Abstract
Abscisic acid (ABA) is one of the most important phytohormones involved in abiotic stress responses, seed maturation, germination, and senescence. ABA is predominantly produced in vascular tissues and exerts hormonal responses in various cells, including guard cells. Although ABA responses require extrusion of ABA from ABA-producing cells in an intercellular ABA signaling pathway, the transport mechanisms of ABA through the plasma membrane remain unknown. Here we isolated an ATP-binding cassette (ABC) transporter gene, AtABCG25, from Arabidopsis by genetically screening for ABA sensitivity. AtABCG25 was expressed mainly in vascular tissues. The fluorescent protein-fused AtABCG25 was localized at the plasma membrane in plant cells. In membrane vesicles derived from AtABCG25-expressing insect cells, AtABCG25 exhibited ATP-dependent ABA transport. The AtABCG25-overexpressing plants showed higher leaf temperatures, implying an influence on stomatal regulation. These results strongly suggest that AtABCG25 is an exporter of ABA and is involved in the intercellular ABA signaling pathway. The presence of the ABA transport mechanism sheds light on the active control of multicellular ABA responses to environmental stresses among plant cells.
Keywords: Arabidopsis, ABCG, transposon-tagged lines
The phytohormone abscisic acid (ABA) plays pivotal roles in various aspects of plant growth and development, including embryo and seed maturation, postgerminative growth, and stress responses to environmental changes (1). Many molecules related to ABA signaling or recognition have been identified (1–3), and the ABA signaling mechanism appears to be a complicated multi-input signaling pathway in which many components directly or indirectly affect each other (2, 3). In particular, multiple receptors that recognize ABA were recently reported, based on various phenomenal characterizations (4–8). In addition, intercellular functioning of ABA is predicted to exist in plants; for example, ABA is predominantly produced in vascular tissues, but acts in distant guard cell responses (9–14). Intercellular ABA function must be integrated with the intracellular signaling triggered by ABA receptors to understand the total ABA regulatory mechanism. The molecular basis of ABA transport is currently unknown.
ATP-binding cassette (ABC) transporters are a highly conserved family of ATP-binding proteins found in both prokaryotes and eukaryotes (15). The largest subfamily in Arabidopsis comprises the half-size ABC transporter genes, represented by a cluster of 28 genes in the AtABCG subfamily, previously called the WBC subfamily (16). The functions of three subfamily members have been reported: CER5/WBC12/AtABCG12 and COF1/WBC11/AtABCG11 are required for wax export and elaboration of the cuticle (17–22), and WBC19/AtABCG19 improves antibiotic resistance (23). The functions of the other AtABCGs remain largely unknown. Here we present evidence that one of the AtABCG genes, AtABCG25, encodes a protein that is responsible for ABA transport and responses in Arabidopsis.
Results and Discussion
Identification of the AtABCG25 Gene and atabcg25 Mutant Alleles.
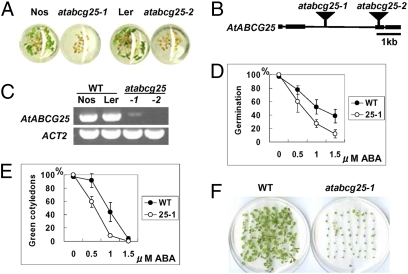
To obtain novel mutants related to ABA responses, we selected ABA-related mutants from our transposon-tagged mutant collection. We previously constructed about 12,000 transposon-tagged lines in Arabidopsis by using the activator (Ac)/dissociation (Ds) system and determined the sequence flanking the Ds element in all independent lines (24). Using this resource, we have been selecting homozygous insertion lines in which the Ds transposon is inserted in coding regions of genes for systematic phenotyping analyses (25). We used high-throughput screening with 96-well multititer plates to screen about 2,000 homozygous mutant lines for ABA-related phenotypes, and isolated one mutant line with an ABA-sensitive phenotype in the germination and seedling stages (Fig. 1A). According to the genomic sequences flanking the Ds insertion in the isolated line (15-0195-1), the Ds element was in the second intron of a predicted open reading frame (ORF) of the At1g71960 gene (Fig. 1B).
Fig. 1.
Identification of the AtABCG25 gene and atabcg25 mutant alleles. (A) Isolation of ABA-sensitive mutants by 96-well multititer plate assay. Compared with wild-types (Nos, Ler), the mutants (atabcg25-1, atabcg25-2) were more sensitive to 1.0 μM ABA solution. The titer plate was incubated in a growth chamber under long-day conditions for 7 days. (B) AtABCG25 gene structure and insertional mutation sites of two atabcg25 alleles. Square boxes represent exons, and black bars represent introns. Transposon insertions in atabcg25-1 and atabcg25-2 are shown as triangles. (C) AtABCG25 transcripts in wild-type plants and mutants identified by RT-PCR analysis. Total RNAs were prepared from two wild types (WT) and two atabcg25 mutants (atabcg25): Nossen (Nos) and Landsberg (Ler), and atabcg25-1 (-1) and atabcg25-2 (-2), respectively. Actin2 (ACT2) was used as a reference. (D–F) ABA-hypersensitive phenotype of atabcg25-1. Seed germination (D) and postgerminative growth (E) were scored for the wild type (WT) and atabcg25-1 mutant (25-1) in different concentrations of ABA at day 2 (D) and day 4 (E). Values are shown as mean ± SD of 50 seeds (obtained from three independent experiments). Photographs were taken of seedlings of wild type (WT) (F, Left) and atabcg25-1 (atabcg25-1) (F, Right) germinated in the presence of 1.0 μM ABA. Fifty seeds of each type were sown and incubated for 18 days.
The At1g71960 gene encodes AtABCG25 (also known as AtWBC26), which is a member of the ABCG subfamily of putative ABC transporters in the Arabidopsis genome (16). Therefore, this mutant was designated as atabcg25-1. The allelic mutant line CSHL_ET7134, designated atabcg25-2, had a Ds insertion in the third exon of AtABCG25 and showed the same phenotype as atabcg25-1 in the multititer plate assay (Fig. 1A). Two additional alleles from T-DNA insertion lines also showed ABA-sensitive phenotypes (Fig. S1), suggesting that the mutation of AtABCG25 corresponds to the ABA-sensitive phenotype. Reverse transcriptase-mediated PCR (RT-PCR) analysis showed that the homozygous line of atabcg25-2 contained no detectable amount of transcripts, indicating that this mutant is a transcriptional knockout (Fig. 1C). Although atabcg25-1 was also a knockout mutant, it showed a very faint band from RT-PCR (Fig. 1C), probably because the insertional mutation was in a relatively long intron (Fig. 1B). All of the atabcg25 mutants displayed ABA-sensitive phenotypes during the early growth stage (Fig. 1 D–F and Fig. S1).
AtABCG25 Gene Expression Patterns in Plant Organs.
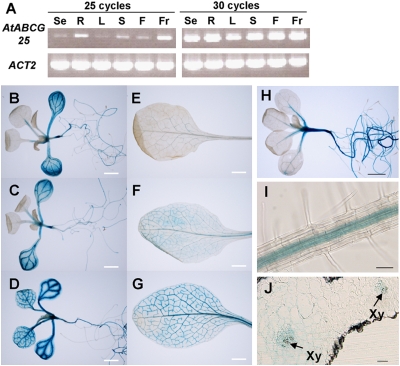
To investigate the gene expression patterns of AtABCG25 in wild-type tissues, we performed semiquantitative RT-PCR. Total RNA was extracted from seedlings, roots, stems, leaves, flowers, and fruits of wild-type plants. Transcripts for AtABCG25 were amplified from the RNA of every tissue (Fig. 2A). For further analysis of tissue-specific expression, ≈2 kb of the AtABCG25 promoter (pAtABCG25) region was used to drive expression of the GUS reporter. In pAtABCG25::GUS transgenic plants, the GUS activity of the transformants was expressed mainly in the hypocotyls, roots, and vascular veins of leaves (Fig. 2 B–G). To check the ABA inducibility of AtABCG25, pAtABCG25::GUS transgenic plants were treated with ABA solution before GUS staining. The expression levels of the GUS reporter in the transformants increased with ABA treatment (Fig. 2 B–G). Additionally, we stained atabcg25-2 mutants, which contained the GUS reporter gene in the Ds element as an enhancer trap system (26). GUS signals in atabcg25-2 were also observed in vascular tissues (Fig. 2H) and were detected along the vascular bundles in the center of roots (Fig. 2I). By cross-sectioning the stained leaves, we determined that the signals were accumulated in an area close to the vascular veins (Fig. 2J). Interestingly, enzymes that biosynthesize ABA are expressed in vascular parenchyma cells, and gene expression is increased under stress conditions in Arabidopsis (9–11). These results suggest that AtABCG25 plays an important role in ABA responses at the site of its biosynthesis.
Fig. 2.
AtABCG25 gene expression patterns in plant organs. (A) RT-PCR analysis of AtABCG25 expression patterns in different organs. Total RNAs were prepared from seedlings (Se), roots (R), leaves (L), stems (S), flowers (F), and fruits (Fr) of wild-type plants. Actin2 (ACT2) was used as a reference. (B–G) Twelve-day-old plants (B–D) and 5-week-old leaves (E–G) were stained without ABA treatment (B and E), after water treatment (C and F), and after 10 μM ABA treatment (D and G). (H–J) atabcg25-2 was used in the GUS staining of 2-week-old plants (H), the roots of 3-week-old stained plants (I), and a longitudinal section of a rosette leaf (J). Xy, Xylem. [Scale bars, (B–G) 2 mm; (H) 1 mm; (I and J) 50 μm.]
Subcellular Localization of AtABCG25 Protein.
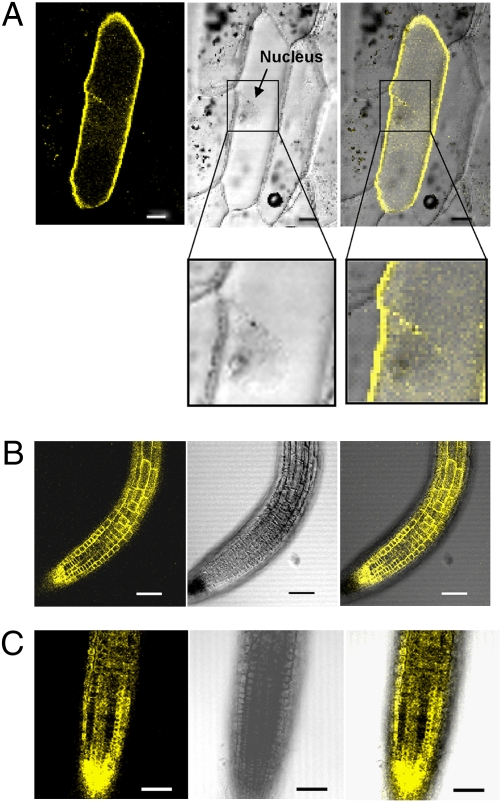
To study the subcellular localization of AtABCG25, we made a construct that produced yellow fluorescent protein (YFP) fused to AtABCG25 protein under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The AtABCG25 ORF was placed downstream of 35S::YFP. The 35S::YFP-AtABCG25 recombinant gene was transiently expressed in onion epidermal cells by particle bombardment. Subcellular localization of the fusion protein was visualized by confocal imaging of the yellow fluorescence signals in the onion cells. The yellow fluorescence of YFP-AtABCG25 recombinant protein was present around the cell surface and outside of the nucleus in these cells (Fig. 3A). Next, the 35S::YFP-AtABCG25 recombinant vector was transformed into Arabidopsis wild-type plants, and fluorescence signals were observed on the cell surface of root tips in transgenic plants expressing YFP-AtABCG25 (Fig. 3B). As root tip cells do not contain a large central vacuole, the yellow fluorescence reflects YFP-AtABCG25 localization in the plasma membrane but not in the tonoplast or cytoplasm (27). To exclude the possibility of cell wall association of YFP-AtABCG25, the root tip cells were observed after plasmolysis under highly osmotic conditions. After plasmolysis, the fluorescence in the root tip cells was internalized, apart from the cell wall (Fig. 3C). These results suggest that AtABCG25 is a plasma membrane-localized protein.
Fig. 3.
Subcellular localization of AtABCG25 protein. (A) Transient expression in onion epidermis. Fluorescence (Left) and bright-field images (Center) were merged (Right). The bottom images are magnifications of the area in the square. (B and C) Subcellular localization in transgenic Arabidopsis plants. The fluorescence signals were observed in root tip cells before (B) and after (C) plasmolysis with 20% (wt/vol) sucrose for 10 min. (Scale bars, 50 μm.)
Functional Analysis of the AtABCG25 Gene Product.
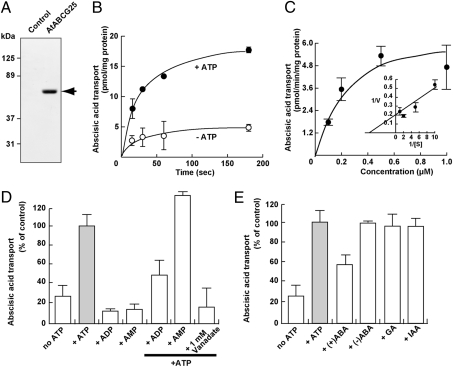
To pursue the possibility that AtABCG25 can transport ABA through the cellular membrane, we performed a vesicle transport assay. Vesicle membranes were generated from Sf9 insect cells (Spodoptera frugiperda) transfected with the virus vector integrated with AtABCG25 cDNA. The expression of AtABCG25 was confirmed by western blotting using anti-AtABCG25 antibodies (Fig. 4A). The efflux activity in plant cells can be detected as ABA uptake of the regenerated membrane vesicles upon the addition of exogenous ATP, because the regenerated membrane includes inside-out vesicles. The uptake of isotope-labeled ABA into the vesicles was significantly facilitated by exogenous ATP (Fig. 4B). The ATP-dependent uptake of ABA exhibited saturation kinetics with apparent Km and Vmax values of 260 nM and 6.9 pmol·min−1·mg−1 protein, respectively (Fig. 4C). In contrast, neither ADP nor AMP facilitated uptake (Fig. 4D). Furthermore, ADP inhibited ATP-dependent ABA uptake, whereas AMP did not show an inhibitory effect (Fig. 4D). Vanadate, an effective inhibitor of ABC transporters, also inhibited the ATP-dependent ABA uptake (Fig. 4D). To evaluate substrate specificity, cis-inhibition was examined (Fig. 4E). The ATP-dependent ABA uptake was sensitive to a 10-fold concentration of (+)ABA, whereas a 10-fold concentration of (−)ABA was not effective. Other plant hormones, such as gibberellic acid and indoleacetic acid, did not inhibit the ATP-dependent ABA uptake (Fig. 4E). These results indicate that AtABCG25 is responsible for ABA uptake, with a preference for (+)ABA rather than (−)ABA.
Fig. 4.
Uptake of isotope-labeled ABA by the AtABCG25 gene product. (A) AtABCG25 protein expression in Sf9 cells. Membranes from AtABCG25-expressing Sf9 cells and from control Sf9 cells (10 μg/lane each) were analyzed by Western blotting. The arrow corresponds to the AtABCG25 protein. (B) ATP-dependent transport of ABA by AtABCG25-expressing membrane vesicles in the presence (closed circles) and absence (open circles) of ATP. (C) Dose dependence of ABA uptake. ATP-dependent ABA uptake at the indicated ABA concentrations was determined at 15 s. Inset, Lineweaver–Burk plot. (D) Energy dependence of ABA uptake. The assay was performed in the presence of the indicated nucleotides at 4 mM. Some experiments were performed in the presence of ATP and 4 mM listed nucleotides or 1 mM vanadate. (E) Cis-inhibition of ABA uptake. ABA uptake was measured in the presence of ATP and 10 μM indicated compounds. Full (100%) activity corresponded to 8.3 pmol/mg protein at 15 s (gray bars). ABA uptake in the absence of ATP is also shown (no ATP). Each value represents the mean ± SD of three determinations. GA, gibberellic acid; IAA, indoleacetic acid.
Overexpression of AtABCG25 and Its Effects on ABA Responsiveness.
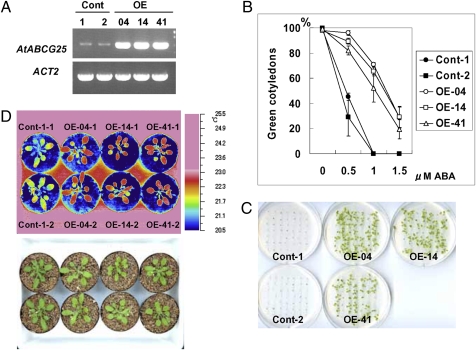
To ascertain whether AtABCG25 is a flux factor in ABA transport, we generated transgenic Arabidopsis plants possessing the 35S::AtABCG25 construct (Fig. 5A) and examined the effect of AtABCG25 overexpression on ABA signaling. To examine ABA responsiveness, T3 seeds from the resultant transgenic lines were tested for ABA inhibition of postgerminative growth. The ratio of the ABA inhibition of postgerminative growth was significantly reduced in three independent transgenic lines expressing the AtABCG25 transgene (Fig. 5 B and C), supporting the conjecture that AtABCG25 functions as a putative efflux factor of ABA.
Fig. 5.
Characterization of AtABCG25-overexpressing plants. (A) RT-PCR analysis of AtABCG25 expression in AtABCG25-overexpressing plants. Total RNAs were prepared from two wild-type plants (Cont-1, -2) and three 35S::AtABCG25 transgenic lines (OE-04, OE-14, and OE-41). Actin2 (ACT2) was used as a reference. (B and C) ABA sensitivity of postgerminative growth of AtABCG25-overexpressing plants. Seedlings of controls (Cont-1 and Cont-2) and three transgenic lines (OE-04, OE-14, and OE-41) expressing the 35S::AtABCG25 transgene were grown for 7 days in different concentrations of ABA (B). Values are shown as mean ± SD for 50 seeds (obtained from three independent experiments). The photographs show seedlings that germinated in the presence of 1.0 μM ABA. Fifty seeds of each type were sown and incubated for 15 days (C). (D) Thermal images of 4-week-old AtABCG25-overexpressing plants (OE-04-1, OE-04-2, OE-14-1, OE-14-2, OE-41-1, and OE-41-2) and control plants (Cont-1-1 and Cont-1-2), captured by an infrared thermography device (air temperature, 22 ± 2 °C; relative humidity, 60–70%).
ABA acts directly on guard cells and induces stomatal closure (28). Thus, we investigated the aerial phenotypes related to stomatal regulation in AtABCG25-overexpressing plants. The leaf temperature of transgenic plants was higher than that of wild-type plants (Fig. 5D), suggesting less transpiration from the leaves of AtABCG25-overexpressing plants. Water loss from detached leaves of the transgenic plants was also slower than that from detached wild-type leaves (Fig. S2). These results are consistent with the idea that AtABCG25 is an ABA exporter which delivers ABA to guard cells. It is possible that ABA is accumulated in the apoplastic area around guard cells in AtABCG25-overexpressing plants.
AtABCG25 Is a Transporter of ABA.
We originally isolated atabcg25 mutants by screening for ABA sensitivity and found that AtABCG25 was expressed mainly in vascular tissues, which is the main area in which ABA is biosynthesized in plants (9–11). Furthermore, the fluorescent protein-fused AtABCG25 protein was subcellularly localized to the plasma membrane in plant cells. Biochemical analyses indicated that AtABCG25 has the capability of transporting ABA molecules. Additionally, AtABCG25-overexpressing plants had a higher leaf temperature and slower rate of water loss from detached leaves, implying that overexpression of AtABCG25 may concentrate ABA in guard cells and enhance stomatal closure. Taken together, these results demonstrate that AtABCG25 is a functional factor in the ABA transport mechanism and probably facilitates the export of ABA from plant cells. Thus, atabcg25 mutants were ABA-hypersensitive under ABA treatment, because the mutants could not remove excess ABA from the cells. On the other hand, AtABCG25-overexpressing plants were resistant to exogenous ABA.
In contrast to AtABCG25-overexpressing plants, atabcg25 knockout mutant lines exhibited no aerial phenotypes. We propose that Arabidopsis has another factor with a redundant function for AtABCG25. In addition to the functional redundancy, the combined actions of AtABCG25 and another half-molecule ABC transporter would be of particular interest, because half-molecule ABC transporters can work as dimer complexes (22, 29). Our results demonstrate that AtABCG25 is a transporter functioning in ABA transport in Arabidopsis and that an ABA transport mechanism exists in plant cells. The identification of AtABCG25 provides a clue to understanding the ABA transport system in plants and gives insight into the intercellular regulation of ABA transport in ABA regulatory networks.
Materials and Methods
Plant Materials and Growth Conditions.
Plants were germinated and grown on MS medium containing 1% (wt/vol) sucrose and 0.8% (wt/vol) agar in a growth chamber or in soil at 22 °C under a 16-h light/8-h dark cycle. The atabcg25-1 (15-0195-1) mutant was isolated from a Ds transposon-tagged mutant population of the Nossen ecotype (25). The atabcg25-2 (CSHL_ET7134) allele was a Ds transposon-tagged mutant of the Landsberg ecotype and was obtained from Cold Spring Harbor Laboratory (26). Genomic DNA of Arabidopsis plants was prepared by using an automatic DNA isolation system (PI-50α; Kurabo). PCR-based genotyping was performed with ExTaq polymerase (Takara Bio). To determine the genotype of atabcg25-1, we used the following primers: 15-0195_5′ (5′-TGTAATGGGTAATGCGATAAAA-3′), 15-0195_3′ (5′-ATCTTTGGTATTGAAACCATGC-3′), and Ds5-3 (5′-TACCTCGGGTTCGAAATCGAT-3′). To determine the genotype of atabcg25-2, we used the following primers: ET7134_3′ (5′-CACGGCTTATGATACATTGCTAA-3′), ET7134_5′ (5′-GAGTGTGTACATACCGGACG-3′), and Ds5-3. The presence of a wild-type allele was detected by PCR using gene-specific primers for the sequences flanking the insertion site (15-0195_5′ and 15-0195_3′, or ET7134_3′ and ET7134_5′), and the mutant allele was detected by a combination of a Ds border primer and one of the gene-specific primers (Ds5-3 and 15-0195_5′, or Ds5-3 and ET7134_5′). For germination and greening assays, 50 sterilized seeds were placed on plates of half-strength MS medium containing 1% sucrose and different concentrations of ABA. After stratification for 4 days at 4 °C, germination was scored based on radicle protrusion, and postgerminative growth was scored by fully green, expanded cotyledons. The means and standard deviations were determined for three independent experiments.
Expression Studies and GUS Staining.
Total RNA from Arabidopsis plants was prepared for RT-PCR by using an RNeasy Plant Mini kit (Qiagen). RT-PCR was performed using a PrimeScript One-Step RT-PCR kit (Takara Bio) with the primers AtABCG25_RT-PCR_5′ (5′-TTTGGTTCTTGATGAGCCTACT-3′) and AtABCG25_RT-PCR_3′ (5′-AAGTACTCCCCAAAAGATGGAT-3′). As a loading control, Actin2 transcripts were amplified with the primers Actin2RT-F (5′-GACCTGCCTCATCATACTCG-3′) and Actin2RT-R (5′-TTCCTCAATCTCATCTTCT TCC-3′). GUS staining was performed according to a standard protocol (26). The observation of GUS-stained plants was conducted under an SZ61 stereo-microscope (Olympus), and digital images were captured using a DS-L1 CCD digital camera (Nikon). Finer images were photographed under a BX60 upright microscope (Olympus) with a VB-7010 charge-coupled-device camera (Keyence). For AtABCG25 promoter-driven GUS expression lines, a 2-kb AtABCG25 promoter region was amplified by using KOD Plus polymerase (Toyobo) with the primers AtABCG25pro_Forward (5′-CACCATCCATATTTTTATCCTGATCGTGTT-3′) and AtABCG25pro_Reverse (5′-AAAGCTGACATTAGTGTTCCTTTGTA-3′); cloned into the pENTR/D/TOPO vector (Invitrogen); and integrated into the GUS-fusion vector pBGGUS (30). For ABA treatment, leaves of 5-week-old pAtABCG25::GUS transgenic plants were soaked in 10 μM ABA for 24 h. Sections were made using a Technovit 7100 plastic embedding kit (Kulzer).
Subcellular Localization.
Full-length cDNAs of the AtABCG25 (At1g71960) gene were obtained from the RIKEN BioResource Center. The 2006-bp AtABCG25 cDNA was amplified by using KOD Plus polymerase with the primers AtABCG25_Forward (5′-CACCATGTCAGCTTTTGACGGC-3′) and AtABCG25_Reverse (5′-CCTCTCCCTCTCTTTATTTAATGTT-3′), and cloned into the pENTR/D-TOPO vector. The sequence of this clone (pENTR-AtABCG25) was confirmed, and the clone was integrated into the YFP-fusion protein vector pH35YG (30) using LR clonase (Invitrogen). To examine transient expression, the surface of the inner part of an onion (Allium cepa) was placed on solid MS medium and bombarded with 0.15 μg of plasmid DNA coated onto 1.5 mg of 1-μm gold particles using a helium biolistic device (PDS-1000; Bio-Rad) at a pressure of 1350 psi (10.7 MPa) according to the manufacturer's instructions. After incubation for about 16 h, the epidermis of the onion was peeled off, and the yellow fluorescence was examined under an LSM 510 META confocal laser scanning microscope (Carl Zeiss). We introduced the YFP-fusion protein construct consisting of pH35YG into Arabidopsis by using an Agrobacterium-mediated transformation system.
Preparation of Membrane Vesicles from AtABCG25-Expressing Sf9 Insect Cells and Immunoblotting.
A BaculoGold baculovirus expression vector system (BD Pharmingen) was used to generate the recombinant baculovirus. Sf9 insect cells (Spodoptera frugiperda) were infected with the virus and cultured with serum-free SF900-SFM medium (Invitrogen) in a shaking incubator at 27 °C for 72 h. The cells were collected by centrifugation at 1100 × g for 10 min and disrupted by nitrogen cavitation in 150 mM NaCl, 3 mM CaCl2, 2 mM MgCl2, 0.1 mM EGTA, and 10 mM Tris-HCl (pH 7.4). Undisrupted cells, nuclear debris, and large mitochondria were removed by centrifugation at 2,600 × g for 10 min. The supernatant was centrifuged for 30 min at 100,000 × g, and the resulting pellet was resuspended in 70 mM KCl, 7.5 mM MgCl2, and 50 mM Mops-Tris (pH 7.0). The membrane vesicles were kept frozen in a deep freezer until use. Protein concentration was determined using a BCA protein assay kit (Pierce), with BSA as a standard. To confirm AtABCG25 protein production in Sf9 cells by western blot analysis, anti-AtABCG25 antibodies were obtained by immunizing a rabbit with three synthetic peptides of 12–14 amino acid residues each (Operon Biotechnologies) representing positions 69–82 (QKPSDETRSTEERT), 132–143 (GKITKQTLKRTG), and 328–340 (GVTEREKPNVRQT) of Arabidopsis AtABCG25 protein. Membrane proteins were solubilized with 4% SDS and subjected to 10% SDS/PAGE. The separated proteins were transferred to a polyvinyldene-difluoride membrane and probed with the rabbit anti-AtABCG25 antibody and a horseradish peroxidase-conjugated donkey anti-rabbit IgG. An enhanced chemiluminescence detection system (ECL-plus; Amersham Biosciences) was used to visualize the specific immunoreactive proteins by exposure to autoradiographic films.
Vesicle Transport Assay.
A membrane transport study was performed using a rapid filtration technique (31). Briefly, 100 μL of transport medium (70 mM KCl, 7.5 mM MgCl2, and 50 mM Mops-Tris, pH 7.0) containing 15 mg of membrane protein, 4 mM ATP, and 1 μM ABA, which included 22 nM DL-cis, trans[G-3H] abscisic acid (GE Healthcare), was incubated at 27 °C. The sample was passed through a 0.45-μm nitrocellulose filter (Millipore), and the filter was washed with 6 mL of ice-cold transport medium. The radioactivity retained on the filter was determined using a liquid scintillation counter (Tri-Carb2800TRs; PerkinElmer). Membrane vesicles from empty vector-containing Sf9 cells were used for basal controls.
Overexpressing Arabidopsis Plants and Thermal Imaging.
To produce the 35S::AtABCG25 plasmid, a clone (pENTR-AtABCG25) containing full-length AtABCG25 cDNA was integrated into the overexpression vector pGWB2, which contained the 35S promoter of pBE2113N at the HindIII-XbaI sites (32). The 35S::AtABCG25 plasmid was electroporated into Agrobacterium GV3101 to generate transgenic plants by floral dipping. From among the T2 plants, lines overexpressing the transgene were selected by examination using RT-PCR. After self-pollination, T3 seeds were used for subsequent experiments. Thermal images were obtained using an infrared camera (Neo Thermo TVS-700; Nippon Avionics) and subsequently analyzed by PE Professional software (Nippon Avionics). Plants were grown on soil under well-watered conditions (22 °C, 60–70% relative humidity, 16-h photoperiod).
Supplementary Material
Acknowledgments
We thank Cold Spring Harbor Laboratory, RIKEN BioResource Center, and Arabidopsis Biological Resource Center for providing the Arabidopsis mutants and cDNAs; Drs. M. Okamoto, M. Kubo, and T. Demura for use of the vectors; T. Kuriyama and Dr. M. Matsui for transgenic experimental support; Dr. T. Sakai for the thermography equipment; and Dr. T. Hirayama for critical reading of the manuscript. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912516107/DCSupplemental.
References
- 1.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Wasilewska A, et al. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 4.Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 6.Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 8.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng WH, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koiwai H, et al. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004;134:1697–1707. doi: 10.1104/pp.103.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo A, et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008;147:1984–1993. doi: 10.1104/pp.108.116632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- 13.Schachtman DP, Goodger JQD. Chemical root to shoot signaling under drought. Trends Plant Sci. 2008;13:281–287. doi: 10.1016/j.tplants.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto M, et al. High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol. 2009;149:825–834. doi: 10.1104/pp.108.130823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins CF. ABC transporters: From microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 16.Verrier PJ, et al. Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Pighin JA, et al. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- 18.Bird D, et al. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- 19.Panikashvili D, et al. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 2007;145:1345–1360. doi: 10.1104/pp.107.105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ukitsu H, et al. Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol. 2007;48:1524–1533. doi: 10.1093/pcp/pcm139. [DOI] [PubMed] [Google Scholar]
- 21.Luo B, Xue XY, Hu WL, Wang LJ, Chen XY. An ABC transporter gene of Arabidopsis thaliana, AtWBC11, is involved in cuticle development and prevention of organ fusion. Plant Cell Physiol. 2007;48:1790–1802. doi: 10.1093/pcp/pcm152. [DOI] [PubMed] [Google Scholar]
- 22.Samuels L, Kunst L, Jetter R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 23.Mentewab A, Stewart CN., Jr Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol. 2005;23:1177–1180. doi: 10.1038/nbt1134. [DOI] [PubMed] [Google Scholar]
- 24.Kuromori T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 25.Kuromori T, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- 26.Sundaresan V, et al. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 27.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 29.Graf GA, et al. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem. 2003;278:48275–48282. doi: 10.1074/jbc.M310223200. [DOI] [PubMed] [Google Scholar]
- 30.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otsuka M, et al. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci USA. 2005;102:17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsuhara I, et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.