Abstract
Very little is known about how animals discriminate pathogens from innocuous microbes. To address this question, we examined infection-response gene induction in the nematode Caenorhabditis elegans. We focused on genes that are induced in C. elegans by infection with the bacterial pathogen Pseudomonas aeruginosa, but are not induced by an isogenic attenuated gacA mutant. Most of these genes are induced independently of known immunity pathways. We generated a GFP reporter for one of these genes, infection response gene 1 (irg-1), which is induced strongly by wild-type P. aeruginosa strain PA14, but not by other C. elegans pathogens or by other wild-type P. aeruginosa strains that are weakly pathogenic to C. elegans. To identify components of the pathway that induces irg-1 in response to infection, we performed an RNA interference screen of C. elegans transcription factors. This screen identified zip-2, a bZIP transcription factor that is required for inducing irg-1, as well as several other genes, and is important for defense against infection by P. aeruginosa. These data indicate that zip-2 is part of a specialized pathogen response pathway that is induced by virulent strains of P. aeruginosa and provides defense against this pathogen.
Keywords: innate immunity, pathogenesis, intestine, host-pathogen interactions
When bacterial pathogens attack host cells, they rapidly set off a myriad of innate immune system responses (1 –3). Important advances have been made in deciphering how different bacterial components trigger these responses. One central finding is that bacteria contain so-called pathogen-associated molecular patterns (PAMPs) that are detected by host cells using pattern recognition receptors (PRRs). Despite their name, PAMPs are molecules that are found in all groups of bacteria, including nonpathogenic species as well as pathogens. For example, the lipopolysaccharide cell wall component of Gram-negative bacteria is a potent PAMP that is detected in mammals by the PRR Toll-like receptor 4 (Tlr4). Activation of Tlr4 leads to up-regulation of antimicrobial peptides and immune-signaling molecules. However, because lipopolysaccharide is found in most or all Gram-negative bacteria, Tlr4 signaling per se, without additional signals, does not allow a host to distinguish pathogens from innocuous bacteria.
Because bacteria of any type should not be present in the metazoan body cavity, it makes sense that professional immune cells such as macrophages, which patrol the body cavity, should express PRRs like Tlr4 that detect broad classes of bacteria. But how do cells that regularly contact benign bacteria, such as the intestinal epithelia, discriminate between pathogens and commensal microbes? This question is important because a hyperinflammatory state could be elicited if epithelia respond inappropriately to nonpathogenic microbes (4). Work in mouse and Drosophila suggests that PRR-dependent signaling may be dampened in intestinal cells to manage responses to innocuous microbes (5, 6). But little is known about the mechanisms that allow a host to discriminate pathogen from nonpathogen.
To address the question of how intestinal cells can discriminate pathogen from nonpathogen, we are investigating bacterial infection in the intestine of the well-studied nematode Caenorhabditis elegans. Under laboratory conditions, C. elegans feeds on a lawn of Escherichia coli, which appear to be relatively nonpathogenic. Therefore, the baseline state of C. elegans in the laboratory is to be in contact with benign bacteria. When C. elegans is fed certain bacterial pathogens, its lifespan is greatly shortened as a consequence of pathogen-mediated killing (7 –9). One of the most well-characterized bacterial pathogens of C. elegans is Pseudomonas aeruginosa, an opportunistic pathogen of humans that is a major cause of death in cystic fibrosis patients (10). Particular P. aeruginosa strains, including the human isolate PA14, cause a lethal intestinal infection in C. elegans that requires some of the same virulence factors important for killing mammalian hosts (11). One of these virulence factors is gacA, a response regulator that is part of a two-component regulatory system (gacS/gacA) that functions as a global regulator of virulence (12). Interestingly, not all P. aeruginosa strains are virulent in C. elegans lifespan assays (11, 13).
Previous work indicated that C. elegans responds to P. aeruginosa PA14 infection by strongly up-regulating many defense-related genes, including those predicted to encode secreted antimicrobial enzymes and peptides (14, 15). The majority of this response appears to be a consequence of pathogenicity rather than direct recognition of PAMPs, because an attenuated P. aeruginosa gacA mutant failed to induce the majority of genes induced by the isogenic wild-type parent (14). Previously published work has shown that a portion of the C. elegans response to wild-type P. aeruginosa strain PA14 infection involves two parallel signaling pathways: the PMK-1 p38 MAPK pathway (14, 16), a highly conserved immune pathway in metazoans, and a pathway containing the leucine-rich repeat G protein-coupled receptor FSHR-1 (17). However, the majority of genes transcriptionally activated by P. aeruginosa PA14 infection are induced independently of the PMK-1 p38 MAPK and FSHR-1 pathways, implicating additional pathways in the infection response.
To identify previously undescribed components of immune signaling pathways that are activated by P. aeruginosa, we made a GFP reporter for the infection response gene irg-1, which is induced by P. aeruginosa independently of the p38 MAPK and FSHR-1 pathways. irg-1::GFP is induced specifically by pathogenic P. aeruginosa, but not by other bacterial pathogens. We describe an RNAi screen for transcription factors that control expression of irg-1::GFP and identify the bZIP transcription factor zip-2. ZIP-2 is required for activation of irg-1::GFP in response to P. aeruginosa infection and zip-2 RNAi knock-down animals are sensitive to killing by P. aeruginosa, but not by another pathogen, Staphylococcus aureus. zip-2 appears to be an important component of a pathway that responds specifically to the virulence of P. aeruginosa. Study of the ZIP-2 immune response pathway may provide insight into how animals discriminate pathogenic bacteria from nonpathogenic bacteria in vivo.
Results
P. aeruginosa-Induced Gene Expression Is Mostly Independent of Known Immunity Pathways.
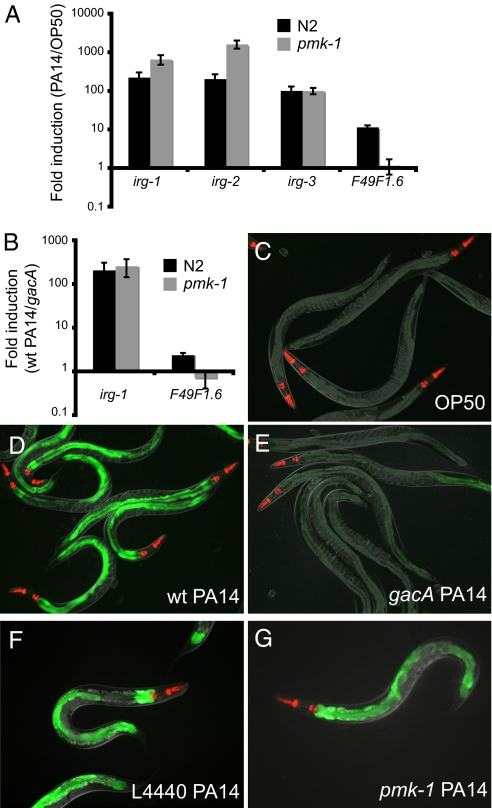
Our previous studies indicated that C. elegans mounts a robust transcriptional response to infection by the bacterial pathogen P. aeruginosa strain PA14. In these studies, we found that the PMK-1 p38 MAPK pathway appears to be required for the induction of a minority of the genes induced by infection with PA14, and that many genes are induced independently of PMK-1 (14) (Fig. 1). To identify signaling components required for the induction of selected PA14 response genes that are activated independently of PMK-1, we focused on the PA14-induced gene C07G3.2, which we named irg-1 (infection response gene 1). irg-1 contains a conserved DUF1768 domain, which is a domain sometimes fused to other proteins, such as the riboflavin biosynthesis protein RibD in rice and Arabidopsis. DUF1768 also appears as a stand-alone protein in several bacterial species, as well as nematodes, but the function of DUF1768 family members has not been clearly defined. Genome-wide transcriptional profiling analysis had shown that irg-1 was highly induced when animals infected with wild-type P. aeruginosa PA14 were compared to animals feeding on nonpathogenic E. coli (14). Like many other P. aeruginosa-response genes, induction of irg-1 mRNA by PA14 is PMK-1 independent, as demonstrated by qRT-PCR (Fig. 1A). Similarly, irg-2 (C49G7.5) and irg-3 (F53E10.4) are PMK-1 independent, whereas F49F1.6, is PMK-1 dependent, as previously shown (Fig. 1A). Because pmk-1(km25) is a null mutation in the terminal kinase of the PMK-1 p38 MAPK pathway, it appears that the PMK-1 p38 MAPK pathway is not required for induction of irg-1.
Fig. 1.
PMK-1 p38 MAPK-independent infection response gene induction by P. aeruginosa. (A and B) qRT-PCR of P. aeruginosa PA14-induced gene expression in wild-type N2 animals and pmk-1(km25) mutants at 4 h postinoculation. Results shown are the average of four (A) or three (B) independent biological replicates. Error bars are SEM. irg-1::GFP expression on E. coli strain OP50 (C), wild-type P. aeruginosa strain PA14 (D), and gacA mutant P. aeruginosa strain PA14 (E). irg-1::GFP expression on P. aeruginosa after treatment with L4440 control RNAi (F) or pmk-1 RNAi (G). Images in C to E were taken at the same time with the same camera exposure. Images F to G were taken at the same time with the same camera exposure. myo-2::mCherry shows red pharyngeal expression as a marker for the presence of the transgene.
The p38 MAPK PMK-1 is activated by an upstream MAPKK called SEK-1 (16). Because SEK-1 might signal through MAPKs other than PMK-1, we also tested whether sek-1 was required for induction of irg-1. Of the infection response genes we tested, sek-1 mutants appeared to have similar effects on gene induction as pmk-1 mutants: a subset of genes were not fully induced by P. aeruginosa in sek-1 mutants, but most genes were still induced, including irg-1, irg-2, and irg-3 (Fig. S1A). Therefore, sek-1 does not appear to be required for the pmk-1-independent-gene induction. We also tested the role of several other pathways implicated in C. elegans immunity, including the TGF-beta pathway, JNK pathway, the G protein-coupled receptor fshr-1, and the beta-catenin homolog bar-1. None of the pathways we tested appeared to be required for induction of irg-1 (see SI Text and Fig. S1).
Consistent with our previously published genome-wide transcriptional profiling data, qRT-PCR analysis showed that irg-1 was also highly induced when animals infected with wild-type P. aeruginosa were compared to animals infected with an attenuated isogenic PA14 gacA mutant (Fig. 1B). Because P. aeruginosa gacA mutants exhibit strongly reduced killing of C. elegans, as well as other hosts, irg-1 induction appears to be a response to some aspect of P. aeruginosa virulence and not a response to the P. aeruginosa bacterial strain itself. However, because many infection response genes are more robustly induced when comparing P. aeruginosa to E. coli than when comparing PA14 wild-type to PA14 gacA (and gacA mutants still exhibit some pathogenicity), we used E. coli instead of gacA as the comparison for subsequent experiments.
To analyze irg-1 tissue expression, and to develop tools for further analysis, we made a fusion between the promoter of irg-1 and GFP. Transgenic animals containing the irg-1::GFP fusion exhibited very little GFP expression when feeding on E. coli (Fig. 1C). However, expression of the irg-1::GFP transgene was robustly induced by infection with pathogenic P. aeruginosa (Fig. 1D). Very little irg-1::GFP expression was induced by infection with P. aeruginosa gacA, although in general there was slightly more GFP expression than in animals feeding on E. coli (Fig. 1E).
To confirm that the irg-1::GFP reporter strain is behaving similarly to endogenous irg-1 mRNA, we tested whether irg-1::GFP induction is independent of the p38 MAPK pathway. We treated irg-1::GFP animals with pmk-1 RNAi or control RNAi, and found that induction was similar under these two conditions (Fig. 1 F and G). Thus, the irg-1::GFP reporter appears to be induced independently of the p38 MAPK pathway, consistent with qRT-PCR experiments.
Specificity of irg-1 Induction.
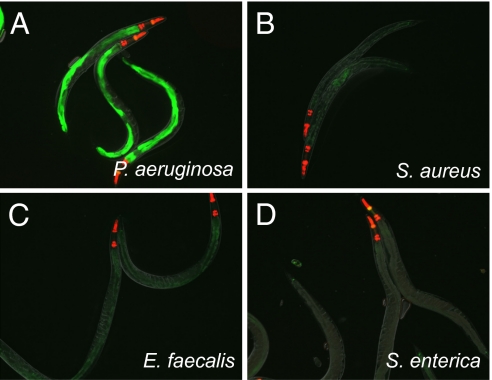
We next tested the specificity of irg-1 induction in response to different pathogens and stressors. irg-1::GFP was not induced by the Gram-positive pathogens S. aureus or Enterococcus faecalis or by the Gram-negative pathogen Salmonella enterica, which are virulent to C. elegans (Fig. 2). In addition, irg-1::GFP was not induced by infection with the natural intracellular microsporidian eukaryotic pathogen Nematocida parisii (n = 50) (18). Therefore, among the pathogens that we examined, irg-1::GFP appears to be specifically induced by virulent P. aeruginosa but not by other pathogens.
Fig. 2.
Specificity of irg-1::GFP induction. irg-1::GFP expression on P. aeruginosa (A), S. aureus (B), E. faecalis (C), S. enterica (D). A low level of GFP is visible because of internal hatching of embryos in adults infected with E. faecalis or S. aureus: irg-1::GFP is expressed constitutively in early larvae. All images were taken at the same time with the same camera exposure. myo-2::mCherry shows red pharyngeal expression as a marker for the presence of the transgene. Number of animals with GFP expression/total number of animals for each condition: P. aeruginosa (167/167), E. coli (1/361), S. aureus (0/187), E. faecalis (0/294 animals), S. enterica (1/181).
To further test the hypothesis that some aspect of P. aeruginosa virulence induces irg-1::GFP, we examined irg-1::GFP expression in animals challenged by a panel of 19 different P. aeruginosa strains, which were isolated previously from both environmental and clinical sources. Previous work had demonstrated that these 19 P. aeruginosa strains exhibited a range of virulence toward C. elegans, with PA14 being one of the most virulent strains (13). We found that virulence correlated well with irg-1::GFP induction, with the most virulent strains inducing the strongest expression of irg-1::GFP, and the least-virulent strains inducing irg-1::GFP very poorly or not at all (Table S1 and Fig. S2 A to D). qRT-PCR analysis of irg-1 mRNA induction in a subset of these strains also indicated that irg-1 induction correlates well with virulence (Fig. S2E). Therefore, consistent with our hypothesis, some aspect of P. aeruginosa virulence appears to be a cue that triggers irg-1::GFP induction.
To determine whether other kinds of stress induce irg-1, we examined published microarray data of the C. elegans response to two other intestinal stressors, the bacterial pore-forming toxin Cry5B and the heavy metal cadmium (19). CdCl2 but not Cry5B treatment was reported to up-regulate irg-1. We used qRT-PCR to confirm that cadmium treatment up-regulates irg-1 mRNA (Fig. S3); however, the level of induction was fairly low. In addition, this induction appeared to be PMK-1-dependent, in contrast to P. aeruginosa-induced induction of irg-1, which is PMK-1-independent. Thus, even though we found another stressor that induces irg-1, this induction appears to be less robust and involves pathways distinct from those responsible for P. aeruginosa-mediated induction of irg-1.
Identification of Transcription Factors Required for irg-1 Induction in Response to P. aeruginosa Infection.
To identify transcription factors that mediate irg-1 induction in response to P. aeruginosa infection, we screened a feeding RNAi library that contains bacterial clones for 345 predicted transcription factors in C. elegans. This comprises about one-third to one-half of the predicted transcription factors in C. elegans (20). We fed irg-1::GFP transgenic animals individual bacterial RNAi clones from the L1 to the L4 stage, then infected these animals with P. aeruginosa and screened for RNAi clones that blocked irg-1::GFP induction. Nine clones were identified that reduced irg-1::GFP expression (Table S2). Based on visual observation, feeding eight of the nine RNAi clones only partially reduced irg-1::GFP expression, including clones corresponding to two genes that have previously been implicated in C. elegans immunity, hsf-1 (21) and elt-2 (15). However, RNAi against these two genes also resulted in obvious morphological or behavioral abnormalities. For example, elt-2 RNAi-treated animals appeared to have greatly reduced intestinal cell volume, thus making it difficult to observe any intestinal fluorescence (Fig S4A), and hsf-1 RNAi caused an uncoordinated phenotype, as previously described (22). Further characterization of the roles of elt-2 and hsf-1 in infection-response gene induction is described in the SI Text.
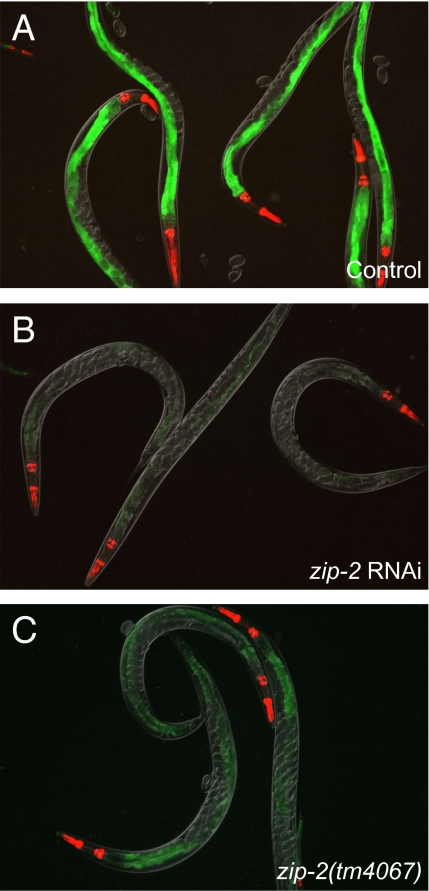
Among the nine genes identified in the screen of 345 transcription factor RNAi clones, only one clone, which corresponds to the bZIP transcription factor zip-2, appeared to completely block irg-1::GFP induction without causing other visual phenotypes (Fig. 3). To confirm that the effects of the zip-2 RNAi clone were specific to knock-down of the zip-2 gene, and not caused by off-target RNAi effects, we analyzed the deletion mutant zip-2(tm4067) from the National Bioresource Project. zip-2(tm4067) contains a deletion and partial insertion that should result in a deletion of 85 amino acids in the middle of the ZIP-2 protein and a frame shift leading to a premature stop. We investigated irg-1::GFP induction in the zip-2(tm4067) mutant background and found that P. aeruginosa induction of irg-1::GFP was greatly reduced. However, there was more GFP expression than in zip-2 RNAi (compare Fig. 3 B and C). The zip-2(tm4067) deletion is not predicted to leave intact the ZIP-2 C-terminal basic region leucine zipper (bZIP) domain, which contains the motif that is predicted to bind DNA and the motif that is predicted to mediate dimerization (Fig. S5). Interestingly, our data suggest that zip-2(tm4067) may nevertheless retain some function. In any case, these data confirm that zip-2 plays an important role in the transcriptional response to P. aeruginosa infection.
Fig. 3.
The bZIP transcription factor zip-2 controls infection-induced irg-1::GFP expression. irg-1::GFP expression on P. aeruginosa after treatment with L4440 control RNAi (A) or zip-2 RNAi (B). irg-1::GFP expression in zip-2(tm4067) mutant on P. aeruginosa (C). myo-2::mCherry shows red pharyngeal expression as a marker for the presence of the transgene. All images were taken at the same time with the same camera exposure.
Transcription Factor zip-2 Is Required for Induction of Several Infection Response Genes.
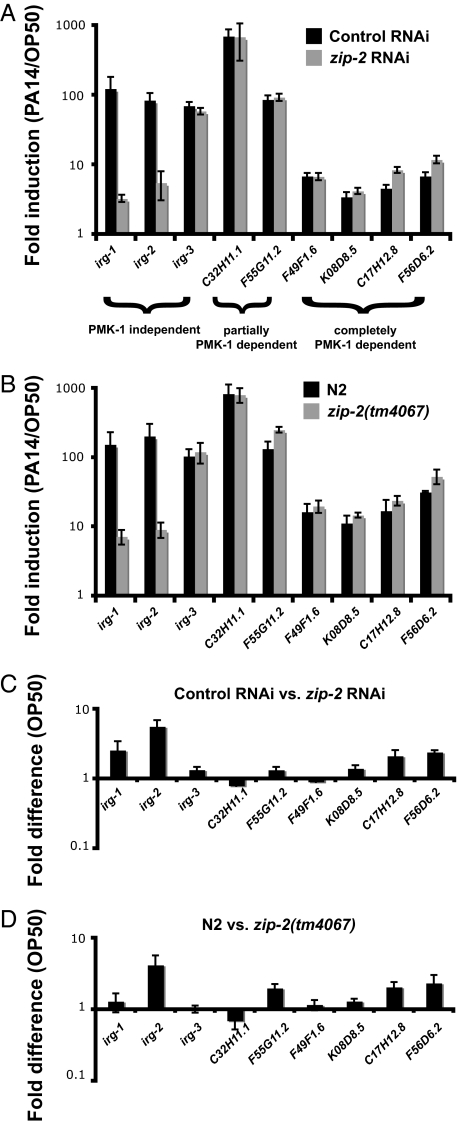
To further investigate the role of zip-2 in the transcriptional response to P. aeruginosa infection, we used qRT-PCR to determine whether zip-2 loss-of-function affects the expression of other P. aeruginosa-induced genes. First, qRT-PCR experiments confirmed that irg-1 mRNA is very poorly induced in animals treated with zip-2 RNAi (Fig. 4A), consistent with the GFP reporter data. irg-1 mRNA was induced ≈100-fold by P. aeruginosa in control-treated animals, compared to about 3-fold in zip-2 RNAi-treated animals. Similarly, irg-2 is also very poorly induced in animals treated with zip-2 RNAi. In contrast, zip-2 RNAi did not have an effect on the induction of genes previously identified to be partially or completely PMK-1-dependent (Fig. 4A). Interestingly, however, ZIP-2 does not appear to be required for all pmk-1-independent gene induction. For example, the pmk-1-independent gene irg-3 is induced in animals treated with zip-2 RNAi. Similar results were obtained with zip-2(tm4067) mutants (Fig. 4B). Therefore, a separate pathway that is independent of both pmk-1 and zip-2 may regulate induction of genes like irg-3, although we have not ruled out functional redundancy between these pathways. Overall, these results demonstrate that zip-2 performs a specific function in infection response gene induction in C. elegans.
Fig. 4.
zip-2 regulates induction of infection response genes. (A and B) qRT-PCR of infection response genes in animals feeding on P. aeruginosa compared to E. coli at 4 h postinoculation. (C and D) Basal expression of infection response genes in animals feeding on E. coli. Results shown are the average of four (A and C) or three (B and D) biological replicates. Error bars are SEM.
To further reduce zip-2 function and to test the idea that zip-2(tm4067) is a hypomorphic allele, we performed zip-2 RNAi in a zip-2(tm4067) mutant background. These animals were strongly impaired for induction of irg-1 and irg-2, similar to or slightly worse than zip-2(tm4067) mutants or zip-2 RNAi alone, and they still induced irg-3 and other genes normally (Fig. S6). These results are consistent with the zip-2(tm4067) mutant being a hypomorph and zip-2 RNAi having stronger effects than zip-2(tm4067). However, further genetic analysis of the full extent of zip-2 effects and interaction with other pathways awaits the isolation of a zip-2 null mutant.
In contrast to knock-down or mutation of the PMK-1 p38 MAPK pathway, neither zip-2 RNAi nor the zip-2(tm4067) mutation had substantial effects on the basal expression of irg-1, irg-2, or other infection-response genes that we examined in animals feeding on E. coli (Fig. 4 C and D). Abrogation of the p38 MAPK pathway has a dramatic effect on basal expression of many infection response genes (14). To examine more broadly whether zip-2 regulates the “basal” expression of genes when the animals are feeding on E. coli, and also to identify additional genes that zip-2 regulates upon infection, we performed genome-wide microarray analysis of animals treated with control or zip-2 RNAi, which were exposed to either E. coli or P. aeruginosa for 4 h. These microarray analyses confirmed the qRT-PCR results: zip-2 is required for induction of irg-1 and irg-2, but not irg-3 or F49F1.6. This transcriptional profiling study also identified 25 additional genes that require zip-2 for full induction upon P. aeruginosa infection (Table S3). These studies indicated that zip-2 mRNA itself is induced 2.7-fold upon P. aeruginosa infection. In addition, we found that 203 genes are down-regulated by zip-2 RNAi in uninfected animals feeding on E. coli, and thus are up-regulated by wild-type zip-2, and 31 genes are up-regulated by zip-2 RNAi, and thus are likely down-regulated by wild-type zip-2 (Table S4). Therefore, zip-2 likely has roles beyond just regulating induction of genes upon P. aeruginosa infection, despite the fact that zip-2-defective animals have no obvious visible phenotype on E. coli.
RNAi Against zip-2 Causes Sensitivity to Killing by P. aeruginosa, but Not by S. aureus.
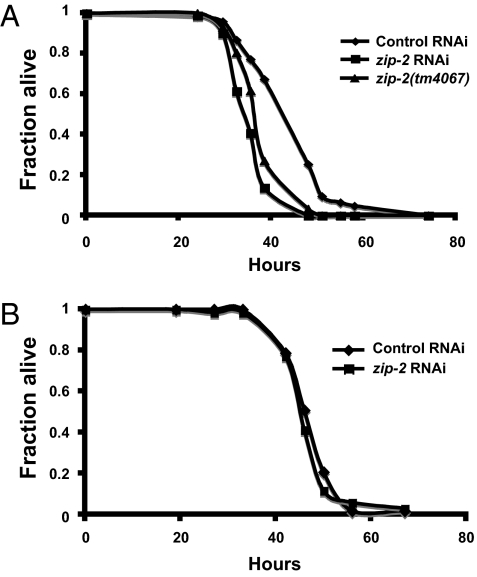
The experiments described above demonstrate that zip-2 is specifically required for inducing a subset of infection response genes up-regulated by P. aeruginosa virulence. To examine whether zip-2 contributes to increased survival upon P. aeruginosa infection, we tested zip-2 mutants and zip-2 RNAi-treated animals. As shown in Fig. 5A, zip-2 RNAi caused enhanced sensitivity to killing by P. aeruginosa compared to control animals. zip-2(tm4067) mutants also exhibited enhanced sensitivity to P. aeruginosa infection (Fig. 5A). In contrast to the experiments with P. aeruginosa, zip-2 RNAi did not increase sensitivity to killing by S. aureus (Fig. 5B). Thus, zip-2-mediated induction of irg-1 appears to be a specific response to P. aeruginosa infection, consistent with the observation that irg-1 expression is not activated by S. aureus.
Fig. 5.
zip-2 is important for defense against killing by P. aeruginosa, but not by S. aureus. (A) zip-2 RNAi and zip-2(tm4067) are more sensitive to killing by P. aeruginosa than wild-type animals. P < 0.0001 for each. (B) zip-2 RNAi does not cause sensitivity to killing by S. aureus. P = 0.24. Results shown are from assays starting with at least 90 animals per condition and are representative of at least three independent assays.
Discussion
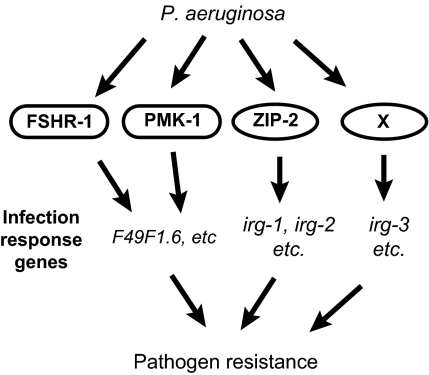
We identified the bZIP transcription factor zip-2 as a key player in the inductive response to infection with P. aeruginosa. zip-2 mediates induction of a discrete subset of infection response genes, and is important for full defense against P. aeruginosa, but not S. aureus. Our data indicate that there are several pathways that mediate the transcriptional response to P. aeruginosa infection (Fig. 6). We previously had shown that the PMK-1 p38 MAPK pathway is responsible for inducing some early infection-response genes, and that the fshr-1 receptor acts in parallel to this pathway to induce common downstream genes. However, the majority of the inductive response to P. aeruginosa infection appeared to be independent of pmk-1, indicating the existence of additional pathways that mediate infection-response gene induction. The data described here demonstrate that the bZIP transcription factor zip-2 is an important component of one of those additional pathways, mediating induction of PMK-1-independent genes, including the genes irg-1 and irg-2. Somewhat surprisingly, there appears to be yet another pathway that mediates early gene induction, as there are infection-response genes like irg-3 that are induced independently of both zip-2 and pmk-1. However, it is formally possible that zip-2 and pmk-1 pathways act redundantly to control induction of genes like irg-3. All together, our data indicate that there are several independent pathways responsible for the transcriptional response to P. aeruginosa infection.
Fig. 6.
Model for C. elegans pathways that control infection response gene induction triggered by P. aeruginosa infection.
What is the function of these downstream infection response genes? Several pathogen-induced genes encode proteins that are predicted to be secreted and have antimicrobial activity. However, many genes induced by pathogen infection have unknown function, including irg-1 and irg-2. Our studies with zip-2 indicate that these genes may have a protective role, because lack of induction of these genes in a zip-2-deficient background leads to increased sensitivity to killing by P. aeruginosa. However, these are only indirect studies and do not show causation between infection response genes and pathogen resistance. The exact role of these genes is uncertain; however, we view them as tools with which we can conveniently dissect early responses to P. aeruginosa infection. Pathogen receptors remain as yet unidentified in C. elegans, and convenient tools like irg-1::GFP can be used to help identify these receptors. The fact that a P. aeruginosa gacA mutant, as well as several P. aeruginosa isolates that are mildly pathogenic in the C. elegans model, only modestly activate defense gene expression compared to wild-type P. aeruginosa, suggests that a substantial portion of the response to P. aeruginosa is not mediated by canonical PRRs that detect highly conserved microbial components.
Much focus has rightly been placed on investigating PRRs in other systems; still, there is much less known about how animals detect virulence. Work in Drosophila has indicated that danger signals, such as proteases secreted by invading bacteria or fungi, can be detected using a proteolytic cascade upstream of the Toll receptor (23, 24). It is possible that animals have evolved multiple such strategies to detect virulence factors or the cellular disruptions they cause, because there are common mechanisms that pathogens use to cause disease. This concept is similar to the “guard hypothesis” in plant immunity (25). Mutations in such host-detection pathways could lead to pathogen-specific defects that would not correlate with standard pathogen phylogenetic classification, such as Gram-positive vs. Gram-negative bacteria. Instead, pathogens that use common virulence-related strategies would exhibit enhanced virulence on particular immune-related mutants. Among the pathogens we tested, the induction of irg-1 by the zip-2 transcription factor appears to be specific for response to P. aeruginosa. However, zip-2 may be important for response to bacterial pathogens we did not test but that use the same virulence mechanism as P. aeruginosa to activate zip-2.
zip-2 is a member of the bZIP transcription factor family, which is a gene family conserved from yeast to humans, and has roles in a wide variety of processes (26). C. elegans appears to have undergone a relatively recent expansion of bZIP genes, some of which do not appear to have direct homologs in mammals, including zip-2. bZIP proteins are one of four transcription factor classes with known roles in mammalian innate immunity, including NF-kappa B, IRF, and Stat (2, 27). In mammals, the bZIP transcription factor heterodimer AP-1 can be activated by JNK and p38 MAPK signaling downstream of interleukin and Tlr receptors. Additional work is required to determine what factors lie upstream of zip-2, and the mechanisms by which zip-2 is activated.
In summary, we have shown that the bZIP transcription factor zip-2 is an important factor in C. elegans innate immunity that mediates a transcriptional response, specifically to virulence by P. aeruginosa. zip-2 is required for induction of a subset of infection-response genes and is required for full resistance to P. aeruginosa infection. Further studies should provide insight into what particular aspect of virulence activates zip-2. More generally, these studies will shed light on the types of virulence factors detected by the innate immune system, what signaling pathways are used to respond to these factors, and how animals discriminate pathogenic from nonpathogenic bacteria.
Materials and Methods
Pathogen Infection Experiments.
Pathogen infection experiments were performed as described (28). In particular, for infection experiments with P. aeruginosa, overnight cultures of P. aeruginosa strain PA14 were seeded onto SK plates, then incubated at 37 °C for 24 h, followed by 25 °C for 24 h. Animals were washed onto plates and were harvested 4 h later for RT-qPCR experiments, or viewed 8 to 20 h later for GFP experiments. Nineteen other P. aeruginosa strains were grown in the same manner for experiments shown in S1 and Fig. S2. See SI Materials and Methodsfor information about experiments with other pathogens and with cadmium chloride.
Gene-Expression Analysis.
RNA extraction, reverse transcription, qRT-PCR, and microarray analysis were performed as described (14). qRT-PCR primer sequences available upon request. For all qRT-PCR experiments, each biological replicate was measured in duplicate and normalized to a control gene, which did not change expression upon infection (nhr-23 or snb-1). All qRT-PCR studies of P. aeruginosa induction were performed at 4 h postinoculation.
C. elegans Strains.
The deletion allele zip-2(tm4067) was a gift of the National BioResource Project, and was back-crossed four times to N2 before experiments. The deletion was confirmed by sequencing a PCR product amplified from this strain. See SI Materials and Methods for details on irg-1::GFP construction.
RNAi Experiments.
All experiments with feeding RNAi used an unc-22 positive control RNAi clone, which resulted in twitching animals in all experiments. The control for efficacy of pmk-1 RNAi was premature death of animals on P. aeruginosa.
Pathogen-Killing Experiments.
P. aeruginosa slow-killing experiments and S. aureus killing experiments were performed as described. Thirty to 50 L4 animals were transferred to a 3.5-cm plate with pathogen, using three plates per strain in each experiment. Survival was monitored over time until all animals had died. Experiments were repeated at least three independent times. Statistical analyses were performed with STATA software using the log-rank method to generate P-values.
Supplementary Material
Acknowledgments
We thank David Simon and Sean Curran for the transcription factor RNAi library, Zhi Yan for excellent technical support, and the National BioResource Project from Japan for the zip-2(tm4067) deletion. We thank the Caenorhabditis Genetics Center for some strains described in this paper. This work was supported by a Leukemia/Lymphoma Society Fellowship and a Moores Cancer Center Institutional Research Grant (to E.R.T.), a National Research Service Award (to J.P.), and National Institutes of Health Grants P01 AI044220 and R01 AI064332 (to F.M.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914643107/DCSupplemental.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.Hornef MW, Frisan T, Vandewalle A, Normark S, Richter-Dahlfors A. Toll-like receptor 4 resides in the Golgi apparatus and colocalizes with internalized lipopolysaccharide in intestinal epithelial cells. J Exp Med. 2002;195:559–570. doi: 10.1084/jem.20011788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryu JH, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila . Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans . Curr Opin Immunol. 2005;17(1):4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol. 2005;7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 9.Schulenburg H, Kurz CL, Ewbank JJ. Evolution of the innate immune system: the worm perspective. Immunol Rev. 2004;198(1):36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- 10.Moreau-Marquis S, Stanton BA, O’Toole GA. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 2008;21:595–599. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa . FEMS Microbiol Rev. 2009;33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee DG, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7(10):R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans . PLoS Genet. 2006;2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapira M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 17.Powell JR, Kim DH, Ausubel FM. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci USA. 2009;106:2782–2787. doi: 10.1073/pnas.0813048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troemel ER, Félix MA, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans . PLoS Biol. 2008;6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huffman DL, et al. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci USA. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reece-Hoyes JS, et al. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6(13):R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci USA. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmer F, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottar M, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amoutzias GD, et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol. 2007;24:827–835. doi: 10.1093/molbev/msl211. [DOI] [PubMed] [Google Scholar]
- 27.Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- 28.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol. 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.