Adaptation to the prevailing environment results from Darwinian natural selection (1). Physical, behavioral, physiological, and biochemical adaptations have been documented in laboratory experiments and in nature (2–7), but it has been more difficult to identify the genes and the molecular mechanisms that underlie adaptation. Recently, however, genes responsible for adaptive evolution have been identified in a wide range of taxa (8, 9).
This recent surge in understanding of the molecular basis of adaptations is remarkable, given the nearly infinite number of possible modifications to the DNA sequence of even “simple” organisms. That some cases of “convergent” evolution (similar traits evolving as adaptations to similar environmental challenges) appear to have arisen by mutation of the same genes is even more remarkable (10, 11). Conservation of molecular mechanisms in these cases has been cited as evidence that adaptive evolution is highly constrained. The argument is that the same genes are used repeatedly because very few mutations can increase adaptation to a new environment without severely compromising the integrity of living systems (12, 13). That claim is controversial, however, in part because there are only a handful of examples where the genetic basis of convergent evolution is known (10). Rarer still are examples where the downstream molecular events caused by an adaptive mutation are understood. Research published in this issue of PNAS addresses both of these issues and illustrates that different mechanisms can underlie similar adaptive phenotypes, even when the causal mutations occur in the same gene (14).
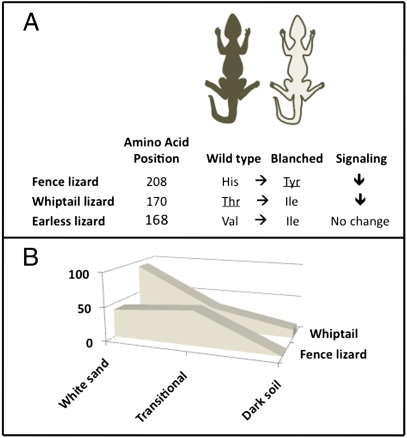
Rosenblum et al. (14) describe an exceptionally detailed study of the molecular and functional basis of convergent evolution. These investigators previously found associations between habitat, skin color, and genotypes at the melanocortin-1 receptor (Mc1r) gene in three species of lizard inhabiting the southwestern United States (15). Spurious associations due to population structure were ruled out by comparing patterns of variation in other genes and by standard tests to detect the signature of natural selection at Mcr1. In this analysis, pale-colored individuals from all three species were collected from the white gypsum soil found in White Sands National Monument in New Mexico, and dark-colored animals from nearby dark-soil habitats. Individuals were also collected from transitional habitats. Highly significant associations between single amino acid substitutions in the Mcr1 gene and dorsal skin color were found for all species (Fig. 1A). The position of the putative causal mutation in the gene differed among species, but all occurred within transmembrane regions of the receptor, which are thought to function in ligand binding, in signaling, and in maintaining structural integrity of the molecule. Because of the limited geographic range of pale (blanched) individuals, and because most other populations and related species are dark, the allele associated with blanched coloration was considered to be derived and the allele associated with darker color was considered to be the ancestral wild-type allele.
Fig. 1.
In each of three species of lizard, the light-colored blanched variety occurs where the soil substrate is pale (in White Sands, NM), although the more highly pigmented wild-type variety occurs where the soil is dark. (A) For each species, color differences are associated with a distinct amino acid substitution in the gene encoding the melanocortin-1 receptor (Mc1r). In the eastern fence lizard (S. undulatus) and the little striped whiptail lizard (A. inornata), the putative causal mutation resulted in lower signal transduction efficiency compared to the wild-type allele when expressed in a mammalian cell. In contrast, the Val-to-Ile mutation in the lesser earless lizard (H. maculata) did not cause decreased signaling efficiency. This result indicates either that the modification to the Mcr1 gene is not the causal mutation in the earless lizard, or that the modification has very different functional effects than in the other two species. Surprisingly, the dominance of the blanched allele differed in the two species for which functional differences were found (dominant alleles are indicated by underlines). (B) The difference in dominance was associated with distinct geographic distributions of alleles, with the blanched allele at nearly 90% in the white sand population of whiptail lizards, but never exceeding 50% in the fence lizards.
Although statistical associations between DNA variants and adaptive phenotypes are suggestive, they do not prove a causal relationship. The gold standard of proof, genetically transforming an individual of one genotype by placing an alternate allele into its genome, is not possible in most organisms. Indeed, many organisms of great evolutionary interest are not even amenable to laboratory rearing and breeding. For these species, other approaches must be deployed to establish causation and to understand function. One relatively powerful method is to place genetic variants into cell cultures that have been developed to allow insertion and expression of genes from many different species. Rosenblum et al. (14) used this approach to determine if the amino acid substitutions they had discovered caused measurable differences in cell function when placed into mammalian cells. By measuring accumulation of intracellular cAMP, the signaling capacity of different Mc1r alleles was tested. In two species, the eastern fence lizard (Sceloporus undulatus) and the little striped whiptail lizard (Aspidoscelis inornata), these assays showed that the derived blanched allele had lower signaling capacity in the presence of a natural agonist of the Mc1r receptor (Fig. 1A). In the third species, however, (the lesser earless lizard, Holbrookia maculata), the blanched and wild-type alleles exhibited no differences in signaling, suggesting that the identified mutation is not causal or that it regulates color in a different manner than the mutations identified in the other two species.
Although similar signaling effects of Mc1r mutations in the fence and whiptail lizards suggest conservation of molecular mechanism, a closer look indicates otherwise. Cellular signaling capacity can be affected by the number of receptors present at the cell surface, or by reduced coupling efficiency of the receptor. The His208Tyr amino acid substitution in the fence lizard caused a 20% reduction in the concentration of Mc1r receptor in the cell membrane, but the Thr170Ile substitution in the whiptail lizard did not cause any change (and neither did the Val168Ile mutation in the earless lizard). The authors conclude that amino acid substitution in the fence lizard leads to low pigmentation because the mutant receptor does not incorporate into membranes of pigment-producing cells as efficiently as the wild-type version, although the mutation in the whiptail lizard must achieve lower signaling capacity through reduced coupling efficiency (14). Moreover, these mechanisms are consistent with observed dominance patterns of the pigmentation phenotypes (Fig. 1A).
Rosenblum et al. describe an exceptionally detailed study of the molecular and functional basis of convergent evolution.
In the fence lizard, the blanched allele is dominant, consistent with the mutant receptor displacing the wild-type version from the cell membrane. In contrast, the blanched allele is recessive in the whiptail lizard, as has been observed for Mc1r mutations that affect signaling efficiency in mice and humans (16).
That details such as dominance are important for understanding the evolutionary dynamics is highlighted by the spatial distribution of allele frequencies in the fence and whiptail lizards (Fig. 1B). In the fence lizard, the blanched allele is dominant, its frequency never exceeds 50% in any habitat, and it is completely absent from the dark-soil area. In contrast, blanched allele is recessive in the whiptail lizard, it is nearly fixed in white-sand habitats, and it persists at low frequency in the dark-soil region. These geographic patterns are partly explained by dominance, but a more complete understanding will require information on the fitness of each genotype, mutation rates, and gene flow. For example, the intermediate frequency of the blanched allele in the fence lizard in the white-sand habitat suggests that homozygotes have reduced fitness or that gene flow from dark-soil areas is high, relative to the situation in the whiptail lizard.
A more profound question arising from this and other studies is “how predictable is the process of adaptation?” The answer at present would have to be “not very.” Mc1r is involved in many but not all cases of vertebrate pigment evolution, and several examples have been attributed to other candidate genes (17, 18). In no case, however, is it understood why a particular gene or mechanism contributes to some cases of adaptation, and not to others. Given the inherently stochastic nature of two major evolutionary forces (genetic drift and mutation), it is not too surprising that our current predictive ability is limited. As examples accumulate, and in particular as more functional approaches are incorporated into evolutionary studies, general patterns might emerge. Indeed, such patterns and evolutionary “rules” have been proposed (9, 11). Time (and more studies like Rosenblum et al. (14)) will tell how well these predictions fare.
Footnotes
The author declares no conflict of interest.
See companion article on page 2113.
References
- 1.Darwin CR. The Origin of Species. London: John Murray; 1859. [Google Scholar]
- 2.Reznick D, Travis J. The empirical study of adaptation in natural populations. In: Rose MR, Lauder GV, editors. Adaptation. San Diego: Academic Press; 1996. p. 511. [Google Scholar]
- 3.Grant PR. Ecology and Evoution of Darwin’s Finches. Princeton: Princeton University Press; 1999. p. 458. [Google Scholar]
- 4.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 5.Reznick DN, Ghalambor CK. Selection in nature: Experimental manipulations of natural populations. Integr Comp Biol. 2005;45:456–462. doi: 10.1093/icb/45.3.456. [DOI] [PubMed] [Google Scholar]
- 6.Buckling A, Maclean RC, Brockhurst MA, Colegrave N. The Beagle in a bottle. Nature. 2009;457:824–829. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- 7.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 8.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 9.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arendt J, Reznick D. Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Gompel N, Prud’homme B. The causes of repeated genetic evolution. Dev Biol. 2009;332:36–47. doi: 10.1016/j.ydbio.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 12.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Harvard University Press; 2002. [Google Scholar]
- 13.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- 14.Rosenblum EB, Rompler H, Schoneberg T, Hoekstra H. The molecular and functional basis of phenotypic convergence. Proc Natl Acad Sci USA. 2010;107:2113–2117. doi: 10.1073/pnas.0911042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenblum EB, Hoekstra HE, Nachman MW. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 2004;58:1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan N, et al. Pleiotropic effects of the melanocortin 1 receptor (Mc1r) gene on human pigmentation. Hum Mol Genet. 2000;9:2531–2537. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 17.Protas ME, Patel NH. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 2008;24:425–446. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- 18.Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]