Abstract
Pancreatic cancer is one of the most lethal malignancies. To discover functionally relevant modulators of pancreatic neoplasia, we performed activity-based proteomic profiling on primary human ductal adenocarcinomas. Here, we identify retinoblastoma-binding protein 9 (RBBP9) as a tumor-associated serine hydrolase that displays elevated activity in pancreatic carcinomas. Whereas RBBP9 is expressed in normal and malignant tissues at similar levels, its elevated activity in tumor cells promotes anchorage-independent growth in vitro as well as pancreatic carcinogenesis in vivo. At the molecular level, RBBP9 activity overcomes TGF-β-mediated antiproliferative signaling by reducing Smad2/3 phosphorylation, a previously unknown role for a serine hydrolase in cancer biology. Conversely, loss of endogenous RBBP9 or expression of mutationally inactive RBBP9 leads to elevated Smad2/3 phosphorylation, implicating this serine hydrolase as an essential suppressor of TGF-β signaling. Finally, RBBP9-mediated suppression of TGF-β signaling is required for E-cadherin expression as loss of the serine hydrolase activity leads to a reduction in E-cadherin levels and a concomitant decrease in the integrity of tumor cell–cell junctions. These data not only define a previously uncharacterized serine hydrolase activity associated with epithelial neoplasia, but also demonstrate the potential benefit of functional proteomics in the identification of new therapeutic targets.
Keywords: functional proteomics, activity profiling, pancreatic cancer TGF-β signaling
Serine hydrolases are central effectors of the proliferative, invasive, and migratory properties of tumors, with roles in growth factor activation, extracellular matrix degradation, and angiogenesis (1 –4). Serine hydrolase transcript and protein levels are elevated in many cancer cell lines and primary tumors, but the functional relevance of these changes remains unknown (3, 5). Classical genomic and proteomic methods are of limited value for the functional characterization of the serine hydrolases because their activities can be tightly regulated by posttranslational modifications such as glycosylation and phosphorylation (2, 3).
Activity-based proteomic profiling (ABPP) is a chemical biology platform that simultaneously determines the functional state of multiple enzymatic activities directly in complex samples such as tumor specimens (3, 5, 6). Bifunctional active site-directed probes allow derivatization of the tumor-associated enzymes: One end of the probe contains a reactive group [e.g., a fluorophosphonate (FP) group will covalently bind the active site of serine hydrolases] and the other end carries a biotin moiety to permit avidin enrichment and molecular identification by a mass-spectrometry (MS)-based method such as Multidimensional Protein Identification Technology (MudPIT) (5, 7). ABPP probes selectively label active enzymes but not their inactive counterparts, allowing characterization of changes in enzyme activities that occur without corresponding alterations in transcript or protein abundance. Importantly, this approach enables rapid detection of tumor-associated enzymatic activities without prior characterization of the malignant proteome; i.e., ABPP can detect changes in the activity of an enzyme that was not previously known to have that activity. Recent studies have demonstrated that serine hydrolase ABPP permits the classification of human cancer lines into functional subtypes on the basis of tissue of origin and state of invasiveness (3). Moreover, this approach has identified tumor-associated activities with roles in cell invasion, migration, and intravasation (1, 3, 4).
In this study, we performed activity-based proteomic analysis of primary pancreatic ductal adenocarcinoma specimens from patients with the disease and identified retinoblastoma-binding protein 9 (RBBP9) as a previously uncharacterized tumor-associated serine hydrolase activity: an “enzyme” that was first described a decade ago as a retinoblastoma-binding protein with no known enzymatic activity (8). In addition to its retinoblastoma-binding function, RBBP9 has been implicated in the cellular response to chronic low-dose radiation (9) and has also been identified as a candidate regulator of aging in hematopoietic stem cells (10). Importantly, from the standpoint of pancreatic cancer, this previously uncharacterized serine hydrolase activity is a purported inhibitor of TGF-β signaling (8), a pathway that is mutated or deregulated in more than half of all pancreatic neoplasms (11 –13). Desensitization to the antiproliferative effects of TGF-β may be central to disease progression as Smad4, a key effector of TGF-β antiproliferative signaling, is lost in approximately half of all pancreatic tumors (14). Here, we provide evidence that RBBP9 activity subverts the antiproliferative function of TGF-β, through its actions on specific modules of the TGF-β signaling pathway, and this in turn can contribute to the development of pancreatic neoplasia without loss of Smad4. These data not only highlight a key role for this serine hydrolase in pancreatic tumorigenesis, but also provide unique insight into the deregulation of TGF-β signaling in malignancy.
Results
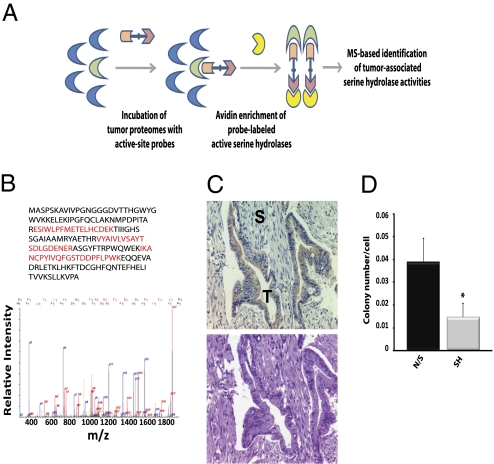
To enable the discovery of biologically relevant effector activities in pancreatic cancer, we analyzed patient pancreatectomy specimens (all stage II ductal adenocarcinoma), using activity-based proteomics coupled to MudPIT as a front-end screening platform (Fig. 1A) (5). In this study, we used a fluorophosphonate probe to interrogate active serine hydrolases in the soluble pancreatic tumor proteome. Surprisingly, we identified RBBP9 (a retinoblastoma-binding protein without known enzymatic activity) as a differentially regulated serine hydrolase that displayed elevated activity in 40% (4/10) of pancreatic tumor biopsies (Fig. 1B and Fig. S1A). By contrast, RBBP9 activity was undetectable in paired adjacent nonneoplastic specimens (0/10) (Fig. S1A). RBBP9 protein levels were equivalent in paired primary tumor and nonneoplastic specimens (Fig. S1B). Hence, the observed differences in enzymatic activity would not have been evident using classical microarray or proteomic approaches. Two previously characterized serine hydrolases, seprase/FAP and complement C1r, were the only other enzymes to display similar activity profiles in the resected clinical specimens. Seprase is frequently overexpressed in carcinoma-associated fibroblasts and has reported roles in tumorigenesis, metastasis, extracellular matrix degradation, and chemoresistance (15 –18), whereas C1r is an established factor in inflammation (19). As such, either of these enzymes could play key roles in pancreatic cancer. However, for the purposes of this study we focused on RBBP9, not only because of its previously uncharacterized serine hydrolase activity but also for its reported role in TGF-β signaling (8), a key pathway in pancreatic neoplasia (11, 13, 14).
Fig. 1.
Activity-based proteomics identified RBBP9 as a pancreatic cancer-associated serine hydrolase. (A) Schematic of the ABPP-MudPIT method. Primary tumor specimens are homogenized and fractionated to yield soluble and insoluble proteomes. Isolated proteomes are chemically labeled with active site-directed probes conjugated to biotin. Avidin beads permit enrichment of biotinylated probe-labeled active hydrolases and MS-based identification of individual hydrolase activities. (B) (Upper) Sequence coverage of RBBP9. Residues in red were identified by automated database searching of the peptides enriched from the tumor proteome that correspond to RBBP9; residues in gray were not observed. (Lower) Tandem mass spectrum of a representative RBBP9 peptide identified by LC/LC MS/MS. (C) (Upper) Immunohistochemical analysis of RBBP9 distribution in pancreatic ductal adenocarcinoma specimens shows strong expression in the neoplastic ductal epithelium. Stromal (S) and tumor (T) subcompartments are labeled. (Lower) Serial sections stained with hematoxylin and eosin. (D) Role of RBBP9 in anchorage-independent growth. Pancreatic carcinoma cells with a stable knockdown of RBBP9 show a significant reduction in colony growth on soft agar. A representative experiment is shown. n = 3 independent experiments. *P < 0.05, as compared to control cells expressing a nonsilencing shRNA, paired t test.
Pancreatic ductal adenocarcinomas (PDAs), the most common form of pancreatic cancer (20), are characterized by an extensive desmoplastic response with the result that large tracts of the tumor are typically composed of collagen-rich stroma (21). Activity-based analysis was performed on whole tumor lysates, meaning that the identified activities may be associated with the stromal and/or tumor subcompartments. To define the distribution of RBBP9 within the tumor microenvironment, we performed immunohistochemical analysis of tumor sections from patients with PDA. RBBP9 was strongly expressed in the neoplastic ductal cells but largely absent from the surrounding stroma, confirming that RBBP9 is enriched in pancreatic carcinoma cells (Fig. 1C). To determine if RBBP9 is commonly expressed in pancreatic tumor cells, we screened a panel of human and mouse pancreatic carcinoma cell lines by immunoblot analysis. RBBP9 was expressed in all lines, confirming that RBBP9 is expressed in the neoplastic epithelial cells of pancreatic cancers (Fig. S2).
To investigate the requirement for RBBP9 in pancreatic carcinoma cell function, we generated tumor cell lines with a stable knockdown of RBBP9 as well as paired control lines that express a nonsilencing shRNA (Fig. S3). We then assessed the impact of altered RBBP9 levels on anchorage-independent growth, a hallmark of cancer cells. Knockdown of RBBP9 in FG human pancreatic carcinoma cells caused a significant reduction (∼60%) in colony formation in soft agar (Fig. 1D), supporting a role for RBBP9 in cell transformation (P < 0.005, paired t test, as compared to cells expressing a nonsilencing shRNA).
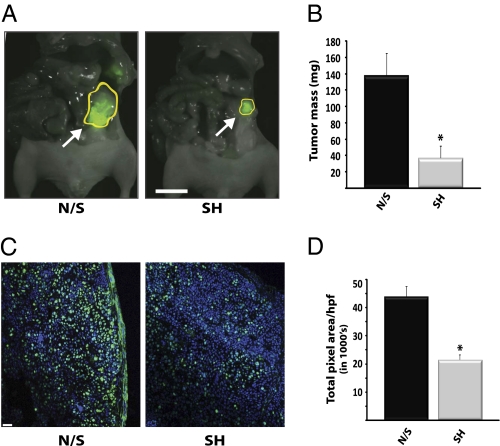
Next, we sought to determine whether the observed changes in soft agar growth in vitro translated to differences in cell tumorigenicity in vivo. To do so, we used FG pancreatic carcinoma cells, a fast growing clone of Colo-357 cells or FGM cells, a metastatic variant of FG cells. FGM or FG cells with a stable knockdown of RBBP9 or paired control cells that stably express a nonsilencing shRNA were orthotopically injected into the pancreas of 8-week-old nu/nu mice (22). Different shRNAs were used to knock down RBBP9 in the different cell lines and the lentiviral constructs used to express the shRNAs in FGM cells also encode GFP, which enables imaging of the resulting pancreatic tumors in vivo. After 21 days, the FGM tumors were analyzed by intravital fluorescence imaging and both the FG and the FGM tumors were resected to assess tumor burden and permit further analysis. Stable knockdown of RBBP9 caused a significant reduction (∼75%) in the burden of tumors derived from each cell line, as was evident from both intravital fluorescence imaging (Fig. 2A, FGM cells) and tumor mass (Fig. 2B, FG cells) (P = 0.0443, n = 8 mice/group, paired t test, as compared to tumors derived from cells expressing a nonsilencing shRNA). These studies support an essential role for RBBP9 in the tumorigenicity of pancreatic carcinoma cells in vivo. To further understand the mechanistic basis of RBBP9-dependent tumorigenesis, we assessed the levels of cellular proliferation and apoptosis by immunofluorescence analysis of sections from the paired orthotopic tumors (RBBP9-sh vs. nonsilencing). Although the numbers of cells undergoing apoptosis were similar between tumors with different levels of RBBP9 (Fig. S4), we noticed a significant reduction (∼50%) in the number of Ki67-positive (proliferating) cells in tumors that lack RBBP9 (P < 0.001, paired t test, as compared to tumors with wild-type levels of the enzyme). These data strongly support a role for RBBP9 in tumor cell proliferation (Fig. 2 C and D).
Fig. 2.
RBBP9 promotes tumor cell proliferation during pancreatic neoplasia. Requirement for RBBP9 in pancreatic tumorigenesis is shown; carcinoma cells (FG or FGM) with a stable knockdown of RBBP9 (SH) developed significantly smaller tumors following orthotopic implantation into the tail of the pancreas in mice, as compared to tumors derived from implanted control cells that express a nonsilencing shRNA (N/S). (A) FGM tumors were visualized by intravital fluorescence imaging (tumor boundary is demarcated in yellow). (Scale bar: 10 mm.) (B) FG tumor burden was assessed following resection. *P < 0.05 compared to tumors with a nonsilencing shRNA, paired t test (n = 8 mice/condition). (C) Loss of RBBP9 led to a significant reduction in tumor cell proliferation as determined by immunofluorescence analysis of the Ki67-positive cells in sections from the FG orthotopic tumors. (Scale bar: 100 μm.) (D) Quantitation of the Ki67-positive cells following analysis of Ki67 signal in 10 high-power fields (hpf’s) per tumor; tumors from 3 mice/condition were analyzed. Data are presented as the mean total pixel area in all hpf’s analyzed for that condition. *P < 0.05 compared to cells expressing a nonsilencing shRNA.
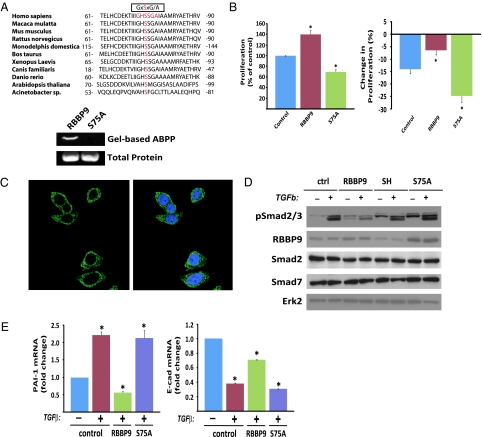
Stable knockdown of RBBP9 demonstrated that the protein is required for tumor cell proliferation. However, these experiments did not address whether the observed effects on proliferation were activity dependent or independent. This question is particularly important because the ABPP-based screen demonstrated that RBBP9 activity was elevated in tumor specimens, despite protein levels being unchanged in the nonneoplastic/neoplastic material (Fig. S1 A and B). Thus, we investigated whether the serine hydrolase activity of RBBP9 is required for the regulation of tumor cell proliferation. The structure of RBBP9 was recently described (23), and from the predicted active site as well as primary sequence alignment of RBBP9 orthologs, we identified Ser-75 as the putative nucleophilic residue in a conserved serine hydrolase motif (Fig. 3A, Upper). To assess the importance of this residue for RBBP9 activity, we generated GST-tagged derivatives of wild-type RBBP9 or mutant RBBP9-S75A and purified the recombinant proteins. Because the substrate is unknown, we assayed serine hydrolase activity using a rhodamine-labeled FP probe in a gel-based activity assay. In this approach, active serine hydrolases irreversibly bind the FP probe and the conjugated rhodamine moiety enables fluorescent visualization of probe-labeled active enzymes following 1D electrophoresis (2). Purified wild-type RBBP9 serine hydrolase bound the FP probe, indicating an active enzyme, but mutagenesis of the putative nucleophillic serine (S75A) abrogated probe binding, demonstrating that this residue is essential for RBBP9 hydrolase activity (Fig. 3A, Lower). To assess the importance of RBBP9 activity for cell proliferation, we stably expressed wild-type RBBP9 or the catalytically inactive RBBP9-S75A in human FG pancreatic carcinoma cells (Fig. S3). RBBP9 expression promoted cellular proliferation and importantly, its serine hydrolase activity was required for this role (Fig. 3B, Left), supporting the notion that the cellular proliferative rate of pancreatic carcinoma cells is dependent on RBBP9 activity.
Fig. 3.
RBBP9 serine hydrolase activity constrains TGF-β-mediated and basal Smad2/3 phosphorylation. (A) (Upper) Primary sequence alignment of RBBP9 orthologs. The canonical serine hydrolase motif is boxed. A conserved serine residue (putative nucleophillic serine) is highlighted in red. (Lower) Gel-based activity proteomics of GST-purified proteins demonstrated that Ser-75 is required for RBBP9 serine hydrolase activity. Total levels of the purified proteins were visualized using Sypro Ruby gel stain. Results represent three similar experiments. (B) (Left) Effect of RBBP9 serine hydrolase activity on cellular proliferation rate. Data are presented as a percentage of the proliferation rate of control cells and are presented as the mean of three separate experiments ± SEM. *P < 0.05 compared to proliferation of control cells. (Right) Change in cell proliferation rate as a result of TGF-β treatment in cells with different levels of RBBP9 activity. Data are presented as the change (%) in proliferation rate of cell lines treated with TGF-β1 compared to vehicle-treated control cells and the impact of wild-type (RBBP9) or catalytically inactive RBBP9 (S75A) expression on TGF-β-driven antiproliferative signaling. Data are presented as the mean of three separate experiments ± SEM. *P < 0.05 compared to proliferation of control cells treated with vehicle. (C) Immunofluorescence analysis demonstrates that RBBP9 (green) is primarily localized external to the nucleus (blue) in pancreatic carcinoma cells. (D) Effect of RBBP9 activity on TGF-β1-mediated phosphorylation of Smad2/3. Results are representative of at least three separate experiments. (E) qPCR analysis of TGF-β target gene expression. Data are presented as transcript levels of PAI-1 (Left) and E-cadherin (Right) in FG carcinoma cells (control) and in cells expressing the wild-type (RBBP9) or catalytically inactive enyzme (S75A) treated with TGF-β1 (+) or vehicle (−) for 24 h. *P < 0.05 compared to control cells that received vehicle (n = 3).
Previous studies have implicated the RBBP9 protein in the regulation of TGF-β-driven antiproliferative signaling (8), yet its role in this process is not clearly understood. Moreover, the role of RBBP9 serine hydrolase activity in TGF-β signaling has not been investigated. Treatment of FG cells with TGF-β1 inhibited cellular proliferation and this antiproliferative effect was reversed by expression of RBBP9 (Fig. 3B, Right). Significantly, RBBP9 serine hydrolase activity was required to counter the effects of TGF-β on cell proliferation, because expression of RBBP9-S75A did not overcome the antiproliferative effects of TGF-β1 (Fig. 3B, Right). In fact, stable expression of RBBP9-S75A actually nullified the inhibitory effects of endogenous RBBP9 on TGF-β1 signaling, commensurate with a role as a dominant negative enzyme, with the result that cells expressing the inactive hydrolase were more sensitive to antiproliferative signaling from the cytokine (Fig. 3B, Right). Thus, the enzymatic activity of RBBP9 activity is required to overcome the TGF-β-mediated antiproliferative response.
Although these findings suggested that RBBP9 activity was required to overcome TGF-β-derived antiproliferative signals, the molecular basis for this antagonistic function was unclear. In early studies on hepatocellular carcinoma cells, RBBP9 was identified as a nuclear protein that modulated cell cycle progression through its association with the retinoblastoma protein (8). However, in our studies on pancreatic carcinoma cells, we found that RBBP9 is primarily restricted to the extranuclear subcompartment (Fig. 3C). Thus, we investigated whether RBBP9 might counter TGF-β-driven antiproliferative signaling by impacting one or more of the key cytoplasmic signaling nodes in the TGF-β/Smad signaling pathway. TGF-β activation of its cognate receptors leads to recruitment and phosphorylation of the receptor (R)-Smads, Smad2 and Smad3. The R-Smads subsequently associate with the co-Smad, Smad4 and this tripartite complex translocates to the nucleus to modulate expression of TGF-β target genes (24).
Treatment of FG cells with TGF-β1 promoted Smad2/3 phosphorylation within 10 min and this status was maintained for 1–2 h (Fig. S5). Surprisingly, expression of RBBP9 diminished Smad2/3 phosphorylation in response to TGF-β1 stimulation (Fig. 3D). RBBP9 has been proposed to regulate TGF-β signaling through its association with the retinoblastoma protein (8), but a functional interaction between RBBP9 and the Smad proteins has not been previously described. Smad2/3 protein abundance remained unchanged, suggesting that RBBP9 regulation of Smad2/3 phosphorylation occurs at the posttranscriptional level (Fig. 3D). Moreover, Smad7 levels were also unchanged, indicating that RBBP9 may attenuate Smad2/3 phosphorylation independent of the inhibitory Smad7 protein (Fig. 3D). Importantly, RBBP9 serine hydrolase activity was required for the inhibition of TGF-β-dependent phosphorylation of the R-Smads, because RBBP9-S75A failed to attenuate Smad2/3 phosphorylation in response to TGF-β stimulation (Fig. 3D). In fact, the catalytically inactive enzyme acted in a dominant negative capacity, because even untreated cells expressing RBBP9-S75A displayed elevated Smad2/3 phosphorylation (Fig. 3D). This prompted the possibility that RBBP9 might actually function to maintain the Smad signaling pathway in the “off state.” To evaluate this, we examined the levels of Smad2/3 phosphorylation in cells with a stable knockdown of RBBP9. Cells lacking RBBP9 also displayed elevated Smad2/3 phosphorylation in the absence of exogenous TGF-β stimulation, highlighting a previously undescribed function for RBBP9 as an inhibitor of basal or autocrine TGF-β signaling (Fig. 3D). Combined, these data support a previously undescribed role for RBBP9 serine hyrolase activity in the suppression of Smad2/3 phosphorylation.
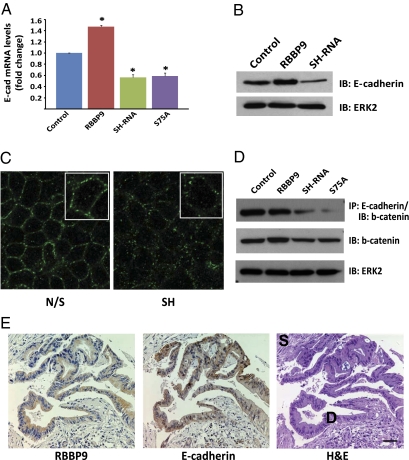
On the basis of the finding that RBBP9 attenuated Smad2/3 phosphorylation, we next examined the impact of elevated RBBP9 activity on TGF-β target gene expression. As expected, treatment with TGF-β promoted expression of the downstream target gene, PAI-1 and notably this effect was reversed by expression of the active, but not the inactive RBBP9 enzyme (Fig. 3E, Left). Conversely and as expected, TGF-β suppressed expression of the downstream target gene, E-cadherin and again, this effect was overcome by expression of the active, but not the inactive RBBP9 enzyme (Fig. 3E, Right). Epithelial cell–cell interactions are mediated by the adherens junction protein E-cadherin and importantly, the aggressiveness of a tumor inversely correlates with the level of cohesiveness between neoplastic ductal cells (25). Because our findings support a role for RBBP9 in the regulation of E-cadherin gene expression, we next considered whether changes in RBBP9 activity might impact E-cadherin levels in pancreatic cancer. Expression of RBBP9 in FG cells resulted in elevated E-cadherin transcript and protein levels, even in the absence of exogenous TGF-β stimulation (Fig. 4 A and B). In contrast, loss of RBBP9 serine hydrolase activity caused a significant reduction in both E-cadherin gene expression and protein abundance (Fig. 4 A and B). In fact, RBBP9 has a key role in the maintenance of E-cadherin levels and function because loss of the serine hydrolase activity led to a dissolution of E-cadherin-containing adherens junctions and changes in gross cellular morphology (Fig. 4C). At the molecular level, RBBP9 activity was required for E-cadherin:β-catenin complex formation because genetic knockdown of RBBP9 or expression of the dominant negative inactive enzyme caused a breakdown of this protein complex (Fig. 4D). Because RBBP9 activity contributes to both E-cadherin expression and the formation of adherens junctions, we next assessed RBBP9 and E-cadherin levels in patient biopsy material. RBBP9 staining was most prominent in tumor ducts with a higher degree of differentiation and notably, the RBBP9-positive ductal structures also exhibited strong E-cadherin staining, suggesting that RBBP9 may contribute to the epithelial phenotype of pancreatic tumor cells (Fig. 4E). Finally, to investigate whether RBBP9 activity might also be relevant to other carcinomas, we performed ABPP coupled to MudPIT on tumor cell lines derived from various extrapancreatic malignancies. RBBP9 activity was detectable in a range of carcinoma cell lines (Table S1), including those derived from cancers of the breast, colon, ovary, and lung, suggesting that this tumor-associated serine hydrolase activity may have broad-ranging implications for epithelial neoplasia.
Fig. 4.
RBBP9 promotes E-cadherin expression and tumor cell cohesiveness in pancreatic neoplasia. (A) qPCR analysis of E-cadherin transcript levels in pancreatic carcinoma cells that express different levels of wild-type or catalytically inactive RBBP9. Data are presented as the fold difference in E-cadherin transcript levels relative to control cells expressing empty vector. *P < 0.05 compared to control cells (n = 3). (B) Immunoblot analysis of E-cadherin abundance in FG pancreatic carcinoma cell lines that stably express different levels of RBBP9 serine hydrolase. (C) Immunofluorescence analysis of E-cadherin in confluent pancreatic carcinoma monolayers. N/S, FG carcinoma cells that stably express a nonsilencing shRNA; SH, FG cells that stably express a RBBP9-targeted shRNA construct. (D) Immunoblot analysis of β-catenin levels from E-cadherin immunoprecipitates (Top) or whole cell lysates (Middle) from FG cells that express different levels of wild-type or catalytically inactive RBBP9. (E) Immunohistochemical analysis of serial sections from well-differentiated ductal adenocarcinoma: RBBP9 (Left), E-cadherin (Center), and H&E (Right). Neoplastic ducts (D) and stroma (S) are labeled. (Scale bar: 50 μm.)
Discussion
Pancreatic cancer remains one of the most lethal malignancies with a median survival of 4–6 months from diagnosis (26, 27). Despite advances in our understanding of the cancer proteome, functional characterization of tumor-associated enzymatic activities is essential if we are to elucidate the molecular basis of this deadly disease and identify bona fide therapeutic targets. Recently, a comprehensive analysis of the pancreatic cancer genome identified a number of new tumor-associated genetic lesions (28). However, classical genomic and proteomic studies have yielded relatively few therapeutic targets in oncology because the expression level/mutational status does not necessarily reflect the activity of a given target.
Here, we report an activity-based profiling study of primary human pancreatic adenocarcinoma specimens. Through this approach we identify RBBP9 as a tumor-associated serine hydrolase that displays elevated activity during neoplasia. That RBBP9 expression is equivalent in normal and malignant specimens indicates that simple analysis of the expression of this protein would not have predicted the relationship between RBBP9 activity and its capacity to influence the growth of pancreatic cancer. To this end, we demonstrated a previously unknown requirement for RBBP9 in anchorage-independent growth in vitro as well as in pancreatic tumorigenesis in an orthotopic mouse model of the disease in vivo. RBBP9 was originally identified more than a decade ago as an Rb-binding protein with no known enzymatic activity (8), but our activity-based proteomics approach detected elevated RBBP9 serine hydrolase activity in tumors. The lack of detectable activity in nonneoplastic specimens may reflect differential posttranslational regulation of activity or may simply be due to a lack of sensitivity in the MS-based assay. To this end, a more traditional substrate-based enzymatic assay would be particularly instructive and efforts toward substrate identification are ongoing. The structure of RBBP9 was recently reported and this hydrolase is predicted to be a member of the DUF1234 superfamily, which includes acyltransferases, lipases, and thioesterases among others (23). The closest known homolog of RBBP9 with structural information is the YdeN protein from Bacillus subtilis (26% sequence identity) (23), but the substrate of this hydrolase also remains unknown (29).
Identification of an RBBP9 substrate(s) will also provide valuable insight into the precise mechanism by which RBBP9 acts as a suppressor of Smad phosphorylation. The TGF-β signaling pathway is particularly relevant to pancreatic cancer because dysregulation of TGF-β signaling has been linked to disease progression (11 –13). Overexpression of the pleiotrophic TGF-β cytokine as well as its cognate receptor has been reported in pancreatic cancer, but loss of Smad4 (a downstream effector of TGF-β signaling) is the most common event in this pathway in patients with the disease (14). This typically occurs through homozygous deletion of the encoding gene, Dpc4 or through intragenic mutations and loss of the second allele (14, 20). The precise role of TGF-β signaling in pancreatic cancer remains to be defined but in preinvasive tumors, a functional TGF-β pathway constrains cellular proliferation through cell cycle control at the G1/S transition (24). Our data support a previously undescribed role for a serine hydrolase activity in the disruption of antiproliferative TGF-β signaling during pancreatic neoplasia. Loss of Dpc4 typically occurs at a later stage in PDA progression and subsequently, the TGF-β pathway may actually promote the invasive phenotype of the advanced neoplasm through a Smad4-independent pathway. Because RBBP9 acts as a suppressor of Smad2/3 phosphorylation, elevated RBBP9 serine hydrolase activity may be most relevant to early stage tumors when Smad4-dependent signaling remains intact.
Pancreatic cancer is characterized by the extensive desmoplastic response that occurs during disease progression (21). Tumors are typically fibrous in nature and have an abundant underlying stroma with extensive collagen deposition and infiltrating fibroblasts (21). TGF-β signaling has been shown to play key roles in both the stromal and the epithelial (tumor) subcompartments in this disease (24). In the present study, activity-based proteomics was performed on heterogeneous tumor biopsies, which included both tumor cells and the associated stroma. Whereas RBBP9 activity was detected in only 40% of the PDA specimens, this is likely to be an underestimate of the actual levels of tumor-associated activity because pancreatic tumors in man are characterized by >80% stroma (21), and the expression of RBBP9 was greatly enriched in the neoplastic ductal epithelium. Thus, RBBP9-dependent regulation of TGF-β signaling in neoplasia may be most relevant to the tumor subcompartment. However, given the heterotypical interactions and cross-talk between tumor/stroma cells in a neoplastic lesion, potentially beneficial effects of RBBP9 inhibition may not be restricted to the ductal carcinoma cells.
In summary, we describe a multidimensional activity-based proteomic analysis of primary pancreatic cancer specimens and have used this approach to not only assign a previously undescribed enzymatic function to the Rb-binding protein, RBBP9 but also identify this hydrolase as a unique carcinoma-associated activity. RBBP9 activity was also detectable in carcinomas of the lung, breast, colon, and ovary, suggesting that this enzyme may impact a wide range of cancers. Significantly, RBBP9 activity contributes to anchorage-independent growth as well as tumorigenesis in vivo. We demonstrate that RBBP9 inhibits neoplastic TGF-β signaling through suppression of Smad2/3 phosphorylation, which in turn promotes proliferation as well as tumor cell cohesiveness, implicating this tumor-associated serine hydrolase as a key enzyme in malignant transformation.
Materials and Methods
ABPP-MudPIT Analysis of Human Pancreatic Tumor Specimens.
Resected patient tumor specimens were accrued in accordance with all local institutional review board guidelines. Proteomes were extracted, fractionated, labeled with a fluorophosphonate probe specific for active serine hydrolases, and analyzed by ABPP coupled to MudPit as described previously (3, 6, 30) and in SI Text. Ms2 spectral data were searched as described in SI Text.
Anchorage-Independent Growth Assays and Orthotopic Pancreatic Carcinoma Model.
Both the anchorage-independent growth assays and the orthotopic pancreatic carcinoma model were performed as described (31). Intravital imaging was performed using an Olympus Ovi100 imaging station. Additional details are in SI Text.
Immunohistochemical and Immunofluorescence Analysis.
Immunostaining was performed on 5-μm sections of primary tumors or human tissue arrays (U.S. Biomax) according to the manufacturer’s recommendations (Vector Labs). Sections were stained with antibodies against E-cadherin (BD Biosciences), RBBP9 (Proteintech), Ki67 (AbCam) or analyzed for apoptosis with the ApopTag kit (Millipore) as described in SI Text.
Immunoprecipitation and Immunoblot Analysis.
Immunoblots were performed using the antibodies RBBP9 (Novus), Erk2, Smad4, β-catenin (Santa Cruz Biotechnology), phospho-Smad2/3, Smad2 (Cell Signaling), Smad7 (Invitrogen), and E-cadherin (BD Biosciences). Immunoprecipitations were performed with E-cadherin antibody (BD Biosciences) as described in SI Text.
Statistical Analysis.
Data presented are means ± SEM. We performed statistical analyses with Prism (GraphPad). Statistical differences for one factor between two groups or more than two groups were determined with a Student’s t test or an analysis of variance (ANOVA) with a posthoc test, respectively. Statistical significance was defined as P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Vicente Torres for valuable discussions; Dana Wu, Charles Yi, and Heather Hoover for excellent technical support; and Gabriel Simon for assistance with data analysis. We acknowledge Dr. Anirban Maitra (Johns Hopkins University) for sharing pancreatic tumor specimens. Panc02 cells were a kind gift from Dr. Keping Xie at M. D. Anderson Cancer Center. This work was supported by National Institutes of Health Grants R21CA104898 and P01-CA078045 (to D.A.C.) and by a Collaborative Translational Research Grant from University of California (San Diego) Moores Cancer Center (to D.A.C. and M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911646107/DCSupplemental.
References
- 1.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Jessani N, et al. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci USA. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci USA. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madsen MA, Deryugina EI, Niessen S, Cravatt BF, Quigley JP. Activity-based protein profiling implicates urokinase activation as a key step in human fibrosarcoma intravasation. J Biol Chem. 2006;281:15997–16005. doi: 10.1074/jbc.M601223200. [DOI] [PubMed] [Google Scholar]
- 5.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 6.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 7.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 8.Woitach JT, Zhang M, Niu CH, Thorgeirsson SS. A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability. Nat Genet. 1998;19:371–374. doi: 10.1038/1258. [DOI] [PubMed] [Google Scholar]
- 9.Cassie S, et al. Novel retinoblastoma binding protein RBBP9 modulates sex-specific radiation responses in vivo. Carcinogenesis. 2006;27:465–474. doi: 10.1093/carcin/bgi261. [DOI] [PubMed] [Google Scholar]
- 10.Geiger H, Rennebeck G, Van Zant G. Regulation of hematopoietic stem cell aging in vivo by a distinct genetic element. Proc Natl Acad Sci USA. 2005;102:5102–5107. doi: 10.1073/pnas.0408654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ijichi H, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truty MJ, Urrutia R. Basics of TGF-beta and pancreatic cancer. Pancreatology. 2007;7:423–435. doi: 10.1159/000108959. [DOI] [PubMed] [Google Scholar]
- 13.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn SA, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 15.Loeffler M, Krüger JA, Niethammer AG, Reisfeld RA. Targeting tumor-associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J Clin Invest. 2006;116:1955–1962. doi: 10.1172/JCI26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JE, et al. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 17.Scanlan MJ, et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA. 1994;91:5657–5661. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng JD, et al. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res. 2002;62:4767–4772. [PubMed] [Google Scholar]
- 19.Gaultier A, et al. Regulation of tumor necrosis factor receptor-1 and the IKK-NF-{kappa}B pathway by LDL receptor-related protein explains the anti-inflammatory activity of this receptor. Blood. 2008;111:5316–5325. doi: 10.1182/blood-2007-12-127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 21.Hernández-Muñoz I, Skoudy A, Real FX, Navarro P. Pancreatic ductal adenocarcinoma: Cellular origin, signaling pathways and stroma contribution. Pancreatology. 2008;8:462–469. doi: 10.1159/000151537. [DOI] [PubMed] [Google Scholar]
- 22.Grimm J, Potthast A, Wunder A, Moore A. Magnetic resonance imaging of the pancreas and pancreatic tumors in a mouse orthotopic model of human cancer. Int J Cancer. 2003;106:806–811. doi: 10.1002/ijc.11281. [DOI] [PubMed] [Google Scholar]
- 23.Vorobiev SM, et al. Crystal structure of human retinoblastoma binding protein 9. Proteins. 2009;74:526–529. doi: 10.1002/prot.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter JM, et al. Absence of E-cadherin expression distinguishes noncohesive from cohesive pancreatic cancer. Clin Cancer Res. 2008;14:412–418. doi: 10.1158/1078-0432.CCR-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 27.Warshaw AL, Fernández-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janda I, et al. Harvesting the high-hanging fruit: The structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:1101–1107. doi: 10.1107/S0907444904007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 31.Desgrosellier JS, et al. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15:1163–1169. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.