Mc1r was amplified using species-specific primers and conditions reported in ref. 6. Using an ABI3730 (Applied Biosystems), we sequenced diploid PCR products in both directions with species-specific primers as well as internal primers universal to reptiles (6). We edited and aligned sequences using Sequencher (Gene Codes). Heterozygous sites were identified by visual inspection of chromatograms and confirmed by sequence from both DNA strands.
Statistical analysis.
We evaluated the distribution of Mc1r alleles in each habitat and the relationship of color phenotypes to Mc1r genotype. For each species, we conducted an ANOVA to compare coloration (i.e., AUC brightness scores) for individuals in each genotypic class (homozygous wild-type, homozygous derived, heterozygous). The goal of this analysis was to determine (i) whether individuals with different Mc1r genotypes exhibit different color phenotypes and (ii) the likely dominance relationship between Mc1r alleles. If an ANOVA was significant, we used post-hoc Tukey HSD tests to determine which genotypic classes differed significantly in brightness. Statistical analyses were executed in Statistica (StatSoft).
Supplementary Material
Acknowledgments
We thank the Museum of Vertebrate Zoology at the University of California, Berkeley for tissues and White Sands National Monument, White Sands Missile Range, Jornada LTER, and the New Mexico Department of Game and Fish for permits. This work was supported by grants from the German Research Foundation (DFG) to H.R. and T.S. and the National Science Foundation to E.B.R. and H.E.H. We thank Luke Harmon, Jonathan Losos, Jim Mallet, and the Rosenblum and Harmon labs for comments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1815.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911042107/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species. London: John Murray; 1959. [Google Scholar]
- 2.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Harvard University Press; 2002. [Google Scholar]
- 3.Arendt J, Reznick D. Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Kocurek G, et al. White Sands Dune Field New Mexico: Age dune dynamics and recent accumulations. Sediment Geol. 2007;197:313–331. [Google Scholar]
- 5.Rosenblum EB. Convergent evolution and divergent selection: Lizards at the White Sands ecotone. Am Nat. 2006;167:1–15. doi: 10.1086/498397. [DOI] [PubMed] [Google Scholar]
- 6.Rosenblum EB, Hoekstra HE, Nachman MW. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 2004;58:1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 7.Weis WI, Kobilka BK. Structural insights into G-protein-coupled receptor activation. Curr Opin Struct Biol. 2008;18:734–740. doi: 10.1016/j.sbi.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John PR, Ramsay M. Four novel variants in MC1R in red-haired South African individuals of European descent: S83P, Y152X, A171D, P256S. Hum Mutat. 2002;19:461–462. doi: 10.1002/humu.9030. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi Y, et al. cDNA cloning of proopiomelanocortin (POMC) and mass spectrometric identification of POMC-derived peptides from snake and alligator pituitaries. Gen Comp Endocrinol. 2007;152:73–81. doi: 10.1016/j.ygcen.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 10.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 11.Ringholm A, et al. Pharmacological characterization of loss of function mutations of the human melanocortin 1 receptor that are associated with red hair. J Invest Dermatol. 2004;123:917–923. doi: 10.1111/j.0022-202X.2004.23444.x. [DOI] [PubMed] [Google Scholar]
- 12.Vidal N, Hedges SB. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C R Biol. 2009;332:129–139. doi: 10.1016/j.crvi.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaumont KA, et al. Altered cell surface expression of human MC1R variant receptor alleles associated with red hair and skin cancer risk. Hum Mol Genet. 2005;14:2145–2154. doi: 10.1093/hmg/ddi219. [DOI] [PubMed] [Google Scholar]
- 15.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization: From ontogeny to signalling regulation. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biebermann H, et al. Autosomal-dominant mode of inheritance of a melanocortin-4 receptor mutation in a patient with severe early-onset obesity is due to a dominant-negative effect caused by receptor dimerization. Diabetes. 2003;52:2984–2988. doi: 10.2337/diabetes.52.12.2984. [DOI] [PubMed] [Google Scholar]
- 17.Li TS, Snyder WK, Olsson JE, Dryja TP. Transgenic mice carrying the dominant rhodopsin mutation P347S: Evidence for defective vectorial transport of rhodopsin to the outer segments. Proc Natl Acad Sci USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada A, Okumoto M, Tsudzuki M. Tawny: A novel light coat color mutation found in a wild population of Mus musculus molossinus, a new allele at the melanocortin 1 receptor (Mc1r) locus. Exp Anim. 1999;48:73–78. doi: 10.1538/expanim.48.73. [DOI] [PubMed] [Google Scholar]
- 19.Flanagan N, et al. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000;9:2531–2537. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 20.Gavrilets S. Perspective: Models of speciation: What have we learned in 40 years? Evolution. 2003;57:2197–2215. doi: 10.1111/j.0014-3820.2003.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 21.Robertson J, Rosenblum EB. Rapid evolution of social signal colouration in White Sands lizards. Biol J Linn Soc Lond. 2009;95:243–255. [Google Scholar]
- 22.Sangkuhl K, Römpler H, Busch W, Karges B, Schöneberg T. Nephrogenic diabetes insipidus caused by mutation of Tyr205: A key residue of V2 vasopressin receptor function. Hum Mutat. 2005;25:505. doi: 10.1002/humu.9337. [DOI] [PubMed] [Google Scholar]
- 23.Schöneberg T, Sandig V, Wess J, Gudermann T, Schultz G. Reconstitution of mutant V2 vasopressin receptors by adenovirus-mediated gene transfer. Molecular basis and clinical implication. J Clin Invest. 1997;100:1547–1556. doi: 10.1172/JCI119678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.