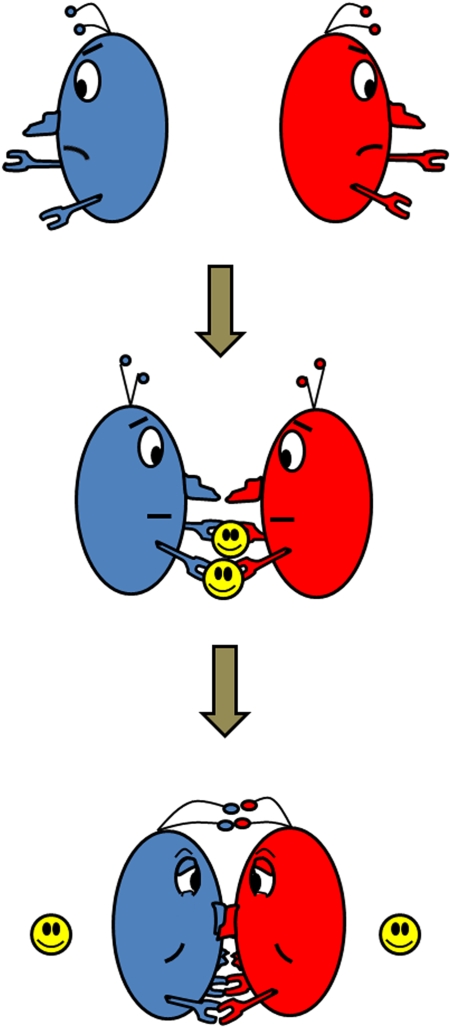
Matchmakers have been shown to be valuable for single men and women in both ancient and modern societies. Matchmakers bring together two people who otherwise have little chance of being associated with each other, nurture the closeness between the two parties by enhancing their commonality and smoothing out their differences, and then quietly disappear after the marriage. In this issue, Salgado et al. (1) describe such a matchmaker for single-domain proteins, with significant implications for protein structural evolution and design (Fig. 1).
Fig. 1.
Metal ions (yellow) as matchmakers for single-domain proteins (blue and red).
Just as families are important units for the functioning of society, multiple-domain proteins are vital for biological functions (2). A well-known example is hemoglobin in red blood cells, which transports O2 from our lungs to other parts of the body. Interfacial interactions between the four domains or subunits of hemoglobin are critical for regulation of the binding and release of O2; a single-residue mutation at the interface can result in diseases such as sickle-cell anemia. It is no wonder that a large number of proteins are multiple-domain proteins. Even single-domain proteins often do not function alone, and their interactions with other proteins play an important role in many biological functions, ranging from electron transfer in photosynthesis and respiration to ligand binding and activation in biocatalysis.
Despite the widespread recognition of the importance of interfacial interactions, there are few clues as to how proteins evolved to form such strong and tightly regulated interfaces. The chance that a single-domain protein will fold into a structure that can match well with that of another protein simply by random collisions is very small. For a long time, scientists have been searching for ways to induce such selective interactions. Several groups have reported de novo design of protein interfaces through docking of two protein structures, making mutations to minimize steric conflicts and to maximize noncovalent interactions, such as hydrogen bonding, hydrophobic and electrostatic interactions (3–7). Despite these successes, few studies have addressed the issue of how two proteins with few interfacial interactions are able to find each other by overcoming the entropic barriers in early events of protein structural evolution. In this issue, Salgado et al. (1) propose and demonstrate that metal ions could possibly fulfill such a role by being a matchmaker. In a strategy called metal templated interface redesign (MeTIR), these investigators first designed histidines into the interface of cytochrome cb562, a monomeric protein even at millimolar concentration, to allow Zn2+–histidine coordination to bring monomeric cytochrome cb562 together. On obtaining crystal structures of such metal-containing multiple domain proteins, they went on to use a variant of the RosettaDesign algorithms to optimize side chain rotamer conformations. With as few as six mutations in cytochrome cb562, the proteins formed dimers at a micromolar concentration in the absence of Zn2+ or other divalent metal ions.
The use of metal ions as matchmakers for single-domain proteins makes sense. About 40% of the natural amino acids are capable of binding metal ions; therefore, the chance for metal-coordinating amino acids to be exposed on the surface is quite high. During the early events of evolution, metal ions were abundant on earth and their presence and concentrations were not as highly regulated as they are now in cells, making it possible for a variety of metal ions to interact with surface-exposed amino acids. Because the metal–ligand bond is much stronger than all noncovalent interactions, such coordination may be strong enough to overcome steric conflicts, electrostatic repulsions, and other noncovalent interactions that prevent two monomeric proteins from associating with each other. Once the two proteins are brought together by metal ions, random mutations in the interface of the two proteins may produce a stronger interface to result in multiple-domain proteins without the help of metal ions. Another support for MeTIR playing a key role in protein structural evolution is the presence of a number of metal-binding sites between domains of proteins, such as the dinuclear copper center in hemocyanin and CuZ center in nitrous oxide reductase (8). Perhaps these metal ions were matchmakers in action, and they have been retained because they can perform additional functions, such as O2 binding or nitrous oxide reduction.
In addition to providing insight into how single-domain proteins are evolved to form multiple domain proteins, this work expands our understanding of the role of metal ions in biological systems. Previous studies have shown that metal ions can help peptides to fold into rigid and predefined structures within a single domain of protein, such as DNA-binding zinc finger proteins (9). In this issue, Salgado et al. (1) indicate that metal ions can play structural roles between domains of proteins. It has been shown that 47% of structurally determined proteins require metals, with 41% containing metals at their catalytic center (10). The work by Salgado et al. (1) suggests that metal ions can play important roles as matchmakers for single-domain proteins, even though metal ions are not present in the final product. Therefore, metal ions manifest their importance in far more than the ∼50% of proteins discovered so far.
This study not only provides fresh perspectives into the past on topics such as protein structural evolution and the roles of metal ions, but opens avenues for the future in terms of protein design. Protein–protein interactions are at the heart of many diverse biological functions, such as regulation of O2 binding in hemoglobin and signal transduction in neurobiology. Understanding these interactions and being able to control the protein–protein interactions rationally and predictably will advance many areas of biological and biomedical sciences. One example is the impact of the yeast two-hybrid system (11) in advancing our understanding of biological systems and in discovering molecules involved in protein–protein interactions. Because of complicated noncovalent interactions, it has been very difficult to control these interactions well. Using the power of metal–amino acid coordination, MeTIR provides a general method for engineering protein interfacial interactions.
This study by Salgado et al. (1) will also have a significant impact in metalloprotein design and engineering. Accounting for almost half of all proteins, metalloproteins catalyze some of the most difficult and important reactions in nature, such as photosynthesis, carbon fixation, and water oxidation. Designing metalloproteins is a touchstone whereby our knowledge can be tested and metalloenzymes with unprecedented properties for biotechnological and pharmaceutical applications can be obtained (12, 13). Most current focus is on designing metal-binding sites within a single domain of proteins.
MeTIR provides a general method for engineering protein interfacial interactions
Because there are many metal-binding sites between domains of proteins, designing those metal-binding sites is just as important but has not been explored extensively. The study by Salgado et al. (1) indicates that the redesigned interface that binds Zn2+ provides sufficient driving force so that when Cu2+ binds to the same site, the site imposes an unsaturated coordination environment around Cu2+, making it possible to design copper enzymes.
Although this work is exciting, the designed interface and metalloprotein are not ready for application, and thus have room for improvement. For example, the strongest dissociation constant (Kd) between the two monomeric proteins achieved in this study is 40 μM; to be useful for practical applications, a Kd in the nanomolar range or tighter is preferred. Furthermore, unlike blue or type 1 copper proteins, where the protein provides a rigid network of noncovalent interactions to enforce a geometry not preferred by Cu2+ (14–16) while also fine-tuning its potential without any perturbation to the primary coordination sphere (17), the designed interface is sufficiently flexible to allow Cu2+ to conform to the square planar geometry it prefers instead of the tetrahedral geometry the study designed. To improve the design, other interactions in the interface, including those in the second shell as demonstrated within single-domain proteins (13, 17, 18), may be considered and improved computing algorithms may be employed. The protein may also be subjected to directed evolution (19, 20).
The next time we see multiple-domain proteins with an intimate interface between them, the work by Salgado et al. (1) may remind us of the possible involvement of metal ions as a matchmaker in helping these single-domain proteins associate into a functional unit. Although it may be debatable whether MeTIR played an important role in the evolution of protein structure and function, it is clear that MeTIR will provide a unique approach for protein designers to engineer protein–protein interfaces for multiple-domain proteins, including many metalloproteins.
Acknowledgments
I thank Kyle Miner and Tiffany D. Wilson for help with the figure and text editing. Work on metalloprotein design and engineering in my group is supported by the National Institutes of Health Grant GM062211 and National Science Foundation Grant CHE 05-52008.
Footnotes
The author declares no conflict of interest.
See companion article on page 1827.
References
- 1.Salgado EN, et al. Metal templated design of protein interfaces. Proc Natl Acad Sci USA. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Harbury PB, Plecs JJ, Tidor B, Alber T, Kimt PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 4.Bryson JW, Desjarlais JR, Handel TM, DeGrado WF. From coiled coils to small globular proteins: Design of a native-like three-helix bundle. Protein Sci. 1998;7:1404–1414. doi: 10.1002/pro.5560070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shukla UJ, Marino H, Huang P, Mayo SL, Love JJ. A designed protein interface that blocks fibril formation. J Am Chem Soc. 2004;126:13914–13915. doi: 10.1021/ja0456858. [DOI] [PubMed] [Google Scholar]
- 6.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertini I, Sigel A, Sigel H. Handbook of Metalloproteins. 2001 (Marcel Dekker, New York) [Google Scholar]
- 9.Berg JM, Shi Y. The galvanization of biology: A growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 10.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: From enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 11.Fields S, Song OK. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 12.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray HB, Malmström BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 15.Solomon EI, Szilagyi RK, DeBeer George S, Basumallick L. Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem Rev. 2004;104:419–458. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y. Cupredoxins. Biocoordination Chemistry. In: Que L Jr, Tolman WB, editors. 2003. Comprehensive Coordination Chemistry II: From Biology to Nanotechnology, eds McCleverty J. Meyer TJ (Elsevier, Oxford), Vol 8, pp 91–122. [Google Scholar]
- 17.Marshall NM, et al. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature. 2009;462:113–116. doi: 10.1038/nature08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y. Biosynthethic inorganic chemistry. Angew Chem Int Ed. 2006;45:5588–5601. doi: 10.1002/anie.200600168. [DOI] [PubMed] [Google Scholar]
- 19.Kolkman JA, Stemmer WPC. Directed evolution of proteins by exon shuffling. Nat Biotechnol. 2001;19:423–428. doi: 10.1038/88084. [DOI] [PubMed] [Google Scholar]
- 20.Bloom JD, Arnold FH. In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci USA. 2009;106:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]