Abstract
Autopsy studies suggest that most aging men will develop lesions that, if detected clinically, would be diagnosed as prostate cancer (PCa). Most of these cancers are indolent and remain localized; however, a subset of PCa is aggressive and accounts for more than 27,000 deaths in the United States annually. Identification of factors specifically associated with risk for more aggressive PCa is urgently needed to reduce overdiagnosis and overtreatment of this common disease. To search for such factors, we compared the frequencies of SNPs among PCa patients who were defined as having either more aggressive or less aggressive disease in four populations examined in the Genetic Markers of Susceptibility (CGEMS) study performed by the National Cancer Institute. SNPs showing possible associations with disease severity were further evaluated in an additional three independent study populations from the United States and Sweden. In total, we studied 4,829 and 12,205 patients with more and less aggressive disease, respectively. We found that the frequency of the TT genotype of SNP rs4054823 at 17p12 was consistently higher among patients with more aggressive compared with less aggressive disease in each of the seven populations studied, with an overall P value of 2.1 × 10−8 under a recessive model, exceeding the conservative genome-wide significance level. The difference in frequency was largest between patients with high-grade, non–organ-confined disease compared with those with low-grade, organ-confined disease. This study demonstrates that inherited variants predisposing to aggressive but not indolent PCa exist in the genome, and suggests that the clinical potential of such variants as potential early markers for risk of aggressive PCa should be evaluated.
Keywords: association, genetics, SNP, Gleason grade, stage
Prostate cancer accounts for one-fourth of all cancer diagnoses in men in the United States, with an estimated 192,280 new cases in 2009 (1). Although most men will have an indolent form of the disease, aggressive prostate cancers are currently the second leading cause of cancer deaths in men in this country. Most cases of prostate cancer are diagnosed as a result of having an elevated serum level of prostate-specific antigen (PSA). PSA-based disease screening leading to early detection and treatment of prostate cancer (PCa) has contributed to the reduction in mortality observed for this disease in the US over the past several years (1). However, results from two large randomized trials in Europe and the US provide strong evidence that PSA-based screening for PCa is associated with a high risk of overdiagnosis (2, 3). In the European trial, PSA screening was associated with decreased PCa related mortality but at a great cost; ∼1,410 men needed to be screened, and 48 additional PCa cases would need to be treated to prevent one death from PCa (2). Although interpretation of these findings is still a subject of discussion, the current inability to accurately distinguish risk for life-threatening, aggressive PCa from the overwhelming majority of indolent cases contributes to the dilemma. Based upon existing evidence for a genetic predisposition to aggressive PCa (4) and PCa-specific death (5), we hypothesized that inherited genetic variants exist that could be used as markers to identify men at risk for more aggressive disease at an early, curable stage of the disease.
Recent breakthroughs in genome-wide association studies (GWAS) have led to the discovery of more than two dozen reported SNPs that are associated with PCa risk by comparing men with or without PCa using case–control study designs (6–25). Unfortunately, none of these PCa risk-associated SNPs consistently distinguish risk for more or less aggressive cancer (26–28), nor are they associated with prostate cancer–specific mortality (29). As a result, there has been much debate regarding the clinical utility of these SNPs as a risk stratification tool (30, 31). Clearly, an alternative approach is needed to search for genetic markers that distinguish those men who are at risk for developing more aggressive PCa. Herein, we report our findings from a systematic evaluation of ∼27,000 SNPs in the genome by comparing 4,829 patients with more aggressive disease and 12,205 patients with less aggressive disease from seven study populations using a case–case study design.
Results
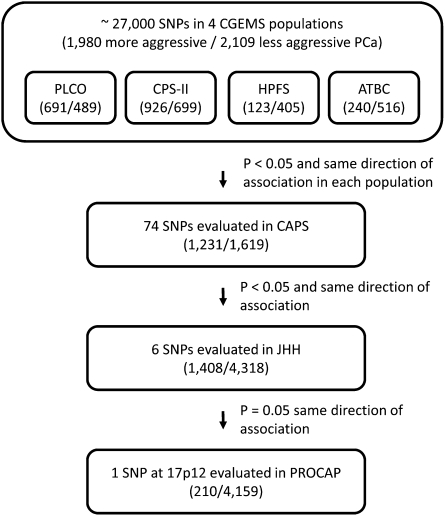
To identify inherited genetic markers that are associated with aggressiveness of PCa, we first analyzed publicly available genotype data for ∼27,000 SNPs across the genome among 1,980 patients with more aggressive disease and 2,109 patients with less aggressive disease from four CGEMS study populations (PLCO, CPS-II, HPFS, and ATBC) using a case–case analysis (Fig. 1, Table 1). Based on the results of a combined allelic test, we selected a subset of SNPs (n = 74) for further evaluation, where P < 0.05 for the difference between more and less aggressive disease, and the direction of association was consistent among the four studies. These SNPs were subsequently evaluated in an independent cohort of 1,231 patients with more aggressive disease and 1,619 patients with less aggressive disease from the CAPS study (Table S1). Six of these 74 SNPs were confirmed; P < 0.05 for the allelic test, with the same direction of association (Dataset S1). We then evaluated these six SNPs in 1,408 patients with more aggressive disease and 4,318 patients with less aggressive disease from the Johns Hopkins Hospital (JHH) study population (Table S2). One SNP (rs4054823 at 17p12) had a marginally different allele frequency between the two types of PCa patients (P = 0.051) with the same direction of association as in the previous studies (Dataset S2). This SNP was further evaluated in an additional independent Swedish PCa patient population (PROCAP), comprising 210 patients with more aggressive disease and 4,159 patients with less aggressive disease. The allelic test confirmed the association (P = 0.01).
Fig. 1.
Flow chart of the study design. Numbers of subjects with more or less aggressive prostate cancer in each study population are indicated in parentheses.
Table 1.
Number of patients with more or less aggressive prostate cancer in each of seven populations
| No. of prostate cancer patients |
||
| Study population | More aggressive | Less aggressive |
| CGEMS* | ||
| PLCO | 691 | 489 |
| ACS(CPS-II) | 926 | 699 |
| HPFS | 123 | 405 |
| ATBC | 240 | 516 |
| Subtotal | 1,980 | 2,109 |
| CAPS† | 1,231 | 1,619 |
| JHH‡ | 1,408 | 4,318 |
| PROCAP§ | 210 | 4,159 |
| Total | 4,829 | 12,205 |
*In the CGEMS study, more aggressive disease is defined as Gleason ≥ 7 or T-stage ≥T3.
†n the CAPS study, more aggressive disease is defined as Gleason ≥ 8 or T-stage ≥T3.
‡In the JHH study, more aggressive disease is defined as Gleason ≥ (4+3) or T-stage ≥T3b or N+.
§In the PROCAP study, more aggressive disease is defined as Gleason ≥ 8 or N+.
As summarized in Table 2, the frequency of allele T of SNP rs4054823 was consistently higher in patients with more aggressive disease compared with patients with less aggressive disease in each of the four CGEMS populations, and was significant in the combined allelic test (P = 9.8 × 10−4). The T allele of rs4054823 was also more frequent in patients with more aggressive disease in each of the three independent populations in the confirmation stage, with a value of P = 5.0 × 10−4 from a combined allelic test. Combining the data from all seven populations, the allelic test of the SNP and aggressiveness of PCa was highly significant (P = 2.1 × 10−6). When genotype frequencies of this SNP between the two types of PCa were tested using dominant and recessive models, the recessive model (allele T) was most significant (P = 2.1 × 10−8). This P value exceeded a study-wide significance level at a 5% false positive rate using a conservative Bonferroni correction (27,000 SNPs and three genetic models). The TT genotype was found in 32% of 4829 cases with aggressive disease and 28% of 12,205 cases with less aggressive disease. Compared with PCa patients who had CC or CT genotypes, patients who had the TT genotype of this SNP had an odds ratio (OR) of 1.26 (95% confidence interval [CI], 1.16–1.36) for aggressive PCa. No heterogeneity was observed in the OR estimates among different populations (P = 0.56, Breslow-Day test).
Table 2.
Association of SNP rs4054823 at 17p12 with aggressiveness of PCa
| Genotype frequency |
Allele test |
Genotype test |
||||||||||||
| Aggressive |
Nonaggressive |
Frequency (T) |
Recessive |
Dominant |
||||||||||
| Study populations | CC | CT | TT | CC | CT | TT | Agg. | Nonagg. | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P |
| CGEMS study | ||||||||||||||
| ACS | 171 | 467 | 275 | 152 | 349 | 183 | 0.56 | 0.52 | 1.15(1.00–1.32) | 0.05 | 1.18(0.95–1.47) | 0.14 | 1.24(0.97–1.58) | 0.09 |
| ATBC | 52 | 119 | 67 | 124 | 253 | 132 | 0.53 | 0.51 | 1.10(0.88–1.37) | 0.39 | 1.12(0.79–1.58) | 0.52 | 1.15(0.80–1.66) | 0.45 |
| HPFS | 29 | 43 | 46 | 75 | 191 | 123 | 0.57 | 0.56 | 1.04(0.78–1.40) | 0.78 | 1.38(0.90–2.12) | 0.14 | 0.73(0.45–1.20) | 0.21 |
| PLCO | 119 | 332 | 233 | 104 | 253 | 126 | 0.58 | 0.52 | 1.28(1.08–1.51) | 3.7E-03 | 1.46(1.13–1.89) | 3.6E-03 | 1.30(0.97–1.75) | 0.08 |
| Sub total | 371 | 961 | 621 | 455 | 1046 | 564 | 0.56 | 0.53 | 1.17(1.06–1.28) | 9.8E-04 | 1.27(1.10–1.47) | 9.1E-04 | 1.18(1.00–1.38) | 0.04 |
| Confirmation | ||||||||||||||
| CAPS | 247 | 589 | 387 | 331 | 841 | 428 | 0.56 | 0.52 | 1.11(1.00–1.24) | 0.04 | 1.27(1.08–1.49) | 4.5E-03 | 1.03(0.86–1.24) | 0.75 |
| JHH | 289 | 662 | 448 | 912 | 2152 | 1217 | 0.56 | 0.54 | 1.09(1.00–1.19) | 0.05 | 1.19(1.04–1.35) | 1.0E-02 | 1.04(0.90–1.21) | 0.61 |
| PROCAP | 35 | 93 | 81 | 853 | 2079 | 1215 | 0.61 | 0.54 | 1.31(1.07–1.61) | 0.01 | 1.53(1.15–2.03) | 3.5E-03 | 1.29(0.89–1.87) | 0.18 |
| Sub total | 571 | 1344 | 916 | 2096 | 5072 | 2860 | 0.56 | 0.54 | 1.12(1.05–1.19) | 5.0E-04 | 1.25(1.13–1.37) | 6.2E-06 | 1.06(0.95–1.18) | 0.32 |
| All populations | 942 | 2305 | 1537 | 2551 | 6118 | 3424 | 0.56 | 0.54 | 1.13(1.08–1.19) | 2.1E-06 | 1.26(1.16–1.36) | 2.1E-08 | 1.09(1.00–1.20) | 0.05 |
Recessive and dominant models are defined in terms of risk allele T. For Subtotal and All populations, the P value and OR (95%CI) were calculated from the CMH test. Breslow-Day P value for all populations / recessive mode is 0.5646.
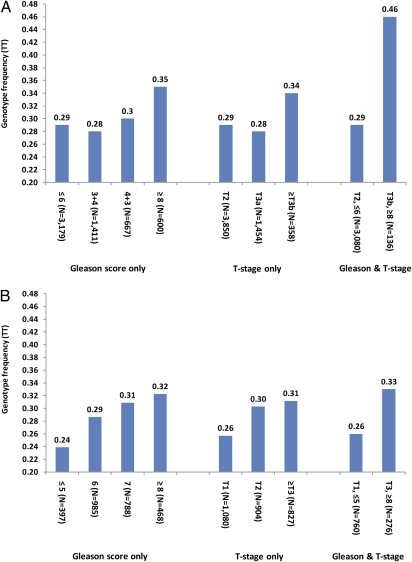
To overcome potential limitations arising from the heterogeneous definitions of aggressive PCa used among these seven study populations, and to more fully characterize the association, we performed an in-depth analysis of the correlation of SNP rs4054823 with specific clinicopathologic variables of PCa including tumor grade as assessed by Gleason score and TNM stage in populations for which this information was available. This analysis was first performed in patients from JHH for the following reasons: (i) a large number of patients (n = 5,955) recruited from the same hospital were available; (ii) all patients were treated with radical prostatectomy and thus, unlike patients receiving either no or nonsurgical treatment, their tumors were available for extensive pathologic evaluation; and (iii) tumors were uniformly graded and staged by pathologists at JHH using the same protocol (32, 33). In this analysis, we found that the frequency of the TT genotype was lower in patients with well- to moderately differentiated cancers (29%, 28%, and 30% in cancers with Gleason scores ≤6, 3+4, and 4+3, respectively) and increased only in patients with more poorly differentiated tumors, i.e., Gleason scores ≥8 (35%), P = 0.002 from a χ2 test comparing patients with Gleason score ≥8 and <8 (Fig. 2A). Similarly, we found that the frequency of the TT genotype was lower in patients with low disease stage (pT2, 29% and pT3a, 28%) and was increased in patients with higher disease stage (≥pT3b, 34%; P = 0.03, from a χ2 test comparing patients with stage ≥pT3b and <pT3b). The difference in TT genotype frequency was largest between the most extreme groups with regard to likelihood of disease progression and lethality: 29% of patients with the least aggressive disease (Gleason score ≤6 and organ-confined stage, pT2, n = 3,080), compared with 46% of patients with the most aggressive PCa (Gleason score ≥8 and non–organ-confined stage, ≥pT3b, n = 136; OR = 2.11;95% CI: 1.507–2.99), P = 1.6 × 10−5.
Fig. 2.
Frequency of TT genotype of rs4054823 at 17p12 among PCa patients for the (A) JHH population and (B) CAPS population in Sweden.
We also examined the association of this SNP with clinicopathologic variables in the Swedish CAPS population, although this population differed from the JHH population in that the treatments included multiple modalities (none, radiation, surgery, and hormonal), resulting in less uniform tumor staging and grading. In this population, the TT genotype frequency also increased with increasing Gleason score and stage; the largest difference was between the most and least aggressive PCa patients (Fig. 2B). The pattern of association, however, differed from that of JHH: a threshold increase of TT genotype frequency in patients with Gleason score ≥8 or stage ≥pT3b was observed in the JHH patients, whereas a gradual increase of TT genotype frequency was observed with increasing Gleason score or stage in CAPS patients. We speculate that this difference is due to the pathologic evaluation of prostatectomy specimens in the JHH study versus the clinical grading of biopsy specimens and clinical staging of the majority of cases in the CAPS study. Typically, a ∼20–30% discrepancy in grading and staging is observed between clinical and pathologic evaluations of the same patient (34).
Discussion
This study reflects an important shift in genetic association studies of PCa. Most studies to date have searched for inherited genetic variants that predispose men to overall PCa risk, by comparing men with and without PCa using a case–control design. In contrast, we strategically set out to identify inherited genetic markers that distinguish between risk for aggressive versus indolent PCa, by comparing SNPs among PCa patients with these two disease phenotypes using a case–case design. The need for this change in approach is supported by several trends, including a concern over increased rates of diagnosis and treatment of indolent disease and the lack of consistently validated markers of aggressive disease identified using currently used case–control study designs (26).
In this study, we have successfully identified a SNP with a genotype frequency that is consistently different between patients with more or less aggressive PCa in each of the seven independent populations studied. The difference between the two types of PCa was statistically significant (P = 2.1 × 10−8), exceeding a conservative study-wide and even genome-wide significance level. More importantly, the difference in frequency was largest between patients with high-grade, non–organ-confined disease and thus at high risk for adverse outcomes compared with patients with low-risk, low-grade, organ-confined disease.
Our finding differs from the two dozen PCa risk–associated SNPs discovered to date from GWAS (6–25). Although the associations for some SNPs, such as those at 8q24, were stronger among patients with aggressive PCa in some studies when each type of case is compared versus unaffected controls (35, 36), these SNPs were not associated with PCa aggressiveness in any of the studies when patients with more or less aggressive PCa were directly compared. Indeed, three large studies that were published recently each concluded the null association of the PCa risk-associated SNPs with PCa aggressiveness (26–28).
The importance of the SNP rs4054823 lies not in its immediate clinical utility to distinguish between these two types of PCa. A single SNP with moderate effect is unlikely to be sufficient in risk prediction. It is clear that multiple genetic and environmental factors need to be assessed to identify individuals with a particularly high risk for complex phenotypes (37). This finding is significant because it serves as a proof of principle that variants predisposing to aggressive but not indolent PCa likely exist in the genome. Therefore, efforts to identify additional variants of this type are warranted.
It is of interest to note that the frequency of the TT genotype of SNP rs4054823 in unaffected controls is similar to that observed in less aggressive cases (Table S3), and is significantly higher only among more aggressive cases. This observation implicates such SNPs as not only being informative of risk for aggressive PCa at the time of diagnosis, but also before diagnosis, to possibly target men for more effective PSA screening based on their risk for clinically important PCa.
The molecular mechanism for the observed association is unknown at this stage. The SNP rs4054823 resides in a region that is evolutionarily conserved but does not contain any known genes. The closest annotated gene is HS3ST3A1, a heparan sulfate biosynthetic enzyme located ∼120 kb telomeric of rs4054823. That heparan sulfate molecules participate as coreceptors for diverse growth factor families, including FGFs, Wnts, Hedgehogs, and BMPs is of possible relevance (38). Additional fine mapping and functional studies are needed to identify the causal genetic variants that increase risk for aggressive PCa.
There are several limitations of this study. The ∼27,000 SNPs evaluated in this study were selected because they had different allele frequencies between cases and controls in the first stage of CGEMS study. Therefore, our study missed potentially important SNPs in the remainder of the genome that are associated with aggressiveness of prostate cancer. GWAS with more complete genome coverage using a case–case design will likely identify additional SNPs associated with PCa aggressiveness. Another limitation is that we were not able to test the association of this SNP with PCa progression because of a lack of long-term follow-up information for the studied patient populations at this time. Testing for the association of this SNP with progression, together with Gleason score and stage, would allow us to assess their independent and joint contributions in predicting for disease outcome. However, an association of a SNP with Gleason score/stage alone, as demonstrated in this study, is important by itself, as Gleason score and stage are informative surrogates of disease outcome, albeit insufficient (39, 40). More importantly, when high-grade/stage tumors are diagnosed, it is often too late for effective treatment. Inherited genetic markers can be used to overcome this limitation because they do not measure a tumor-derived product and thus can be measured and can be informative years before PCa develops. Based on our study, we speculate that a panel of SNPs with characteristics similar to the one described here could be an important part of a genetic-based, targeted PSA screening strategy that is effective in reducing the number of men requiring disease screening, thereby reducing overdiagnosis while also decreasing mortality by facilitating identification of those men at risk for aggressive PCa at a stage when the disease is potentially curable. Further studies will be necessary to assess this possibility.
Methods
Study Subjects.
Seven independent populations were included in this study (Table 1). The first four populations were from the publicly available CGEMS study, and include the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial, the American Cancer Society Cancer Prevention Study II (CPS-II), the Health Professionals Follow-up Study (HPFS), and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), as described in detail elsewhere (9, 11). PCa aggressiveness was defined by the CGEMS study as follows: patients with clinical stage T3/T4 or Gleason score of 7 or higher (stage and grade designations as described below) based on biopsy specimens were classified as having more aggressive disease, whereas the remaining patients were classified as having less aggressive disease.
The other three populations were from our collaborative research group, including a hospital-based case series from the JHH, and two population-based studies based on the National Prostate Cancer Register of Sweden; a case-control study; CAncer Prostate in Sweden (CAPS) (41, 26), and a case series of PCa patients treated for localized PCa (PROCAP) (42, 43).
PCa patients from the CAPS study were identified and recruited from four regional cancer registries in Sweden, diagnosed between July 2001 and October 2003. Patients were classified as having more aggressive disease if their cancers met any of the following criteria: advanced stage as evidenced by disease spread outside of the prostate; presence of cancer in the lymph nodes or other metastatic sites (clinical stage T3/T4, N+, M+, respectively); presence of poorly differentiated cancer at biopsy as indicated by a high Gleason score (i.e., 4+4 = 8 or higher; Gleason scores are the sum of the two most prevalent histologic patterns, rated on a scale of 1–5, with 5 being the most poorly differentiated); or a serum PSA level associated with a high likelihood of extensive disease (>50 ng/mL (n = 1,231). Otherwise, the patients were classified as having less aggressive disease (n = 1,619) (Table S1).
The PCa patients from the JHH study were men who underwent radical prostatectomy for treatment of PCa at JHH from January 1, 1999, through December 31, 2008. Because of the non-JHH populations analyzed in this study including only individuals of European descent, the JHH population was similarly confined. Tumors were graded and staged after resection; those with Gleason scores of 7, with the most prevalent pattern being 4, or higher, or stage T3b or higher, or N+ or M+ were defined as more aggressive disease (n = 1,408). Tumors with Gleason score of 7 with most prevalent pattern 3, or lower and no evidence of disease dissemination (pathologic stage T2/N0/M0) were defined as having less aggressive disease (n = 4,318) (Table S2).
The PROCAP study was a cohort of PCa patients diagnosed predominantly with clinically localized disease between 1997 and 2002 and recruited from the National Prostate Cancer Register of Sweden. Among 4,356 patients, 210 were classified as having more aggressive disease (clinical stage T3/T4, N+, M+, Gleason Score ≥8, or pretreatment serum PSA ≥50 ng/mL). The remaining 4,159 patients were classified as having less aggressive disease.
This study received institutional approval at Wake Forest University School of Medicine, Johns Hopkins University School of Medicine, and the Karolinska Institutet.
SNPs and Genotyping Methods.
The genotyping data for ∼27,000 SNPs in four CGEMS study populations (PLCO, CPS-II, HPFS, and ATBC) were publically available. These SNPs were genotyped because they were significantly associated with PCa risk in the first-stage GWAS of the CGEMS study (PLCO) using a case–control analysis (11). Individual genotype data from PLCO were obtained through an approved data request application. Summary genotype information from CPS-II, HPFS, and ATBC were downloaded from a publicly accessible CGEMS website (http://cgems.cancer.gov/data/).
SNP genotyping in the CAPS, JHH, and PROCAP were performed using the MassARRAY iPLEX genotyping system (Sequenom) at Wake Forest University. Duplicate test samples and two water samples (PCR negative controls) that were blinded to the technician were included in each 96-well plate. The rate of concordant results between 100 duplicate samples was >99%.
Statistical Analysis.
Allele frequency differences between two groups of patients were tested for each SNP using a χ2 test with 1 degree of freedom within each population. The allelic odds ratio (OR) and 95% confidence interval (95% CI) were estimated based on a multiplicative model. Genotype frequency differences between two groups of patients were also tested using both a dominant and a recessive model for SNPs that were confirmed in an allele test from multiple populations. Results from multiple populations were combined using a Mantel-Haenszel model in which the populations were allowed to have different allele frequencies but were assumed to have a common OR. The homogeneity of ORs among different study populations was tested using Breslow-Day χ2 test.
For SNPs that were confirmed to be significantly associated with aggressiveness of PCa, a χ2 test using a 2 × K table was performed for Gleason scores and T-stage, in which K is the number of possible categories within each variable. All reported P values were based on a two-sided test.
Supplementary Material
Acknowledgments
The authors thank the study subjects who participated in this study and the Regional Cancer Registries and the Cancer Prostate in Sweden steering committee, including Dr. Eberhart Varenhorst. Marta Gielzak and Guifang Yan are acknowledged for expert assistance. The authors also thank the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) for making the data from this study publicly available. The study is supported by National Cancer Institute CA129684, CA106523, CA105055, and CA95052 (to J.X.), CA112517 (to W.B.I.), and CA 133009 and CA131338 (to W.B.I., and S.L.Z); Department of Defense Grants PC051264 (to J.X.) and W81XWH-09-1-0488 (to J.S.); and support from Swedish Cancer Society (Cancerfonden), Swedish Research Council, and Linneus grant (to H.G.). The support of William T. Gerrard, Mario Duhon, Jennifer and John Chalsty, and P. Kevin Jaffe (to W.B.I.) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914061107/DCSupplemental.
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Schröder FH, et al. ERSPC Investigators. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Andriole GL, et al. PLCO Project Team. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaid DJ, et al. Investigators of the International Consortium for Prostate Cancer Genetics. Pooled genome linkage scan of aggressive prostate cancer: Results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 5.Lindström LS, et al. Familial concordance in cancer survival: A Swedish population-based study. Lancet Oncol. 2007;8:1001–1006. doi: 10.1016/S1470-2045(07)70282-6. [DOI] [PubMed] [Google Scholar]
- 6.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 7.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 10.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 11.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eeles RA, et al. UK Genetic Prostate Cancer Study Collaborators; British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 14.Duggan D, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 15.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng SL, et al. Additive effects of two unlinked loci at 8q24 are associated with a considerable fraction of prostate cancer among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang BL, et al. Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet. 2009;18:1368–1375. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu FC, et al. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69:2720–2723. doi: 10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng SL, et al. Two independent prostate cancer risk-associated loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eeles RA, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators; PRACTICAL Consortium. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al Olama AA, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK Prostate testing for cancer and Treatment study (ProtecT Study) Collaborators. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 26.Kader AK, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kote-Jarai Z, et al. PRACTICAL Consortium. Multiple novel prostate cancer predisposition loci confirmed by an international study: The PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzgerald LM, et al. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: Associations with family history and clinical features. Clin Cancer Res. 2009;15:3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiklund FE, et al. Established prostate cancer susceptibility variants are not associated with disease outcome. Cancer Epidemiol Biomarkers Prev. 2009;18:1659–1662. doi: 10.1158/1055-9965.EPI-08-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelmann EP. Complexities of prostate-cancer risk. N Engl J Med. 2008;358:961–963. doi: 10.1056/NEJMe0708703. [DOI] [PubMed] [Google Scholar]
- 31.Witte JS. Prostate cancer genomics: Towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein JI, Allsbrook WC., Jr Amin MB, Egevad LL ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 33.Hoedemaeker RF, Vis AN, Van Der Kwast TH. Staging prostate cancer. Microsc Res Tech. 2000;51:423–429. doi: 10.1002/1097-0029(20001201)51:5<423::AID-JEMT4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Lotan TL, Epstein JI. Gleason grading of prostatic adenocarcinoma with glomeruloid features on needle biopsy. Hum Pathol. 2009;40:471–477. doi: 10.1016/j.humpath.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng I, et al. 8q24 and Prostate cancer: Association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helfand BT, et al. Tumor characteristics of carriers and noncarriers of the deCODE 8q24 prostate cancer susceptibility alleles. J Urol. 2008;179:2197–2201. doi: 10.1016/j.juro.2008.01.110. discussion 2202. [DOI] [PubMed] [Google Scholar]
- 37.Kraft P, Hunter DJ. Genetic risk prediction—are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 38.Cooperberg MR, Broering JM, Carroll PR. Risk assessment for prostate cancer metastasis and mortality at the time of diagnosis. J Natl Cancer Inst. 2009;101:878–887. doi: 10.1093/jnci/djp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 40.Stephenson AJ, et al. Prostate cancer-specific mortality after radical prostatectomy for patients treated in the prostate-specific antigen era. J Clin Oncol. 2009:27. doi: 10.1200/JCO.2008.18.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 42.Adolfsson J, et al. Clinical characteristics and primary treatment of prostate cancer in Sweden between 1996 and 2005. Scand J Urol Nephrol. 2007;41:456–477. doi: 10.1080/00365590701673625. [DOI] [PubMed] [Google Scholar]
- 43.Stattin P, et al. National Prostate Cancer Register. Surveillance and deferred treatment for localized prostate cancer. Population based study in the National Prostate Cancer Register of Sweden. J Urol. 2008;180:2423–2429. doi: 10.1016/j.juro.2008.08.044. discussion 2429–2430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.