Abstract
Myosin light chain kinase is a Ca2+/calmodulin-dependent protein kinase which exhibits a very high degree of protein substrate specificity. The regulatory light chain of myosin is the only known physiological substrate of the enzyme. Based upon epitope mapping of monoclonal antibodies which inhibit kinase activity competitively with respect to the light chain substrate, residues 235–319 of the rabbit skeletal muscle kinase have been proposed to contain a light chain-binding site (Herring, B. P., Stull, J. T., and Gallagher, P. J. (1990) J. Biol. Chem. 265, 1724–1730). With the expression of a truncated kinase, we have further localized this putative binding site to residues 235–294. Mutation of acidic residues at positions 269 and 270 of the kinase resulted in a 10-fold increase in the Km value for the myosin light chain, with no significant change in the Vmax value. In contrast, altering a cluster of acidic amino acids at positions 261–263 had little effect on the Km value for the myosin light chain. These results suggest that residues 269 and 270 may be involved in protein-substrate binding. Interestingly, these residues, located amino-terminal of the homologous catalytic core (positions 302–539), are in a region which is highly conserved among myosin light chain kinases, but not other protein kinases. It is probable that the homologous catalytic core contains structural elements required for phosphotransferase activity. The catalytic domain of myosin light chain kinase would therefore include these conserved elements together with additional specific substrate-binding residues.
Phosphorylation of myosin regulatory light chains by myosin light chain kinases causes potentiation of contraction in skeletal muscle (Stull et al., 1986; Sweeney and Stull, 1990) and is responsible for the initiation of contraction in smooth muscle (Kamm and Stull, 1985). Myosin light chain kinases are completely dependent on Ca2+/calmodulin for activation (see Stull et al. (1986) for review). Distinct forms of the kinase exist in smooth and skeletal muscle (Stull et al., 1985). Skeletal muscle isoforms of the enzyme have been particularly well-characterized biochemically and physically. The complete amino acid sequences of the rabbit (607 residues; Takio et al., 1986; Herring et al., 1990), rat (610 residues; Roush et al., 1988), and chicken (825 residues; Leachman et al., 1989) skeletal muscle myosin light chain kinases have been deduced by direct amino acid sequencing and cDNA cloning. A shape model of the kinase has been proposed in which the amino terminus of the kinase forms an asymmetric, rod-like tail with no enzymatic activity (Mayr and Heilmeyer, 1983). The carboxyl-terminal portion of the kinase forms a globular structure containing the catalytic and regulatory domains.
The globular region of the myosin light chain kinase is highly conserved among different animal species. In addition, residues 302–539 are homologous to the catalytic domains of other protein kinases. These residues have therefore been referred to as the catalytic domain of the skeletal muscle myosin light chain kinase (Hanks et al., 1988; Takio et al., 1986). Consistent with this hypothesis, an active proteolytic fragment comprising residues 256–584 of the rabbit kinase can be generated following limited proteolytic digestion with chymotrypsin (Edelman et al., 1985). Results from these proteolysis studies together with synthetic peptide studies have demonstrated that the calmodulin-binding domain of the kinase is located at the extreme carboxyl terminus of the enzyme between residues 577 and 593 (Edelman et al., 1985; Blumenthal et al., 1985).
The catalytic domain of myosin light chain kinase has been further defined using monoclonal antibodies (Nunnally et al., 1987; Herring et al., 1990). The epitopes for several monoclonal antibodies (12a, 9a, and 2a) which inhibit kinase activity competitively with respect to the myosin light chain substrate have been localized to residues 235–319. We have proposed that a light chain-binding site may be located within this region, immediately amino-terminal of the homologous catalytic core of the kinase (Herring et al., 1990). However, residues 235–319 overlap with a small part of the catalytic core (positions 302–539) and include residues known to be involved in ATP binding (Hanks et al., 1988; Takio et al., 1986). Thus, to identify more precisely the localization of the proposed light chain-binding site, we have further mapped the epitopes for the monoclonal antibodies to residues 235–294 which are amino-terminal of the homologous catalytic region. We demonstrate that alteration of acidic amino acids at positions 269 and 270 within the proposed light chain-binding domain results in a 10-fold increase in the Km for the light chain substrate. These data suggest that acidic residues 269 and 270 are part of the light chain substrate-binding site on skeletal muscle myosin light chain kinase.
EXPERIMENTAL PROCEDURES
Protein Purification and Kinase Assays
Rabbit skeletal muscle myosin light chain kinase was purified as described previously (Nunnally et al., 1985) with the modifications of Herring et al. (1990). Rabbit skeletal muscle myosin light chains were purified according to Blumenthal and Stull (1980). Calmodulin was purified from bovine testes (Blumenthal and Stull, 1982). COS cell lysates were prepared as described previously (Herring et al., 1990). Myosin light chain kinase activity, present in the COS cell lysates, was measured as 32P incorporation into the rabbit skeletal muscle regulatory light chain (Blumenthal and Stull, 1980). Lysates were diluted 100–500-fold into the assay mixture. Under these conditions, the activity of all the expressed myosin light chain kinases was completely Ca2+/calmodulin-dependent; in addition, lysates prepared from nontransfected COS cells had no detectable kinase activity (using either rabbit skeletal muscle light chains or the synthetic peptide KKRAARATSNVFA as a substrate). The light chain concentration present in each assay was calculated from the measured specific activity of the ATP and the maximum 32P incorporation. The quantity of myosin light chain kinase present in COS cell lysates was determined by immunoblotting with purified rabbit skeletal muscle myosin light chain kinase as a standard (Herring et al., 1990).
Oligonucleotide-directed Mutagenesis
Oligonucleotide-directed mutagenesis was carried out as described by Zoller and Smith (1984). The oligonucleotide 5′-TGGAAACAGTTCTGCTGCCTGGCT-3′ was used to generate the mutant LCB1 (Fig. 1), and the oligonucleotide 5′-ACAGTTCTCCTGCCTGGCT-3′ was used to generate LCB2 (Fig. 1). The LCB3 mutant was generated with LCB2 as a template for the mutagenic LCB1 oligonucleotide. All mutations were confirmed by DNA sequencing.
Fig. 1. Domain organization of and proposed light chain-binding site on rabbit skeletal muscle myosin light chain kinase.
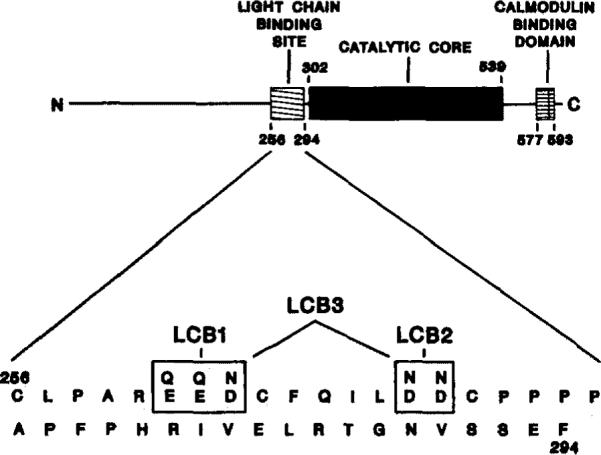
A schematic representation of the linear amino acid sequence and domain organization of rabbit skeletal muscle myosin light chain kinase is shown (top). The catalytic core is the region homologous to other protein kinases (Hanks et al., 1988); the calmodulin-binding domain was defined by Blumenthal et al. (1985); a light chain-binding region is defined from results presented in this paper. The amino acid sequence of the proposed light chain-binding region is shown (bottom). The acidic residues (positions 261–263 and 269 and 270) which were altered in this present study are boxed. The substituted amino acids are indicated above the native residues. The nomenclature of the mutated kinases is also indicated.
Expression of Wild-type and Mutant Myosin Light Chain Kinases
Wild-type and mutated myosin light chain kinase cDNAs were subcloned into a pCMV2 expression vector (Andersson et al., 1989). Expression was then performed in COS cells as described previously (Herring et al., 1990).
Deletion Mutagenesis
A truncated kinase encoding residues 1–293 was constructed by subcloning the EcoRI restriction fragment of the 5′-cDNA described previously (Herring et al., 1990) into the pCMV2 vector.
RESULTS
Mutant Kinases
We previously proposed (Herring et al., 1990) that, since monoclonal antibodies 12a, 9a, and 2a inhibited kinase activity competitively with respect to the light chain substrate, the epitopes for these antibodies may constitute a myosin light chain substrate-binding site on myosin light chain kinase. The epitopes for these antibodies were localized to residues 235–319. The catalytic core of the kinase begins at residue 302 with a conserved hydrophobic residue followed by the glycine cluster and the nucleotide-reactive lysine at position 325 and extends carboxyl-terminal to an invariant arginine at position 539. To identify more precisely the localization of the antibody epitopes and thus the proposed light chain-binding site, an additional truncated kinase was constructed. The truncated kinase (residues 1–293) was expressed in COS cells and analyzed by immunoblot. Each of the inhibitory monoclonal antibodies (12a, 2a, and 9a) reacted with this truncated kinase (Table I). This result further restricts the epitopes for these antibodies to residues 235–293 and indicates that the proposed myosin light chain-binding site is located amino-terminal of the catalytic core of the kinase.
Table I. Kinetic and immunological properties of wild-type and mutant myosin light chain kinases.
Kinetic parameters were determined from double-reciprocal plots. Each assay was performed in duplicate; the means ± S.E. obtained from multiple assays are presented. Immunoblots were performed as described previously (Herring et al. 1990). Examples of the blots are shown in Fig. 3. The reactivity of each monoclonal antibody to the mutant kinases is summarized; a plus sign indicates detectable reactivity, and a minus sign indicates that no reactivity was detected.
| Kinase | Km | Vmax | Antibody reactivity |
||||
|---|---|---|---|---|---|---|---|
| 16a | 14a | 12a | 9a | 2a | |||
| μM | pmol32P/min/ng | ||||||
| WTa | 3.5 ± 0.6 (n = 5) | 22.8 ± 4.6 | + | + | + | + | + |
| LCB1 | 9.4 ± 0.9 (n = 12) | 18.3 ± 1.4 | − | + | + | − | + |
| LCB2 | 43.7 ± 4.6 (n = 5) | 32.1 ± 3.9 | + | + | − | + | − |
| LCB3 | 30.7 ± 2.3 (n = 6) | 20.9 ± 1.4 | − | + | − | − | − |
| Nt | 0 | + | + | + | + | + | |
WT, wild-type myosin light chain kinase expressed in COS cells; Nt, truncated kinase (residues 1–294).
The substrate determinants on the myosin light chain are two groups of basic residues located amino-terminal of the phosphorylatable serine residue (Michnoff et al., 1986). These residues probably form ionic interactions with acidic residues within the substrate-binding site on the kinase. Three mutant myosin light chain kinases were constructed in which acidic amino acids within the proposed light chain-binding region were altered to neutral residues. For the LCB1 mutant, acidic residues EED (positions 261–263) were mutated to QQN; and for the LCB2 mutant, acidic residues DD (positions 269 and 270) were changed to NN (Fig. 1). The double-mutant LCB3 has both groups of acidic residues altered (Fig. 1). The expressed mutant kinases were quantitated in the COS cell lysates by denistometric scanning of immunoblots with purified myosin light chain kinase standards. The level of expression of each mutant was similar to the wild-type kinase.
Myosin Light Chain Kinase Activity
Km determinations were made by varying the concentrations of skeletal muscle myosin light chain in kinase assays. The Km and Vmax values for each mutant and wild-type kinase expressed in COS cells were determined from double-reciprocal plots (Fig. 2 and Table I). The Km value (±S.E.) determined for the myosin light chain with the wild-type kinase was 3.5 ± 0.6 μM; for LCB1, 9.4 ± 0.9 μM; for LCB2, 43.7 ± 4.6 μM; and for LCB3, 30.7 ± 2.3 μM. The Km values for all the mutant kinases were significantly greater than that of the wild-type enzyme. Furthermore, the mutant kinase LCB2 had a Km value of 40 μM for the synthetic peptide substrate (KKRAARATSNVFA), compared to a value of 8.6 μM reported for the native tissue purified kinase (Michnoff et al., 1986).
Fig. 2. Kinetic analysis of mutant myosin light chain kinases.
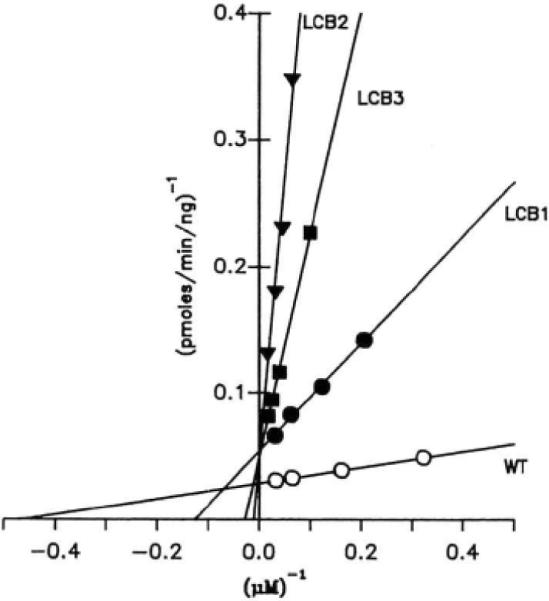
Kinase assays were performed in duplicate as described under “Experimental Procedures.” Results from four individual assays are plotted with linear regression lines drawn through the data. A summary of multiple analyses is presented in Table I.
There were no significant differences in the Vmax values of the expressed mutant kinases (LCB1, LCB2, and LCB3) compared to the wild-type enzyme (Table I). The truncated kinase (residues 1–293) lacking the catalytic domain, as expected, had no enzyme activity.
Immunoblots of Mutant Myosin Light Chain Kinases
Wild-type and mutant myosin light chain kinases were analyzed for their ability to react with each of the inhibitory monoclonal antibodies. Monoclonal antibody 14a reacted with the wild-type kinase together with all the mutant kinases. This was used as a control antibody as its epitope had previously been localized to residues 165–173 (Herring et al., 1990). Antibodies 12a and 2a bound only to the LCB1 mutant in addition to the native enzyme, whereas antibody 9a bound only to the LCB2 mutant (Fig. 3 and Table I). A fifth noninhibitory antibody (16a) reacted with the mutant myosin light chain kinases in a manner identical to antibody 9a (Table I).
Fig. 3. Immunoblot analysis of mutant myosin light chain kinases.
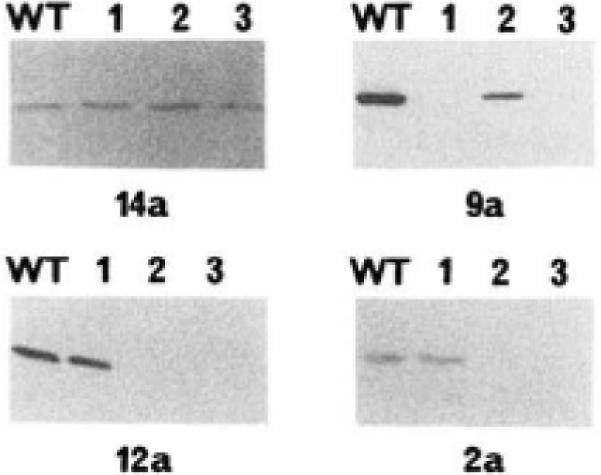
Immunoblots were performed as described previously (Herring et al., 1990); ~20 ng of kinase were electrophoresed in each lane. The order of loading of each immunoblot was identical. Lane WT, wild-type kinase; lane 1, mutant LCB1; lane 2, mutant LCB2; lane 3, double-mutant LCB3. The immunoblots were reacted with the monoclonal antibody indicated below each blot.
DISCUSSION
We have postulated that antibodies 2a, 9a, and 12a may bind to a light chain-binding site, located between residues 235 and 319, on rabbit skeletal muscle myosin light chain kinase (Herring et al., 1990). This region overlaps part of the catalytic core (residues 302–539) and includes residues known to be important for ATP binding (Hanks et al., 1988). Immunoblotting of a truncated kinase (residues 1–294), expressed in COS cells, demonstrated that the epitope for the antibodies and hence the proposed light chain-binding site is located amino-terminal of these conserved residues in the catalytic core. All of the monoclonal antibodies reacted with the truncated kinase; thus, the proposed light chain-binding region can be further localized to residues 235–294 (Table I).
A catalytically active 35-kDa chymotryptic fragment of the rabbit skeletal muscle myosin light chain kinase (residues 256–584) has a similar Km value for the light chain substrate as compared to the intact kinase (Edelman et al., 1985; Mayr and Heilmeyer, 1983). Thus, it is likely that binding sites for the light chain are located within the chymotryptic fragment. These data together with the experiments performed with the truncated mutant are consistent with the hypothesis that a light chain-binding site may be located between residues 256 and 294 (Fig. 1). In support of this hypothesis, the marked similarity (~95%) among the catalytic core regions (residues 302–539) of the skeletal muscle myosin light chain kinases from rabbit, rat, and chicken (Takio et al., 1986; Roush et al., 1988; Herring et al., 1989; Leachman et al., 1989) also extends amino-terminal to residue 260 of the rabbit kinase and would therefore include this proposed light chain-binding region.
Synthetic peptide studies have shown that two groups of basic residues located amino-terminal of the phosphorylatable serine residue in the myosin light chain are important for the interaction of the kinase with its substrate (Michnoff et al., 1986). It is likely that these basic residues form ionic interactions with acidic residues within the kinase. Within the proposed light chain-binding region there are two groups of acidic amino acids, EED (positions 261–263) and DD (positions 269 and 270); other acidic residues are only found singularly (Fig. 1). We tested the hypothesis that these two groups of acidic amino acids were involved in substrate binding. The acidic residues were altered to neutral amino acids, and the effect of the mutations on the Km values for the myosin light chain were determined. If either group of acidic residues form ionic interactions with the light chain substrate, alteration of their ionic charge would be expected to weaken this interaction. This would be predicted to cause an increase in the Km for the light chain substrate. In addition, if one or both of these groups comprise part of the epitopes of the monoclonal antibodies, the mutations would be predicted to decrease the affinity of the antibodies for the mutant kinases. If this decrease in affinity is sufficiently large, the antibodies may no longer be able to detect the mutant kinases by immunoblot analysis.
The mutant kinase LCB2 and the double-mutant LCB3 have Km values which are ~10-fold greater than that of the wild-type kinase. This is consistent with the hypothesis that residues 269 and 270 are important for substrate binding. Monoclonal antibodies 12a and 2a no longer detect the LCB2 mutant kinase or the double mutant (LCB3) on immunoblots, although they readily react with both the wild-type and LCB1 kinases. Thus, residues 269 and 270 comprise part of the epitope for these antibodies. This observation together with the kinetic data support the hypothesis that the epitopes for antibodies 12a and 2a include part of the light chain-binding site on the myosin light chain kinase.
The mutant kinase LCB1 has a slightly higher Km value (2.5-fold greater) for the isolated myosin light chain than the wild-type kinase. However, the double mutant (LCB3) does not have a greater Km than the mutant kinase LCB2 (Fig. 2 and Table I). These data suggest that residues 261–263 are of either minor or no importance in light chain binding. It is possible that the altered residues in LCB1 may have produced a structural change which has been manifest as a small increase in the Km value for the light chain substrate. Monoclonal antibody 9a reacts with mutant LCB2 and the wildtype kinase, but not with LCB1 or LCB3, on immunoblots (Fig. 3 and Table I). Thus, residues 261–263 comprise part of the epitope for this antibody. However, an additional antibody (16a), which does not inhibit kinase activity, also did not react with the LCB1 mutant (Table I). Thus, this antibody also includes residues 261–263 as part of its epitope. The binding of antibody 9a to these residues alone is therefore unlikely to account for the inhibitory activity of this antibody. It is probable that antibody 9a exerts its inhibitory effect by sterically blocking substrate binding to residues 269 and 270. The epitope for antibody 9a may extend, either in the primary sequence or in the tertiary structure of the kinase, closer to residues 269 and 270 than the epitope for the noninhibitory antibody, 16a.
From our model it would be predicted that acidic residues 269 and 270 are interacting with the basic substrate determinants of the myosin light chain. To evaluate which of the basic residues in the substrate may be involved in this interaction, a kinetic analysis of the LCB2 mutant was performed with a synthetic peptide substrate modeled after the smooth muscle light chain. When the peptide KKRAARATSNVFA was used as a substrate as compared to the skeletal muscle light chain (amino-terminal sequence PKKAKRRAAAEGGSSNVFS), identical Km values were obtained. The determinants which are common to this peptide and the skeletal muscle light chain are the basic residues at positions 5–7 of the skeletal light chain. Previously, it has been shown (Michnoff et al., 1986) that substitution of the basic residue at position 1 of the synthetic peptide had little effect on the Km value of the mutant peptide. Thus, it would be predicted that it is the basic residues at positions 6 and 7 of the skeletal muscle light chain that are interacting with residues 269 and 270 in the kinase. Additional mutations together with chemical cross-linking studies will be required to test further this prediction.
In summary, the data obtained from our mutagenesis studies provide evidence that residues 269 and 270 comprise part of the substrate-binding site on rabbit skeletal muscle myosin light chain kinase. These residues are located amino-terminal of the catalytic core of the kinase (defined by its homology to the catalytic region of all other protein kinases). Similarly, it has recently been proposed that a group of acidic residues located outside the catalytic core of cAMP-dependent protein kinase (carboxyl-terminal) is involved in substrate binding (Buechler and Taylor, 1990). Thus, we propose that the catalytic core of protein kinases contains the residues required to catalyze the phosphotransferase reaction, but the catalytic domain of the kinase includes residues which may be outside the catalytic core. The organization of these additional residues may be unique to each protein kinase and may contribute to the substrate specificity of each kinase.
Acknowledgments
This work was supported by National Institutes of Health Grant HL06296.
REFERENCES
- Andersson S, Davis DL, Dahlbäch H, Jörnvall H, Russell DW. J. Biol. Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- Blumenthal DK, Stull JT. Biochemistry. 1980;19:5608–5614. doi: 10.1021/bi00565a023. [DOI] [PubMed] [Google Scholar]
- Blumenthal DK, Stull JT. Biochemistry. 1982;21:2386–2391. doi: 10.1021/bi00539a017. [DOI] [PubMed] [Google Scholar]
- Blumenthal DK, Takio K, Edleman AM, Charbonneau H, Titani K, Walsh KA, Krebs EG. Proc. Natl. Acad. Sci. U. S. A. 1985;82:3187–3191. doi: 10.1073/pnas.82.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler JA, Taylor SS. Biochemistry. 1990;29:1937–1943. doi: 10.1021/bi00459a039. [DOI] [PubMed] [Google Scholar]
- Edelman AM, Takio K, Blumenthal DK, Hansen RS, Walsh KA, Titani K, Krebs EG. J. Biol. Chem. 1985;260:11275–11285. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Herring BP, Nunnally MH, Gallagher PJ, Stull JT. Am. J. Physiol. 1989;256:C399–C404. doi: 10.1152/ajpcell.1989.256.2.C399. [DOI] [PubMed] [Google Scholar]
- Herring BP, Stull JT, Gallagher PJ. J. Biol. Chem. 1990;265:1724–1730. [PMC free article] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. Am. J. Physiol. 1985;249:C238–C247. doi: 10.1152/ajpcell.1985.249.3.C238. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Herring BP, Gallagher PJ, Stull JT. J. Cell Biol. 1989;107:678a. abstr. [Google Scholar]
- Mayr GW, Heilmeyer LMG., Jr. FEBS Lett. 1983;157:225–231. doi: 10.1016/0014-5793(83)80552-3. [DOI] [PubMed] [Google Scholar]
- Michnoff CH, Kemp BE, Stull JT. J. Biol. Chem. 1986;261:8320–8326. [PubMed] [Google Scholar]
- Nunnally MH, Rybicki SB, Stull JT. J. Biol. Chem. 1985;260:1020–1026. [PubMed] [Google Scholar]
- Nunnally MH, Hsu LC, Mumby MC, Stull JT. J. Biol. Chem. 1987;262:3833–3838. [PubMed] [Google Scholar]
- Roush CL, Kennelly PJ, Glaccum MB, Helfman DM, Scott JD, Krebs EG. J. Biol. Chem. 1988;263:10510–10516. [PubMed] [Google Scholar]
- Stull JT, Nunnally MH, Moore RL, Blumenthal DK. Adv. Enzyme Regul. 1985;23:123–140. doi: 10.1016/0065-2571(85)90043-3. [DOI] [PubMed] [Google Scholar]
- Stull JT, Nunnally MH, Michnoff CH. In: The Enzymes. Krebs EG, Boyer PD, editors. Academic Press; Orlando, FL: 1986. pp. 113–166. [Google Scholar]
- Sweeney HL, Stull JT. Proc. Natl. Acad. Sci. U. S. A. 1990;87:414–418. doi: 10.1073/pnas.87.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takio K, Blumenthal DK, Walsh KA, Titani K, Krebs EG. Biochemistry. 1986;25:8049–8057. doi: 10.1021/bi00372a038. [DOI] [PubMed] [Google Scholar]
- Zoller MJ, Smith M. DNA (N. Y.) 1984;3:479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]


