Abstract
Sexual dimorphism is a widespread phenomenon and contributes greatly to intraspecies variation. Despite a long history of active research, the genetic basis of dimorphism for complex traits remains unknown. Understanding the sex-specific differences in genetic architecture for cranial traits in a highly dimorphic species could identify possible mechanisms through which selection acts to produce dimorphism. Using distances calculated from three-dimensional landmark data from CT scans of 402 baboon skulls from a known genealogy, we estimated genetic variance parameters in both sexes to determine the presence of gene-by-sex (G × S) interactions and X-linked heritability. We hypothesize that traits exhibiting the greatest degree of sexual dimorphism (facial traits in baboons) will demonstrate either stronger G × S interactions or X-linked effects. We found G × S interactions and X-linked effects for a few measures that span the areas connecting the face to the neurocranium but for no traits restricted to the face. This finding suggests that facial traits will have a limited response to selection for further evolution of dimorphism in this population. We discuss the implications of our results with respect to the origins of cranial sexual dimorphism in this baboon sample, and how the genetic architecture of these traits affects their potential for future evolution.
Keywords: Evolution, genetic correlation, genetic variance, heritability, morphometrics, X-linkage
Sexual dimorphism is common in many animal and plant species and often constitutes a major source of variation both within and between species. Given the importance of the effects of variation on morphological evolution, studies of sexual dimorphism and its impact on trait variation provide clues to the underlying processes of morphological evolution. Although we have a relatively detailed understanding of the phenotypic differences between sexes for many species, few studies have focused on the genetic underpinnings for these morphological differences and their evolution. To understand how a species will respond to selection, as well as other evolutionary mechanisms, we need to know the genetic architecture of the traits of interest.
The aim of this study is to measure the genetic variation associated with cranial dimorphism in the baboon. Baboons exhibit marked sexual dimorphism for most bodily systems, including aspects of cranial morphology (Schultz 1960; Sigg et al. 1982; Leigh and Cheverud 1991; Plavcan 2001, 2003). We use a sample of baboons of known pedigree from the captive baboon population housed at the Southwest National Primate Research Center (SFBR http://www.sfbr.org/pages/snprc_index.php) to investigate differences in the genetic architecture of cranial traits between the sexes. Differences in gene effects between the two sexes are the genetic source of variation in, and evolution of, phenotypic sexual dimorphism (Lande 1980).
Sexual dimorphism evolves through the differential response of males and females to selection but the exact mechanisms by which sexual dimorphism evolves remains elusive. Theory predicts that the evolution of sexual dimorphism can occur in at least two ways: gene-by-sex (G × S) interactions involving the autosomes and X-linked effects (Lande 1980; Fairbairn and Roff 2006). G × S interaction encompasses both variance dimorphism and the autosomal genetic correlation between the sexes (Lande 1980; Leutenegger and Cheverud 1982, 1985). G × S interaction can be perceived as a special case of gene-by-environment interaction (Falconer 1952; Roff 1997) where the sex of an individual is the “environment” in which its genes are expressed (Towne et al. 1992). The G × S interaction approach considers homologous traits as the same trait in males and females but expressed in a male or female environment. For both G × S interactions and X-linked effects, the genetic architecture of males and females differs, allowing each sex to respond differently to selection.
Our study uses both the G × S interaction approach and analysis of the effects of the X chromosome to examine the genetic architecture of sexual dimorphism in the baboon skull. We hypothesize that traits that show the greatest degree of phenotypic dimorphism will also exhibit significant G × S interaction and/or significant X-linkage. Although all cranial traits measured are significantly dimorphic in this baboon sample, facial traits measured on the muzzle display the greatest magnitude of dimorphism (K. Willmore, J. Rogers, J. Cheverud, and J. Richtsmeier, unpubl. ms.). Therefore, we expect these traits of the lower face to have significant G × S interactions and/or show significant X-linkage, but expect smaller G × S interactions or X effects for other suites of cranial traits measured on the orbit (upper face) and basicranium.
Materials and Methods
COMPOSITION OF THE SAMPLE
Our sample consists of 402 adult baboon skulls (113 males and 289 females) from the Southwest Foundation for Biomedical Research (SFBR) in San Antonio, TX. The baboons in the sample are Papio hamadryas, including two subspecies P. h. anubis (olive baboons) and P. h. cynocephalus (yellow baboons) and their hybrids, although most animals are pure P. h. anubis. The colony as a whole has about 300 founding animals and substantial levels of molecular polymorphism (Rogers et al. 2000). All of the animals used in this study are members of a known pedigree permitting analyses using quantitative genetic methods. Only adult baboons were used; the age range for our sample is 6.04 to 33.7 years. Throughout life the baboons have ad libitum access to commercial monkey chow, and are housed outside in social groups. Dietary and housing conditions are the same for males and females. In accordance with the Guide for the Care and Use of Laboratory Animals (Council 1996), animal care personnel and staff veterinarians provide daily maintenance and health care to all animals. All procedures are performed following the policies established by the Southwest Foundation Institutional Animal Care and Use Committee.
DATA ACQUISITION
Baboon heads collected from the SFBR animals at necropsy were macerated in a water bath. Computed tomography (CT) images were then acquired for each macerated skull at Washington University in St. Louis' Mallinkrodt Institute of Radiology using a medical CT scanner (Siemens Medical Systems, Erlangen, Germany). Pixel size ranged from 0.20 to 0.61 mm depending upon the size of the specimen and slice thickness was 0.75 mm. Three-dimensional (3D) reconstructions were produced from the CT image data and subsequently digitized to obtain 3D landmark coordinate data using eTDIPS, a multidimensional volume visualization and analysis software, codeveloped by the National Institutes of Health and the National University of Singapore. Three-dimensional coordinates of 30 biological landmarks were recorded from each skull that included 10 bilateral and 10 midline landmarks. We did not record calvarial landmarks as most of the individuals in the sample were necropsied without regard to the specific placement of the cut when the brain was removed. Supporting Figure S1 illustrates the landmark positions and Supporting Table S1 provides an anatomical description of each landmark (see Supporting information and our website www.getahead.psu.edu/hominid_index.html). Further information on landmarking can be found on the landmarks page at www.getahead.psu.edu. Measurement error of the data collected from CT images was evaluated using methods previously described by Valeri et al. (1998), and our repeatabilities varied from 0.936 to 0.998 with an average of 0.981. Landmarks were chosen for analyses based on accuracy of digitizing and to provide as complete coverage of the skull as possible. Measurement error was further minimized for those landmarks chosen for analysis by landmarking each skull twice, checking for gross errors such as side reversal and using the average of the two data collection trials in analyses.
DATA ANALYSIS
Interlandmark distances were estimated for each animal from the 3D landmark coordinate data. A subset of 16 interlandmark distances chosen on the basis of their effective sample size from an initial set of 35 interlandmark distances (Supporting Table S2) are used in our analysis. The effective sample size is the effective number of independent breeding values represented in the pedigree data (Cheverud 1996) or the number of individuals that make an independent genetic contribution to the next generation (Roff 1997, p. 401). Because of the pedigree structure of our baboon sample, the effective sample size for each distance is much lower than the 402 individuals for whom we have data. As such, we used only those distances that had an effective sample size of at least 25 (8 males and 17 females) to ensure the accurate estimation of genetic correlations and adequate power to detect genetic correlations between sexes that are less than one (Cheverud 1995, 1996). Another interpretation of this value is that it represents the number of infinite-sized sibship equivalents represented in the complex baboon genealogy. Each of the 16 linear distances was identified as belonging to the face, the basicranium, or the areas joining the face to the neurocranium (Supporting Table S2). Given the large difference in size between sexes, all distances were log-transformed before analyses to account for possible scaling effects.
GENE-BY-SEX INTERACTION
A significant G × S interaction for a given quantitative trait indicates that sexual dimorphism of that trait is heritable (Towne et al. 1992). We follow the method of Towne et al. (1992) to test for G × S interactions. This method involves three assumptions. First, the traits must be heritable. Using the program Sequential Oligogenic Linkage Analysis Routines (SOLAR; Almasy and Blangero 1998), we estimated the heritability of the 16 traits using a maximum-likelihood approach with sex and age as covariates. Second, variation in the quantitative characters has a polygenic basis. We are considering continuous traits that are generally accepted as having a polygenic basis (Frankham 1968; Wright 1968; Falconer and Mackay 1996; Roff 1997). Third, the loci influencing the traits are autosomal. To examine this last assumption, we tested for X-linked heritability using SOLAR and algorithms described by Kent et al. (2005a).
A brief description of the models used for SOLAR likelihood estimates and the estimation of the genetic covariance between the sexes is presented in the Supporting Information. We test the eight models described by Towne et al. (1992), allowing us to determine if there is a significant G × S interaction for each trait as well as whether the G × S interaction is due to a genetic correlation less than 1.0 and/or sex differences in the level of genetic variance. Supporting Table S3 outlines the parameter assumptions involved with each model. Model 1, considered the general model, estimates all of the parameters and allows for unequal genetic variation (σ2GM ≠ σ2GF), unequal environmental variation (σ2EM =σ2EF) and a genetic correlation of less than one between the sexes (ρG(M,F) < 1.0). Models 1 through 6 allow for G × S interaction, whereas models 7 and 8 do not permit G × S interactions. Each of these models were tested using a maximum-likelihood method with the program SOLAR with sex and age as covariates. Potential variance differences between sexes related to their differences in means were removed by log-transforming the distance data. Additionally, the data were Z-transformed to reduce issues of convergence encountered with untransformed data.
Because all parameters were estimated in the general model (Model 1) it is assumed to most closely match reality, therefore the rest of the models were compared with the general model and tested for goodness of fit. Two statistics were used to compare each model with the general model, a chi-square and Akaike's Information Criterion (AIC) test (see Towne et al. 1992). For each trait tested, the model with the most optimal fit was the one that had the lowest AIC value and according to the chi-square approximation is not significantly different from the general model. Additionally, we tested the optimal model with models that differed by only one parameter to test the significance of each estimated parameter. These additional comparisons were also done using chi-square and AIC tests.
X-LINKED EFFECTS
The test for X-linkage developed by Kent et al. (2005a) tests the fit of five models to the actual data using restricted maximum-likelihood estimates. The simplest or null model tests for the likelihood of no significant X effects. Because Kent et al. (2005b) found that constraining the environmental variances can decrease the sensitivity to detect a significant X effect, it is important to determine if environmental variances are affecting the genetic architecture before testing for X effects. Consequently, the second model allows the environmental variances to vary. The third model tests for mitochondrial linkage as mitochondrial effects can create asymmetric patterns of allele transmission and can be confounded with G × S interactions (Kent et al. 2005b). The fourth model tests for X-linkage with dosage compensation, whereas the fifth model tests for X-linkage with no dosage compensation. These tests are done sequentially for each trait, and at each step the more restricted model is retained unless a likelihood of a more complex model was significantly greater than the restricted model (P < 0.05) (Kent et al. 2005a,b). This analysis uses the pedigree data and estimates the X effect as a single parameter for both sexes rather than separately for each sex, thereby simplifying our interpretation of results.
Results
Heritability (h2) was estimated for 35 interlandmark distances and was statistically significant for all but one of the distances (Supporting Table S4). The average h2 for all distances is 0.45 with heritabilities ranging from 0.108 to 0.831, which is consistent with estimates for craniofacial h2 for other primate species (Cheverud 1982, 1995, 1996; Carson 2006). Linear distances with low heritability and an effective sample size < 25 (Cheverud 1995, 1996) were excluded from further analysis to assure sufficient information to test G × S interaction. The average h2 for the remaining subset of 16 distances is 0.58 with heritabilities ranging from 0.48 to 0.83 (Supporting Table S4 and Table 1).
Table 1.
Heritability estimates, estimated parameter values, and significant G×S interactions for the 16 distances. Narrow-sense heritabilities are represented by h2 and probabilities by P h2, ρG is the genetic correlation between sexes, σ2 GM and σ2 GF are the male and female genetic variances, respectively, and σ2 EM and σ2 EF are the male and female environmental variances, respectively. Statistically significant results are highlighted in bold.
| Distance | h2 | P h2 | ρG | σ2 GM | σ2 GF | σ2 EM | σ2 EF | G×S interaction description |
|---|---|---|---|---|---|---|---|---|
| Mda to mxt | 0.547±0.143 | 5.0×10−7 | 1.0 | 0.237±0.017 | 0.240±0.007 | 0.262±0.011 | 0.165±0.006 | |
| mda to zmi | 0.482±0.138 | 3.0×10−7 | 0.933±0.273 | 0.211±0.017 | 0.205±0.009 | 0.259±0.012 | 0.177±0.006 | |
| mxt to pns | 0.483±0.124 | 1.4×10−6 | 1.0 | 0.210±0.022 | 0.421±0.010 | 0.543±0.008 | 0.202±0.012 | |
| ans to nal | 0.675±0.129 | 3.0×10−11 | — | 0 | 0.253 | 0.408 | 0.091 | |
| zms to nal | 0.562±0.139 | 1.0×10−7 | 0.869±0.225 | 0.133±0.004 | 0.113±0.004 | 0.047±0.007 | 0.106±0.003 | |
| nal to vsj | 0.502±0.121 | 2.4×10−10 | — | 0 | 0 | 13.53 | 0.259 | |
| pns to vsj | 0.506±0.130 | 2.2×10−8 | 0.866±0.274 | 0.223±0.014 | 0.169±0.006 | 0.146±0.015 | 0.164±0.004 | ρG(M,F) < 1.0 |
| fzj to nas | 0.598±0.156 | 1.2×10−5 | 1.0 | 0.436 | 0.179 | 0 | 0.104 | and σGM ≠ σGF |
| cgp to fzj | 0.624±0.133 | 1.1×10−8 | 1.0 | 0.329±0.011 | 0.153±0.004 | 0.054±0.035 | 0.124±0.003 | |
| fzj to pmm | 0.544±0.139 | 1.0×10−6 | — | 0 | 0.162 | 0.256 | 0.077 | |
| fzj to zas | 0.624±0.111 | 1.2×10−12 | 1.0 | 0.406±0.014 | 0.253±0.004 | 0.115±0.029 | 0.186±0.004 | σGM ≠ σGF |
| bas to vsj | 0.676±0.121 | 3.7×10−9 | 1.0 | 0.481±0.013 | 0.328±0.008 | 0.184±0.019 | 0.135±0.011 | |
| bas to opi | 0.618±0.127 | 9.6×10−11 | 0.960±0.204 | 0.574±0.054 | 0.507±0.010 | 0.429±0.051 | 0.266±0.011 | |
| bas to pns | 0.831±0.10 | 9.2×10−31 | 1.0 | 0.425±0.002 | 0.249±0.002 | 0 | 0.070±0.004 | |
| bas to car | 0.514±0.115 | 3.1×10−9 | 1.0 | 0.239±0.009 | 0.235±0.007 | 0.193±0.008 | 0.259±0.005 | |
| cga to nas | 0.553±0.120 | 3.5×10−9 | — | 0 | 0.361 | 0.690 | 0.272 |
GENE-BY-SEX INTERACTION
Genetic correlations between sexes and genetic variances for males and females for each trait are listed in Table 1. Four traits failed to produce an estimate of the male genetic variance (ans to nal, nal to vsj, fzj to pmm, cga to nas) whereas only one trait failed to produce a female genetic variance (nal to vsj), most likely reflecting the higher percentage of females in our sample. Estimates of the genetic correlation between sexes for these traits are unreliable due to a lack of male genetic variance.
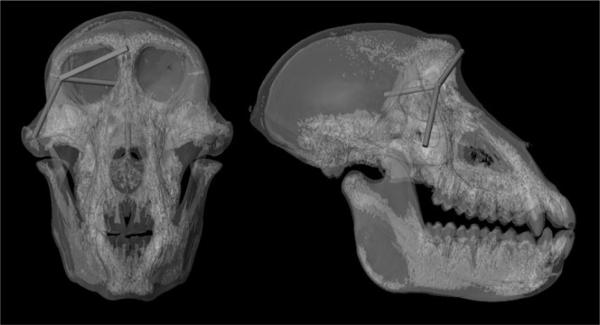
Over all traits in which sex-specific variances could be estimated, eight show higher genetic variances in males, one in females, and three have subequal variances, but genetic variance differences are only significant for two traits. Two traits linking the face and neurocranium have significant G × S interactions (Table 1, Fig. 1 distances represented by red lines) due to greater genetic variance in males.
Figure 1.
Illustration of the four distances that showed either a G×S interaction or X-linkage in this sample of baboon skulls from an anterior view (left-hand side) and a right lateral view (right-hand side). Red lines represent distances that had a significant G×S interaction, green lines represent distances with significant X-linkage.
Only the distance between the posterior nasal spine and the vomer-sphenoid junction (pns to vsj) has an intersex genetic correlation significantly less than 1.0 (ρG = 0.87). This is also the lowest correlation estimate obtained. The average intersex genetic correlation over the 12 traits with estimable correlations is 0.97. Genetic correlation between sexes for craniofacial morphology is quite high.
X-LINKED EFFECTS
X-linked h2 estimates range from 0.00 to 0.18, with an average of 0.04 and most fail to reach statistical significance. The two distances with significant X-linked heritability are frontal-zygomatic junction to nasion (fzj to nas), an orbital distance, and the posterior aspect of crista galli to the frontal-zygomatic junction (cgp to fzj), a joining distance between the face and the basicranium. The distance between basion to opisthion (bas to opi, measuring the anteroposterior diameter of the foramen magnum) is nearly significant with P = 0.07 (Table 2).
Table 2.
Narrow-sense X-linked heritability estimates (X-linked h2) and their probabilities (X-linked P h2), narrow-sense autosomal heritability estimates (Autosomal h2) and their probabilities (Autosomal P h2) as well as the probabilities of the five models tested for the X-linkage analysis (Kent et al. 2005a). Statistically significant results are highlighted in bold.
| Distance | X-linked h2 | X-linked P h2 | Autosomal h2 | Autosomal P h2 | No. of X Effect | Environmental effect | Mitochondrial effect | X effect Dosage compensation | X effect No dosage compensation |
|---|---|---|---|---|---|---|---|---|---|
| mda to mxt | 0.000 | 0.500 | 0.538±0.148 | 3.2×10−6 | 2.3×10−6 | 0.084 | 0.469 | 0.500 | 0.500 |
| mda to zmi | 0.000 | 0.500 | 0.493±0.143 | 5.0×10−7 | 2.7×10−7 | 0.123 | 0.366 | 0.500 | 0.500 |
| mxt to pns | 0.022±0.026 | 0.409 | 0.481±0.125 | 2.1×10−6 | 6.5×10−7 | 0.054 | 0.134 | 0.408 | 0.500 |
| ans to nal | 0.000 | 0.500 | 0.676±0.129 | 3.8×10−11 | 2.1×10−11 | 0.184 | 0.500 | 0.500 | 0.500 |
| zms to nal | 0.000 | 0.500 | 0.477±0.131 | 8.0×10−7 | 5.0×10−7 | 0.325 | 0.500 | 0.500 | 0.419 |
| nal to vsj | 0.000 | 0.500 | 0.532±0.126 | 8.9×10−11 | 4.4×10−11 | 0.564 | 0.500 | 0.500 | 0.500 |
| pns to vsj | 0.000 | 0.500 | 0.506±0.130 | 2.4×10−8 | 2.2×10−8 | 0.536 | 0.500 | 0.500 | 0.500 |
| fzj to nas | 0.159±0.067 | 0.026 | 0.587±0.158 | 1.8×10−5 | 1.8×10−5 | 0.065 | 0.443 | 0.028 | 0.496 |
| cgp to fzj | 0.183±0.071 | 0.016 | 0.609±0.134 | 3.4×10−8 | 3.1×10−8 | 0.156 | 0.090 | 0.016 | 0.382 |
| fzj to pmm | 0.000 | 0.500 | 0.553±0.142 | 1.5×10−6 | 7.7×10−7 | 0.184 | 0.346 | 0.500 | 0.500 |
| fzj to zas | 0.036±0.034 | 0.314 | 0.620±0.112 | 3.5×10−12 | 3.5×10−12 | 0.525 | 0.185 | 0.314 | 0.500 |
| bas to vsj | 0.000 | 0.500 | 0.698±0.121 | 1.9×10−9 | 2.1×10−9 | 0.024 | 0.077 | 0.500 | 0.500 |
| bas to opi | 0.139±0.075 | 0.076 | 0.644±0.130 | 1.3×10−10 | 7.7×10−11 | 0.053 | 0.500 | 0.074 | 0.500 |
| bas to pns | 0.000 | 0.500 | 0.867±0.103 | 4.5×10−29 | 9.1×10−30 | 0.321 | 0.500 | 0.500 | 0.500 |
| bas to car | 0.000 | 0.500 | 0.511±0.115 | 3.7×10−9 | 4.1×10−9 | 0.312 | 0.351 | 0.500 | 0.500 |
| cga to nas | 0.059±0.047 | 0.228 | 0.554±0.120 | 5.1×10−9 | 4.2×10−9 | 0.660 | 0.176 | 0.227 | 0.389 |
For all of the distances, the simplest model of no X-linkage is significant by log-likelihood (Table 2). The model that allows for environmental variances to vary (Model 2) is significant for the basicranial distance basion to vomer-sphenoid junction (bas to vsj) (Table 2), however, including this environmental effect did not uncover a significant X effect. Mitochondrial linkage is insignificant for all of the traits, as is X-linkage with no dosage compensation (Table 2). The two distances that show significant X-linkage with dosage compensation are the same distances that have significant X-linked h2: fzj to nas, and cgp to fzj (Fig. 1, distances represented by green lines).
Discussion
The aim of our study was to determine the underlying genetic architecture of cranial traits from a sample of baboons to gain an understanding of the mechanisms responsible for the evolution of cranial sexual dimorphism. Evolution of sexual dimorphism is constrained by the fact that males and females share the same autosomes over generations. Therefore, we hypothesized that cranial traits with the greatest degree of sexual dimorphism (i.e., facial traits in baboons) would also show G × S interactions for autosomes and/or evidence of X-linkage. Our results do not support our hypothesis; rather we find that distances that join the face to the neurocranium show evidence of G × S interactions and X-linkage (Fig. 1) whereas those showing the greatest dimorphism (facial traits) do not. Even so, the relative lack of significant G × S interactions, which is inversely related to the heritability of sexual dimorphism, indicates that sexual dimorphism heritability is quite low in this population despite moderate levels of heritability for variation in the traits themselves.
Our results counter theoretical predictions that sexually dimorphic traits should show evidence of the potential for evolution of sexual dimorphism (Lande 1980; Charlesworth and Charlesworth 1980; Rice 1984; Charlesworth et al. 1987; Fairbairn and Roff 2006). A possible explanation for these unexpected results is our relatively small sample size of 402 baboon skulls with a sex ratio (2♀:1♂) that reflects the demography of adult animals in the SFBR pedigreed colony. This sample is reduced to 25 effective independent individual breeding values (17 female and 8 male) even after limiting the traits analyzed to those with relatively high heritability (from 35 traits to 16 traits). Although our effective sample size is seemingly small (Ne = 25), it is fairly high relative to other published studies of heritability of cranial traits in primates and humans (Cheverud 1988, 1995, 1996; Carson 2006).
Although it is difficult to determine the exact effect that management of this colony has on the genetic architecture of our sample, the colony at SFBR has been maintained for about 40 years (approximately six to seven generations) and patterns of mating are determined by colony managers. We propose that the relatively short period of time that these baboons have been captive has not dramatically altered their genetic architecture.
Although our results differed from our expectations, they are consistent with results of similar studies reported in the literature. Only one trait in these baboons had an intersex correlation significantly less than 1.0 and that correlation is still quite high (ρG = 0.87). All other estimable correlations were greater than this, limiting the potential for the evolution of dimorphism through differences in selection between the sexes in this population (Charlesworth and Charlesworth 1980; Lande 1980; Rice 1984; Charlesworth et al. 1987; Towne et al. 1992; Havill et al. 2004; Fairbairn and Roff 2006). Our results support previous studies that have generally reported high intersex correlations for morphological traits in mammals (Eisen and Legates 1966; Rogers and Mukherjee 1992), invertebrates (Cowley and Atchley 1988; Reeve and Fairbairn 1996), birds (Merilä et al. 1998; Jensen et al. 2003), and even plants (Meagher 1994; Ashman 2003). Differences in genetic variances between sexes have been commonly reported in the literature for a variety of animals, suggesting that variance dimorphism might be the common mechanism for the evolution of sexual dimorphism (Shaklee et al. 1952; Eisen and Legates 1966; Cowley et al. 1986; Rogers and Mukherjee 1992; Jensen et al. 2003). Only two traits had statistically significantly higher genetic variance in males than in females in our study. Although few individual differences were statistically significantly different between the sexes, male genetic variance estimates were generally higher than female estimates with an average increase in variance of 38% across all traits.
We found only two instances of significant X-linked heritability. The average X-linked heritability for the traits was only 0.04 whereas the average autosomal heritability is 0.55 (all were significant). Again, most genetic variation in craniofacial traits is autosomal (93%) and only a small portion is X-linked (7%). Based on the human genome and assuming a similar proportional relationship, the X-chromosome contains about 5% of the base pairs in the primate genome so that the X-linked percentage of genetic variance for cranial traits analyzed here is approximately proportional to the size of the X chromosome.
In summary, our analysis reveals a limited capacity for the evolution of cranial dimorphism in this population. All intersex genetic correlations are 0.87 or greater and X-linked genetic variation is small, proportional to its contribution to total genome size. Male genetic variance estimates were at least 10% greater than female genetic variance estimates for 75% of the traits studied, but only two traits showed significantly larger genetic variances in males. Because cranial sexual dimorphism is marked in the baboon lineage, either the genetic architecture is different now than in the past, or the evolution of dimorphism has been very slow to occur in this lineage. Given that the genetic architecture of this population is similar to what is commonly found for morphological traits in most mammals (high intersex genetic correlations, larger male genetic variances), we hypothesize that the evolution of cranial dimorphism in baboons has been facilitated by genetic variance differences between the sexes.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Hildebolt for advice and aid in preparing the CT scans of the baboon skulls and J. Kent for providing us with the programs used to run the X-linked analyses in SOLAR. We would also like to thank all of the members of the Genomics of Cranial Morphology Consortium for their insightful comments and suggestions that greatly improved this study. Funding for this study was provided by NSF grants BCS 0522112, BCS 0523305, BCS 0523637, BCS 0725031, BCS 0725068, and BCS 0725227. The research used facilities and/or resources supported by the base grant for the Southwest National Primate Research Center (P51-RR013986) and facilities improvement grants C06-RR014578 and C06-RR013556 from the National Center for Research Resources, NIH.
Footnotes
Supporting Information The following supporting information is available for this article:
Supporting Information may be found in the online version of this article.
(This link will take you to the article abstract).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting informations supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
LITERATURE CITED
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman T-L. Constraints on the evolution of males and sexual dimorphism: field estimates of genetic architecture of reproductive traits in three populations of gyndodioecious Fragaria virginiana. Evolution. 2003;57:2012–2025. doi: 10.1111/j.0014-3820.2003.tb00381.x. [DOI] [PubMed] [Google Scholar]
- Carson AE. Maximum likelihood estimation of human craniometric heritabilities. Am. J. Phys. Anthropol. 2006;131:169–180. doi: 10.1002/ajpa.20424. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Sex-differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 1980;35:205–214. doi: 10.1017/s0016672300014051. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Cheverud JM. Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution. 1982;36:499–516. doi: 10.1111/j.1558-5646.1982.tb05070.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. A comparison of genetic and phenotypic correlations. Evolution. 1988;42:958–968. doi: 10.1111/j.1558-5646.1988.tb02514.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Morphological integration in the saddle-back tamarin (Saguinus fuscicollis) cranium. Am. Nat. 1995;145:63–89. [Google Scholar]
- Cheverud JM. Quantitative genetic analysis of cranial morphology in the cotton-top (Saguinus oedipus) and saddle-back (S. fuscicollis) tamarins. J. Evol. Biol. 1996;9:5–42. [Google Scholar]
- Council NR. Guide for the care and use of laboratory animals. National Academy of Sciences; Washington, DC: 1996. [Google Scholar]
- Cowley DE, Atchley WR, Rutledge JJ. Quantitative genetics of Drosophila melanogaster. I. Sexual dimorphism in genetic parameters for wing traits. Genetics. 1986;114:549–566. doi: 10.1093/genetics/114.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley DE, Atchley WR. Quantitative genetics of Drosophila melanogaster. II. Heritabilities and genetic correlations between sexes for head and thorax traits. Genetics. 1988;119:421–433. doi: 10.1093/genetics/119.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen EJ, Legates JE. Genotype-sex interaction and the genetic correlation between the sexes for body weight in Mus musculus. Genetics. 1966;54:611–623. doi: 10.1093/genetics/54.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Roff DA. The quantitative genetics of sexual dimorphism: assessing the importance of sex-linkage. Heredity. 2006;97:319–328. doi: 10.1038/sj.hdy.6800895. [DOI] [PubMed] [Google Scholar]
- Falconer DS. The problem of environment and selection. Am. Nat. 1952;86:293–298. [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4th ed. Pearson Prentice Hall; London, UK: 1996. [Google Scholar]
- Frankham R. Sex and selection for a quantitative character in Drosophila: II. The sex dimorphism. Aust. J. Biol. Sci. 1968;21:1225–1237. doi: 10.1071/bi9681225. [DOI] [PubMed] [Google Scholar]
- Havill LM, Mahaney MC, Rogers J. Genotype-by-sex and environment-by-sex interactions influence variation in serum levels of bone-specific alkaline phosphatase in adult baboons (Papio hamadryas) Bone. 2004;35:198–203. doi: 10.1016/j.bone.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Jensen H, Sæther BE, Ringsby TH, Tufto J, Griffiths SC, Ellegren H. Sexual variation in heritability and genetic correlations of morphological traits in house sparrow (Passer domesticus) J. Evol. Biol. 2003;16:1296–1307. doi: 10.1046/j.1420-9101.2003.00614.x. [DOI] [PubMed] [Google Scholar]
- Kent JW, Jr., Dyer TD, Blangero J. Estimating the additive genetic effect of the X chromosome. Genet. Epidemiol. 2005a;29:377–388. doi: 10.1002/gepi.20093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JW, Jr., Lease LR, Mahaney MC, Dyer TD, Almasy L, Blangero J. X chromosome effects and their interactions with mitochondrial effects. BMC Genet. 2005b;6(Suppl 1):S157. doi: 10.1186/1471-2156-6-S1-S157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Leigh SR, Cheverud JM. Sexual dimorphism in the baboon facial skeleton. Am. J. Phys. Anthropol. 1991;84:193–208. doi: 10.1002/ajpa.1330840209. [DOI] [PubMed] [Google Scholar]
- Leutenegger W, Cheverud JM. Correlates of sexual dimorphism in primates: ecological and size variables. Int. J. Primatol. 1982;3:387–402. [Google Scholar]
- Leutenegger W, Cheverud JM. Sexual dimorphism in primates: the effects of size. In: Jungers WL, editor. Size and scaling in primate biology. Plenum Press; New York, NY: 1985. pp. 33–50. [Google Scholar]
- Meagher TR. The quantitative genetics of sexual dimorphism in Silene latifolia (Caryophllaceae). II. Response to sex-specific selection. Evolution. 1994;48:939–951. doi: 10.1111/j.1558-5646.1994.tb05284.x. [DOI] [PubMed] [Google Scholar]
- Merilä J, Sheldon BC, Ellegren H. Quantitative genetics of sexual size dimorphism in the collared flycatcher, Ficedula albicollis. Evolution. 1998;52:870–876. doi: 10.1111/j.1558-5646.1998.tb03711.x. [DOI] [PubMed] [Google Scholar]
- Plavcan JM. Sexual dimorphism in primate evolution. Yrbk. Am. J. Phys. Anthropol. 2001;44:25–53. doi: 10.1002/ajpa.10011.abs. [DOI] [PubMed] [Google Scholar]
- Plavcan JM. Scaling relationships between craniofacial sexual dimorphism and body mass dimorphism in primates: implications for the fossil record. Am. J. Phys. Anthropol. 2003;120:38–60. doi: 10.1002/ajpa.10154. [DOI] [PubMed] [Google Scholar]
- Reeve JP, Fairbairn DJ. Sexual size dimorphism as a correlated response to selection on body size: an empirical test of the quantitative genetic model. Evolution. 1996;50:1927–1938. doi: 10.1111/j.1558-5646.1996.tb03580.x. [DOI] [PubMed] [Google Scholar]
- Rice WM. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Roff DA. Evolutionary quantitative genetics. Chapman and Hall; New York, NY: 1997. [Google Scholar]
- Rogers AR, Mukherjee A. Quantitative genetics of sexual dimorphism in human body size. Evolution. 1992;46:226–234. doi: 10.1111/j.1558-5646.1992.tb01997.x. [DOI] [PubMed] [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriquez LA, Rice KS, Slifer SH, Perelygin A, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatelite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- Schultz AH. Age changes and variability in the skulls and teeth of the Central American monkeys Alouatta, Cebus, and Ateles. Proc. Zool. Soc. Lond. 1960;133:337–390. [Google Scholar]
- Shaklee WE, Knox CW, Marsden SJ. Inheritance of the sex difference of body weight in turkeys. Poultry Sci. 1952;31:822–825. [Google Scholar]
- Sigg H, Stolba A, Abegglen JJ, Dasser V. Life history of hamadryas baboons: physical development, infant mortality, reproductive parameters and family relationships. Primates. 1982;23:473–487. [Google Scholar]
- Towne B, Blangero J, Mott GE. Genetic analysis of sexual dimorphism in serum apo A1 and HDL-C concentrations in baboons. Am. J. Primatol. 1992;27:107–117. doi: 10.1002/ajp.1350270206. [DOI] [PubMed] [Google Scholar]
- Valeri CJ, Cole TM, III, Lele S, Richtsmeier JT. Capturing data from three-dimensional surfaces using fuzzy landmarks. Am. J. Phys. Anthropol. 1998;107:113–124. doi: 10.1002/(SICI)1096-8644(199809)107:1<113::AID-AJPA9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and genetics of populations: genetic and biometric foundations. Volume 1. Univ. of Chicago Press; Chicago: 1968. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.