Abstract
Genome-wide association (GWA) studies have identified multiple loci at which common variants modestly influence the risk of developing colorectal cancer (CRC). To enhance power to identify additional loci with similar effect sizes, we conducted a meta-analysis of two GWA studies, comprising 13,315 individuals genotyped for 38,710 common tagging SNPs. We undertook replication testing in up to eight independent case-control series comprising 27,418 subjects. We identified four previously unreported CRC risk loci at 14q22.2 (rs4444235, BMP4; P = 8.1 × 10−10), 16q22.1 (rs9929218, CDH1; P = 1.2 × 10−8), 19q13.1 (rs10411210, RHPN2; P = 4.6 × 10−9) and 20p12.3 (rs961253; P = 2.0 × 10−10). These findings underscore the value of large sample series for discovery and follow-up of genetic variants contributing to the etiology of CRC.
Whereas inherited susceptibility is responsible for ~35% of all CRC1, high-risk germline mutations in APC, the mismatch repair (MMR) genes, MUTYH (MYH), SMAD4, BMPR1A and STK11/LKB1 account for <6% of all cases2. Recent GWA studies have validated the hypothesis that part of the heritable risk is caused by common, low-risk variants, identifying CRC susceptibility loci mapping to 8q24 (rs6983267)3,4, 8q23.3 (rs16892766, EIF3H)5, 10p14 (rs10795668)5, 11q23 (rs3802842)6, 15q13 (rs4779584)7 and 18q21 (rs4939827, SMAD7)6,8.
GWA studies are not contingent on prior information concerning candidate genes or pathways, and thereby have the ability to identify important variants in hitherto unstudied genes. However, the effect sizes of individual variants, the need for stringent thresholds for establishing statistical significance, and financial constraints on numbers of variants that can be followed up inevitably constrain study power. We recently published two separate GWA studies for CRC. To augment the power to detect additional CRC risk loci, we have conducted a meta-analysis of data from these studies and followed up the best supported associations in large sample sets. This analysis, in conjunction with a replication study using eight independent case-control series, has enabled us to identify four new loci predisposing to CRC. This brings to ten the number of independent loci conclusively associated with CRC risk, and provides additional insight into the genetic architecture of inherited susceptibility to CRC.
RESULTS
Meta-analysis of genome-wide association scans
The GWA studies were both conducted by centers in London and Edinburgh, and were both based on designs involving two-phase strategies and using samples from UK populations (Table 1 and Supplementary Table 1 online). The London phase 1 was based on genotyping 940 cases with familial colorectal neoplasia and 965 controls ascertained through the Colorectal Tumour Gene Identification (CoRGI) consortium for 555,352 SNPs using the Illumina HumanHap550 BeadChip Array. Phase 1 in the Edinburgh study consisted of genotyping 1,012 early-onset (aged ≤55 years) Scottish CRC cases and 1,012 controls for 555,510 SNPs using the Illumina HumanHap300 and HumanHap240S arrays. After applying quality control filters, the following data were available: London phase 1, 547,487 SNP genotypes from 922 familial neoplasia cases (614 with CRC and 308 with high-risk colorectal adenomas) and 927 controls; Edinburgh phase 1, 548,586 SNP genotypes from 980 CRC cases and 1,002 controls.
Table 1.
Overview of study design
| Study | Cases | Controls | Number of SNPs genotyped |
|---|---|---|---|
| GWA series | |||
| London phase 1 | 922 | 927 | 547,487 |
| Edinburgh phase 1 | 980 | 1,002 | 548,586 |
| London phase 2 | 2,854 | 2,822 | 38,715 |
| Edinburgh phase 2 | 2,024 | 2,092 | 38,710 |
| Replication series | |||
| London replication | 3,286 | 3,017 | 9 |
| Edinburgh replication | 676 | 842 | 8 |
| VCQ58 | 1,543 | 2,236 | 9 |
| FCCPS | 962 | 846 | 7 |
| Canada | 1,175 | 1,184 | 3 |
| DACHS | 1,373 | 1,480 | 3 |
| Kiel | 2,169 | 2,145 | 3 |
| SEARCH | 2,222 | 2,262 | 3 |
| Total | 20,186 | 20,855 | |
London phase 2 was based on genotyping 2,873 CRC cases and 2,871 controls ascertained through the National Study of Colorectal Cancer Genetics (NSCCG), whereas Edinburgh phase 2 was based on genotyping 2,057 cases and 2,111 controls. For phase 2, the London and Edinburgh samples were genotyped for a common set of SNPs: the 14,982 SNPs most strongly associated with colorectal neoplasia from London phase 1; the 14,972 most strongly associated SNPs from Edinburgh phase 1 (432 of these SNPs were common to both the London and Edinburgh lists of most strongly associated SNPs); and 13,186 SNPs showing the strongest association with CRC risk from a joint analysis of all CRC cases and controls from both phase 1 data sets (that were not already included in any of the preceding categories). Therefore, phase 2 was based on genotyping 42,708 SNPs in total. After applying quality control filters, the following data were available: London phase 2, 38,715 polymorphic SNPs in 2,854 cases and 2,822 controls; Edinburgh phase 2, 38,710 polymorphic SNPs in 2,024 cases and 2,092 controls. Overall, there were 38,710 polymorphic SNPs common to all four data sets (phases 1 and 2 in London and Edinburgh).
Prior to undertaking the meta-analysis of phases 1 and 2, we searched for potential errors and biases in the four case-control series. Comparison of the observed and expected distributions showed little evidence for an inflation of the test statistics in any of the data sets (inflation factor λ = 1.02 and 1.05 for London phases 1 and 2, and 1.02 and 1.08 for Edinburgh phases 1 and 2, based on the 90% least significant SNPs; Supplementary Fig. 1 online), thereby excluding the possibility of significant hidden population substructure, cryptic relatedness among subjects or differential genotype calling. Using data on all CRC cases and controls from the four series, we derived joint odds ratios (ORs) and confidence intervals (CIs) under a fixed-effects model for each SNP, and associated P values from the standard normal distribution. The distribution of the association P values was significantly skewed from the null distribution: 76 of the SNPs had a P value < 10−4, greater than the 54 conservatively expected under the null hypothesis (P = 2 × 10−3, binomial test).
Of the 23 SNPs associated with CRC risk at P < 10−5, 14 map to regions that have been the subject of previous fast-tracking replication analyses: 8q24 (rs6983267, rs7014346, rs7837328, rs10808555)3,4, 11q23 (rs3802842, rs11213809, rs10749971)6, 18q21 (rs12953717, rs4939827)8, 8q23 (rs16892766, rs11986063, rs6983626)5 and 15q13 (rs4779584, rs10318)9 (Supplementary Table 2 online). We therefore focused on the nine remaining SNPs from five distinct genomic loci that were associated with CRC risk at P < 10−5 (Table 2), and that potentially represented previously unreported disease-associated loci. This threshold for follow-up did not exclude the possibility that other SNPs represented genuine association signals, but was simply a pragmatic strategy for prioritizing replication.
Table 2.
Summary of results for nine SNPs selected for replication
| Phases 1 and 2 meta |
Replicationb |
All phases |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr. | Position (bp) | Major allele | Minor allele | MAFa | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | Phet |
| rs10411210 | RHPN2 | 19 | 38,224,140 | C | T | 0.10 | 0.79 | 4.9 × 10−8 | 0.90 | 7.0 × 10−4 | 0.87 | 4.6 × 10−9 | 0.02 |
| (0.72–0.86) | (0.85–0.96) | (0.83–0.91) | |||||||||||
| rs961253 | 20 | 6,352,281 | C | A | 0.36 | 1.13 | 8.9 × 10−7 | 1.11 | 4.6 × 10−5 | 1.12 | 2.0 × 10−10 | 0.70 | |
| (1.08–1.19) | (1.06–1.17) | (1.08–1.16) | |||||||||||
| rs355527 | 20 | 6,336,068 | G | A | 0.33 | 1.13 | 1.2 × 10−6 | 1.11 | 3.7 × 10−5 | 1.12 | 2.1 × 10−10 | 0.95 | |
| (1.08–1.19) | (1.06–1.17) | (1.08–1.17) | |||||||||||
| rs9929218 | CDH1 | 16 | 67,378,447 | G | A | 0.29 | 0.88 | 1.4 × 10−6 | 0.93 | 3.5 × 10−4 | 0.91 | 1.2 × 10−8 | 0.56 |
| (0.83–0.92) | (0.90–0.97) | (0.89–0.94) | |||||||||||
| rs4444235 | BMP4 | 14 | 53,480,669 | T | C | 0.46 | 1.12 | 1.8 × 10−6 | 1.10 | 9.4 × 10−5 | 1.11 | 8.1 × 10−10 | 0.64 |
| (1.07–1.18) | (1.05–1.16) | (1.08–1.15) | |||||||||||
| rs1862748 | CDH1 | 16 | 67,390,444 | C | T | 0.31 | 0.88 | 2.6 × 10−6 | 0.93 | 7.1 × 10−4 | 0.91 | 2.9 × 10−8 | 0.71 |
| (0.84–0.93) | (0.90–0.97) | (0.88–0.94) | |||||||||||
| rs4951291 | 1 | 202,273,161 | C | T | 0.14 | 0.85 | 5.3 × 10−6 | 1.02 | 0.6 | 0.93 | 4.1 × 10−3 | 0.01 | |
| (0.79–0.91) | (0.95–1.09) | (0.88–0.98) | |||||||||||
| rs7259371 | RHPN2 | 19 | 38,226,481 | G | A | 0.18 | 0.86 | 5.7 × 10−6 | 0.91 | 5.2 × 10−3 | 0.89 | 2.2 × 10−7 | 0.03 |
| (0.81–0.92) | (0.86–0.97) | (0.85–0.93) | |||||||||||
| rs4951039 | 1 | 202,273,220 | A | G | 0.14 | 0.85 | 5.8 × 10−6 | 1.09 | 0.05 | 0.94 | 2.4 × 10−2 | ||
| 1.8 × 10−3 | |||||||||||||
| (0.79–0.91) | (1.00–1.19) | (0.89–0.99) | |||||||||||
MAF in controls in weighted average of MAFs from each replication series
Based on replication data from the following series: for rs10411210 and rs9929218, London replication, Edinburgh replication, VCQ58, FCCPS, Canada, DACHS, Kiel and SEARCH; for rs1862748, London replication, Edinburgh replication, VCQ58, Canada, DACHS, Kiel and SEARCH; for rs4951039, London replication and CVQ58; for all other SNPs, London replication, Edinburgh replication, VCQ58 and FCCPS.
Replication analyses
To identify the true risk alleles among these nine SNPs, we conducted a replication study initially based on four independent case-control series (London replication, Edinburgh replication, VCQ58 and Finnish Colorectal Cancer Predisposition Study) involving a total of 13,408 individuals (Table 1, Supplementary Table 1 and Supplementary Table 3 online). On the basis of the combined analyses, we found that signals from seven SNPs, representing four loci, reached strong levels of evidence (i.e., P < 5.0 × 10−7) for an association with CRC risk (Table 2 and Fig. 1), with four SNPs (three genomic regions) satisfying the proposed threshold for genome-wide statistical significance (i.e., P < 10−8).
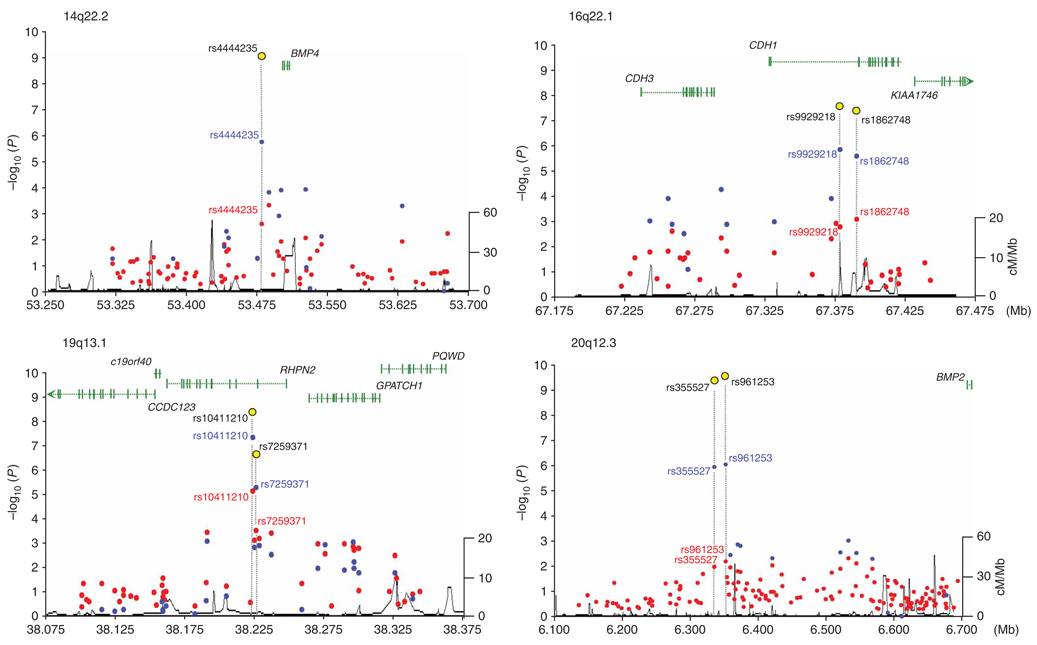
Figure 1.
Regional plots of the four confirmed associations (14q22.2, 16q22.1, 19q13.1 and 20p12.3). Each panel shows single-marker association statistics (as −log10 P) from the combined analysis of phase 1 (red), phases 1 and 2 (blue) and all phases (yellow) as a function of genomic position (NCBI build 36.1). Recombination rate across each region in HapMap CEU shown in black (right y axis). Also shown are the relative position of genes mapping to each region of association.
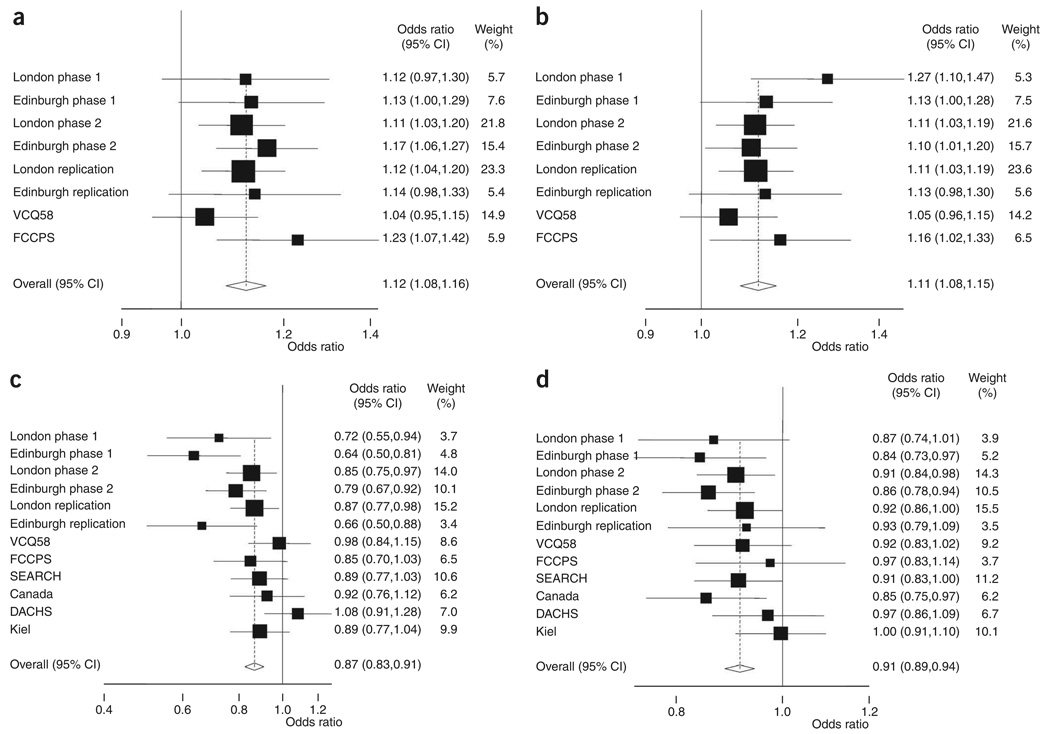
The strongest statistical evidence for a new CRC risk locus was provided by two SNPs: rs961253 (combined OR = 1.12, 95% CI 1.08–1.16, P = 2.0 × 10−10, Phet = 0.70, I2 = 0%; Fig. 2) and rs355527 (combined OR = 1.12, 95% CI 1.08–1.17, P = 2.1 × 10−10, Phet = 0.95, I2 = 0%; Table 2). The corresponding statistics excluding the discovery phase 1 of the GWA studies were OR = 1.12, 95% CI 1.08–1.16; P = 4.1 × 10−9 and OR = 1.13, 95% CI 1.08–1.17; P = 2.8 × 10−9, respectively. These two SNPs are in strong linkage disequilibrium (LD) (r2 = 0.82) and map to a 38-kilobase (kb) region of LD at 20p12.3 (6,316,089–6,354,440 base pairs (bp)), a region bereft of genes or predicted protein-encoding transcripts (Fig. 1 and Supplementary Fig. 2 online). Furthermore, there are no predicted genes or micro-RNAs in the vicinity of the markers (i.e., 250-kb flanking sequence). However, BMP2 maps 342 kb telomeric to the locus, and this may have relevance due to homology and functional similarity to another associated locus (see below).
Figure 2.
Forest plot of effect size and direction for the four SNPs associated with CRC. (a) rs961253. (b) rs4444235. (c) rs10411210. (d) rs9929218. Boxes denote allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% CIs. The diamond (and broken line) represents the summary OR computed under a fixed-effects model, with the 95% CI given by its width. The unbroken vertical line is at the null value (OR = 1.0).
The second strongest statistical evidence for an association was for rs4444235, which maps to a 16-kb region of LD at 14q22.2 (53,477,192–53,494,200 bp; combined OR = 1.11, 95% CI 1.08–1.15, P = 8.1 × 10−10, Phet = 0.64, I2 = 0%; Table 2 and Fig. 2; excluding phase 1, OR = 1.10, 95% CI 1.06–1.14, P = 2.0 × 10−7). This SNP is 9.4 kb from the transcription start site of the gene encoding bone morphogenetic protein 4 preproprotein (BMP4; Fig. 1 and Supplementary Fig. 2). Like BMP2, BMP4 is a member of the transforming growth factor-β family of signal transduction molecules that play an important role in CRC10. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling11. Intriguingly, inactivating mutations in the BMP receptor subunit BMPR1A are one cause of the rare juvenile polyposis syndrome12,13, which carries a very high risk of CRC, and we have previously found evidence that SNPs close to the BMP antagonist GREM1 are associated with CRC risk9.
The third strongest evidence for an association was shown by rs10411210 and rs7259371, two SNPs in moderate LD (r2 = 0.54), which map to a 96-kb block of LD (38,203,614–38,300,573 bp) on 19q13.1 encompassing Rho GTPase binding protein 2 (RHPN2) (Fig. 1 and Supplementary Fig. 2). rs10411210 showed the strongest evidence of association in this region, with an OR of 0.83 (95% CI 0.78–0.88; P = 1.1 × 10−9; Phet = 0.05, I2 = 50%; excluding phase 1, OR = 0.85, 95% CI 0.80–0.91; P = 1.0 × 10−6), whereas for rs7259371 the OR was 0.89 (95% CI 0.85–0.93, P = 2.2 × 10−7, Phet = 0.03, I2 = 55%; excluding phase 1, OR = 0.90, 95% CI 0.86–0.95, P = 2.8 × 10−5). RHPN2 encodes a Rho GTPase involved in the regulation of the actin cytoskeleton and cell motility14. Expression of RhoA has been implicated in the biology of several cancers, including CRC, promoting invasiveness15 through downregulation of adherens junction formation16.
The fourth locus displaying strong statistical evidence for association was rs9929218 (combined OR = 0.90, 95% CI 0.87–0.94, P = 1.2 × 10−7, Phet = 0.79, I2 = 0%), which localizes to intron 1 of the gene encoding cadherin 1 (CDH1, also known as E-cadherin) and maps to a 110-kb region of LD at 16q22.1 (67,286,613–67,396,803 bp) (Fig. 1 and Supplementary Fig. 2). Another SNP in CDH1, rs1862748 (r2 = 0.91 with rs9929218) also showed strong evidence for association (P = 4.9 × 10−7). CDH1 has an established role in CRC: aberrant activation of Wnt–β-catenin signaling is an initiating event in the development of CRC17, and somatic inactivation of CDH1 by mutation or promoter methylation occurs frequently18, leading to increased activity of the β-catenin–TCF transcription factor. Furthermore, germline mutations in CDH1 can cause familial diffuse-type gastric cancer19, occasionally in association with early-onset CRC20,21.
Because the association between rs10411210 (RHPN2) and CRC risk showed evidence of between-study heterogeneity, and the association between rs9929218 (CDH1) was of borderline genome-wide significance, we genotyped both SNPs in four additional independent replication case-control series from populations of European ancestry (Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH), Canada, Darmkrebs: Chancen der Verhütung durch Screening (DACHS) and Kiel). After pooling data from these studies with the previously genotyped data sets, the association for rs9929218 attained genome-wide significance (OR = 0.91, 95% C: 0.89–0.94, P = 1.2 × 10−8, Phet = 0.56, I2 = 0%; excluding phase 1, P = 4.8 × 10−7; Table 2 and Fig. 2). The association for rs10411210 was still highly significant, but there remained evidence of between-study heterogeneity, albeit nonsignificant after correction for multiple testing (OR = 0.87, 95% CI: 0.83–0.91, P = 4.6 × 10−9, Phet = 0.02; I2 = 52%; excluding phase 1, P = 1.4 × 10−6; Table 2 and Fig. 2).
We assessed in more detail the pattern of the risks associated with each of the four newly identified loci, as defined by rs961253, rs4444235, rs10411210 and rs9929218. For rs961253, the minor allele was associated with an increased risk of CRC in a dose-dependent manner, with a higher risk in homozygous than heterozygous carriers (ORhet = 1.14, 95% CI 1.08–1.20 and ORhom = 1.24, 95% CI 1.15–1.34), supporting a multiplicative model (Supplementary Table 4 online). A similar risk pattern was observed for rs4444235 (ORhet = 1.13, 95% CI 1.07–1.20 and ORhom = 1.23, 95% CI 1.15–1.32). For rs10411210, the minor allele was associated with a decreased risk of CRC in a dose-dependent manner (ORhet = 0.87, 95% CI 0.83–0.92 and ORhom = 0.72, 95% CI 0.59–0.89). There was little difference in the fit provided by multiplicative and dominant models due to the very low frequency of rare homozygotes, although a recessive model could be excluded (Supplementary Table 4). For rs9929218, genotype-specific ORs were most compatible with a multiplicative model (ORhet = 0.92, 95% CI 0.89–0.96 and ORhom = 0.82, 95% CI 0.76–0.89; Supplementary Table 4).
Genotype-phenotype correlations
We assessed associations between clinicopathological variables and genotypes through case-only logistic regression. The association between rs4444235 (BMP4) and CRC was significantly stronger in cases with microsatellite stable tumors compared with microsatellite instability (MSI) cases (P = 2.1 × 10−3; based on 2,074 cases from London phase 2 and London replication series; Supplementary Table 5 online). There was also some evidence of an association with gender for rs9929218 (P = 0.02, based on 17,152 cases from all series bar VCQ58 and FCCPS), with the susceptibility allele more common in females than males (Supplementary Table 5). We did not find any other significant associations between SNP genotype and clinicopathological data (specifically, age at diagnosis, site of tumor or family history of CRC; Supplementary Table 5).
Architecture of genetic susceptibility to colorectal cancer
We estimate the contribution of each of the ten loci identified to date to the familial risk of CRC to be <1%. As epistasis could affect the overall contribution of the loci to the genetic susceptibility of CRC, we examined for pairwise interactions. Because the six previously validated SNPs had not been genotyped in all replication series from the current study, the number of individual genotypes available for each calculation varied. However, all comparisons were based on at least 13,000 individuals. This analysis had >80% power to detect an interaction between the two SNPs that have the lowest minor allele frequency (MAF) and smallest effect size, namely rs16892766 and rs10411210. We found no evidence of interactive effects between any of the CRC disease loci identified thus far (Supplementary Table 6 online), suggesting an independent role for each locus in CRC development. Counting two for a homozygote, the risk of CRC increased with possession of an increasing numbers of risk alleles for the ten loci (Ptrend = 5.4 × 10−44; based on 9,692 cases and controls from London and Edinburgh phase 2; Supplementary Table 7 online). On the basis of an additive model, the ten loci so far identified are likely to collectively account for ~6% of the excess familial risk.
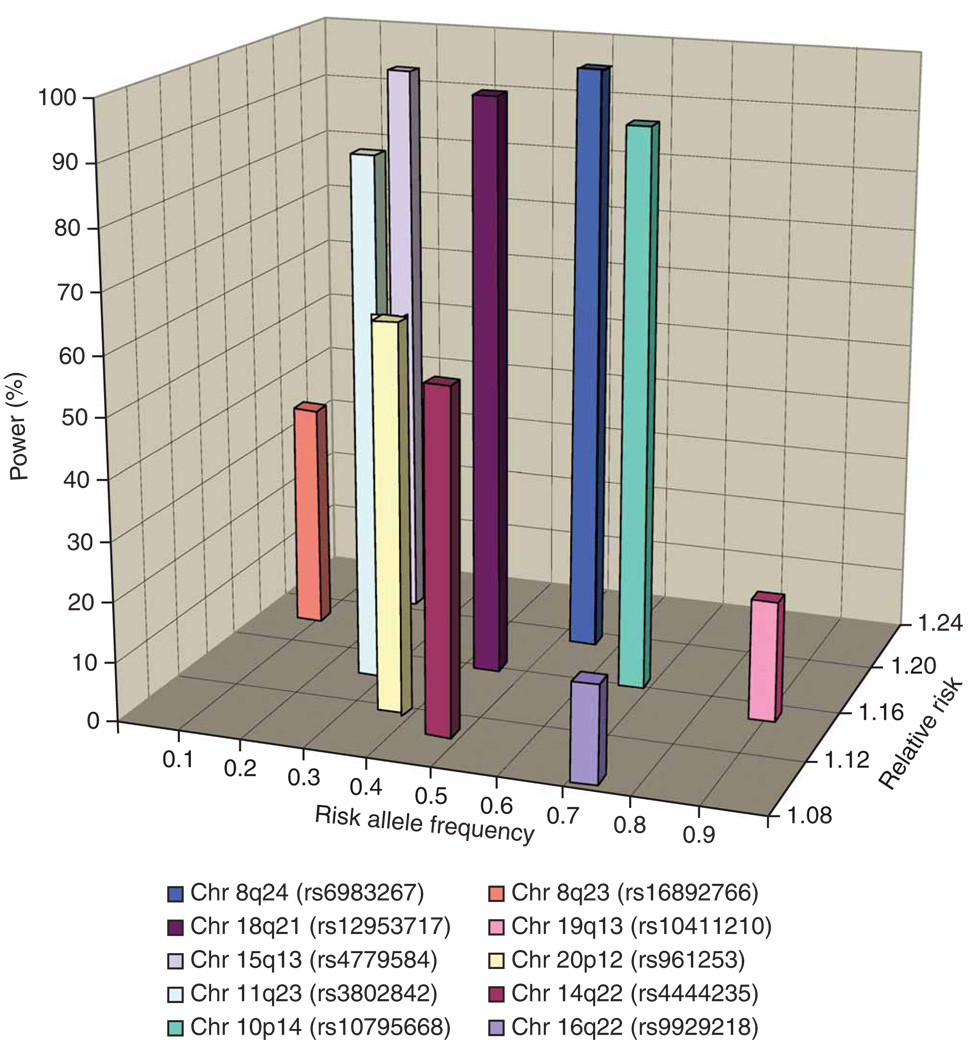
To gain insight into the basis of the residual familial risk of CRC, we estimated the power of our meta-analysis to identify each of the ten disease-associated loci on the basis of their MAF and associated genotypic risk (Fig. 3). Although our combined analysis was well powered to identify common variants (MAF > 0.3) such as those at 8q24, the power to identify those loci with lower MAF or smaller genotypic risk was limited.
Figure 3.
Power of the meta-analysis to identify each of the ten colorectal cancer susceptibility alleles, stipulating a P value of 10−5. Genotypic risks associated with each locus derived from replication data.
Although fine-mapping and resequencing are required to identify the specific variants underlying each of the ten associations so far identified, we conducted two analyses to investigate the basis of causality. Accepting the caveat that HapMap is not comprehensive, we interrogated HapMap to identify nonsynonymous SNPs highly correlated (i.e., r2 > 0.8) with the most strongly associated SNP within each region of association. The only nonsynonymous SNPs showing strong LD was rs17563 (BMP4 A152V), which is correlated with rs4444235 (D′ = 0.96, r2 = 0.81; Supplementary Table 8 online). However, genotyping both phase 2 case-control series for rs17563 provided no support for A152V as the basis for the 14q22.2 association (P = 0.027 compared with P = 5.90 × 10−4 for rs4444235). These data suggest many of the associations identified so far are mediated through LD with sequence changes that influence gene expression rather than protein sequence, or through LD with low frequency variants (i.e., variants with MAF of 0.001–0.01) that are not cataloged by HapMap. The C-160A promoter variant (rs16260), which is in strong LD with rs9929218 (D′ = 0.95, r2 = 0.92), has been previously documented to influence CDH1 transcription22. We have previously reported allele-specific expression associated with SMAD7 variants8. To explore whether any of the other eight associations reflect similar cis-acting regulatory effects on a nearby gene, we searched for genotype–expression correlations in 90 lymphoblastoid cell lines using previously described data23,24. We did not, however, find any significant relationship between SNP genotype and gene expression (Supplementary Fig. 3 online).
DISCUSSION
By pooling data from two GWA studies and conducting replication analyses, we have identified four previously unreported variants influencing CRC susceptibility, in addition to the six variants we have previously shown to be associated with CRC risk. The new loci identified in the current study are of modest effect size, which is unsurprising given that those with a larger impact on CRC were discovered in previously reported fast-tracking analyses3,4,6,8,9. Collectively, these data are thus consistent with a model in which the loci readily detectable through current GWA studies of 1,000–2,000 CRC cases and controls are associated with modest effects (i.e., those with MAF > 0.2 and conferring genotypic risks of ~1.2).
Our estimate of the contribution of the ten loci to excess familial risk of CRC is likely to be conservative as the effect of the causal variant will typically be larger than the association detected through a tag SNP. In addition, multiple causal variants may exist at each locus, including low-frequency variants with significantly larger effects on risk. Indeed, our analysis provides evidence that CDH1 may be a locus where a spectrum of common and rare alleles contributes to CRC risk, as reported in other complex traits.
Our GWA data and the results from similar gene discovery efforts in other tumors are proving to be highly informative regarding the allelic architecture of cancer susceptibility in general. First, the number of common variants explaining more than 1% of inherited risk is very low, and only a small proportion of the heritability of any cancer can be explained by the currently identified loci. Second, the genetic landscape defined by the common risk loci identified to date does not feature significant epistatic effects. Third, few of the observed associations seem to be due to correlation with common coding variants, and furthermore, many of the loci map to regions bereft of genes or protein-coding transcripts. It is therefore likely that much of the common variation in cancer risk is mediated through sequence changes influencing gene expression, perhaps in a subtle fashion. Therefore, in addition to searching for cis-regulatory correlations, expression quantitative trait locus analyses may aid elucidation of causality. Whereas examination of genotypic effects on expression in lymphoblastoid cell lines can be informative if genes are ubiquitously expressed, it is likely to be limited by tissue-specific effects. Furthermore, the target tissue need not even be the organ or cell type from which cancer develops. Hence, although we failed to demonstrate effects of cis-regulatory polymorphisms in LD with the variants identified, lymphocytes show either low levels of gene expression (CDH1, CDH3, RHPN2) or absent expression (BMP4, BMP2) of target genes, making analyses based on colorectal or other tissues desirable.
Many mendelian cancer predisposition genes influence the risk of more than one tumor type. Pleiotropic effects are also a feature of 8q24 variants such as rs6983267, which affects the risk of CRC, prostate and ovarian cancer25,26. To examine whether any of the four variants we have identified influences the risk of breast or prostate cancer, we interrogated GWA data from the Cancer Genetic Markers of Susceptibility (CGEMS) breast and prostate cancer study. Although requiring replication, there was evidence from CGEMS that variation at 20p12 (rs355527) influenced the risk of developing breast cancer.
The power of our study to detect the major common loci conferring risks of 1.2 or greater (such as the 8q24 variant) was high. Hence, we consider that there are unlikely to be many additional CRC SNPs with similar effects for alleles with frequencies >0.2 in populations of European ancestry. However, it is notable that the 10p14 variant rs10795668, which we have previously robustly shown5 to be associated with CRC risk (P = 6.9 × 10−12), was not captured by our meta-analysis, which stipulated a threshold of <10−5 (P value in meta-analysis 3.1 × 10−4). In contrast, we had low power to detect alleles with smaller effects and/or MAFs < 0.1. By implication, variants with such profiles are likely to represent a much larger class of susceptibility loci for CRC, whether because of truly small effect sizes or submaximal LD with tagging SNPs. In addition to sample size considerations, the 550K tagging SNPs we used for the GWA studies capture on average ~80% of common SNPs in the European population (i.e., r2 > 0.8), but only ~12% of SNPs with MAFs of 5–10% are tagged at this level, limiting power to detect this class of susceptibility allele. Furthermore, GWA-based strategies are not optimally configured to identify low-frequency variants with potentially stronger effects. In addition, these arrays are not ideally formatted to capture copy number variants, which may also affect CRC risk. Thus, it is highly likely that a large number of low-penetrance variants remain to be discovered. This assertion is supported by the continued excess of associations observed over those expected, in addition to the regions studied herein. Further efforts to expand the scale of GWA meta-analyses, in terms of both sample size and SNP coverage, and to increase the number of SNPs taken forward to large-scale replication, may identify additional variants for CRC.
Irrespective of the nature of the causal variants, a high proportion of the population carries at-risk genotypes. Whereas individual alleles exert only small effects, much larger risks are seen in carriers of multiple risk alleles. Therefore, the ten SNPs identified thus far potentially have public health relevance as further susceptibility loci are identified. Additional studies are required to characterize genetic variation at these loci and determine their relationship to the functional consequences that lead to CRC. Furthermore, it will also be intriguing to examine how our findings translate to non-European populations, some of which are typified by a considerably lower prevalence of CRC.
METHODS
Study participants
Supplementary Table 1 provides a summary of all cases and controls in the study.
London genome-wide association study
Phase 1 comprised 940 cases with colorectal neoplasia (443 males, 497 females) ascertained through the CoRGI consortium. All had at least one first-degree relative affected by CRC and one or more of the following phenotypes: CRC at age 75 or less; any colorectal adenoma at age 45 or less; three or more colorectal adenomas at age 75 or less; or a large (>1 cm diameter) or aggressive (villous and/or severely dysplastic) adenoma at age 75 or less, Controls (n = 965; 439 males, 526 females) were spouses or partners unaffected by cancer and without a personal family history (to second-degree relative level) of colorectal neoplasia. All cases and controls were of European ancestry and from the UK. Phase 2 consisted of 2,873 CRC cases (1,199 males, 1,674 females; mean age at diagnosis 59.3 years; s.d. ± 8.7) ascertained through two ongoing initiatives at the Institute of Cancer Research/Royal Marsden Hospital NHS Trust (from 1999 onwards: the NSCCG27 and the Royal Marsden Hospital NHS Trust/Institute of Cancer Research Family History and DNA Registry. A total of 2,871 healthy individuals were recruited as part of ongoing National Cancer Research Network genetic epidemiological studies, NSCCG (n = 1,235), the Genetic Lung Cancer Predisposition Study28 (1999–2004; n = 917) and the Royal Marsden Hospital NHS Trust/Institute of Cancer Research Family History and DNA Registry (1999–2004; n = 719), These controls (1,164 males, 1,707 females; mean age 59.8 years; s.d. ± 10.8) were the spouses or unrelated friends of individuals with malignancies. None had a personal history of malignancy at time of ascertainment. All cases and controls were British of recent European ancestry, and there were no obvious differences in the demography of cases and controls in terms of place of residence within the UK. Genotyping data for these samples have been presented previously5.
Edinburgh genome-wide association study
Phase 1 included 1,012 CRC cases (518 males, 494 females; mean age at diagnosis 49.6 years; s.d. ± 6.1) and 1,012 age- and gender-matched cancer-free population controls (518 males, 494 females; mean age 51.0 years; s.d. ± 5.9). Cases were enriched for genetic etiology by early age at onset (age ≤ 55 years). Known dominant polyposis syndromes, hereditary nonpolyposis colrectal carcinoma/Lynch syndrome or bi-allelic MUTYH mutation carriers were excluded. Control subjects were population controls, matched by age (± 5 years), gender and area of residence within Scotland. Phase 2 comprised 2,057 CRC cases (1,249 males, 808 females; mean age at diagnosis 65.8 years; s.d. ± 8.4) and 2,111 population controls (1,257 males, 854 females; mean age 67.9 years; s.d. ± 9.0) ascertained in Scotland. Cases were taken from an independent, prospective, incident CRC case series and aged <80 years at diagnosis. Control subjects were population controls matched by age (± 5 years), gender and area of residence within Scotland. Genotyping data for phase 1 and for 15,008 SNPs in phase 2 have been presented previously6.
Replication series
London replication: 3,286 CRC cases (2,158 males, 1,128 females; mean age at diagnosis 59.1 years; s.d. ± 8.1) and 3,017 controls (1,212 males, 1,805 females; mean age 61.7 years; s.d. ± 11.4) ascertained through NSCCG post 2005. Edinburgh replication: 676 CRC cases (335 males, 341 females; mean age at diagnosis 53.2 years; s.d. ± 15.4) and 842 cancer-free population controls (394 males, 448 females; mean age 51.8 years; s.d. ± 11.5). Controls were recruited as part of the Generation Scotland study29. VCQ58: 1,543 CRC cases (925 males, 618 females, mean age of diagnosis 62.4 years; s.d. ± 10.7), consisting of 1,234 (2 SNPS typed for 1,310) samples from the VIOXX in Colorectal Cancer Therapy: Definition of Optimal Regime (VICTOR)/QUASAR2 trials and 309 CRC cases collected through the CoRGI study. There were 2,236 controls (1,007 males, 1,226 females, three unknown) consisting of 1,438 population controls from the Wellcome Trust Case Control Consortium 1958 birth cohort (58C, also known as the National Child Development Study, which included all births in England, Wales and Scotland during a single week in 1958 (ref. 30), 418 cancer-free controls collected through the CoRGI study and 380 European Collection of Cell Cultures samples. All cases and controls were of European origin and from the UK. FCCPS: 962 CRC cases (509 males, 452 females, one unknown; mean age at diagnosis 66.9 years; s.d. ± 12.2) and 846 controls (randomly selected anonymous Finnish blood donors) ascertained in southeastern Finland. SEARCH: 2,222 CRC cases (1,278 males, 944 females; mean age at diagnosis 59.2 years; s.d. ± 8.1) and 2,262 controls (949 males, 1,313 females; mean age 57,6 years; s.d. ± 15.1. Samples were ascertained through the SEARCH study based in Cambridge, UK. Recruitment of CRC started in 2000; initial patient contact was though the general practitioner. Control samples were collected post 2003. Eligible individuals were sex and frequency matched in 5-year age bands to cases. DACHS: 1,373 CRC cases (790 males, 583 females; mean age at diagnosis 68.1 years; s.d. ± 10.4) and 1,480 controls (719 males, 761 females; mean age 68.0 years; s.d. ± 9.9) ascertained through DACHS, a population-based case-control study of incident CRC in the Rhine-Neckar-Odenwald region around Heidelberg between 2003 and 2006. Kiel: 2,169 CRC cases (1,089 males, 1,080 females; mean age at diagnosis 60.9 years; s.d. ± 8.8) and 2,145 controls (1,059 males, 1,086 females; mean age 64.7 years; s.d. ± 10.0) ascertained through the PopGen and SHIP population-based biobank projects based in Kiel and Greifswald, Germany. Canada: 1,175 CRC cases (503 males, 672 females; mean age at diagnosis 60.3 years; s.d. ± 8.7) and 1,184 controls (667 males, 517 females; mean age 60.9 years; s.d. ± 8.1) ascertained through the Ontario Familial Colorectal Cancer Registry.
In all cases, CRC was defined according to the ninth revision of the International Classification of Diseases31 by codes 153–154, and all cases had pathologically proven adenocarcinoma. Collection of blood samples and clinicopathological information from cases and controls was undertaken with informed consent and ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Genotyping
DNA was extracted from samples using conventional methodologies and quantified using PicoGreen (Invitrogen). The London phase 1 GWA study was conducted using the Illumina HumanHap550 Bead Arrays, and the Edinburgh phase 1 GWA study was conducted using Illumina HumanHap300 and HumanHap240S according to the manufacturer’s protocols (Supplementary Methods online). DNA samples with GenCall scores <0.25 at any locus were considered ‘no calls’. In London and Edinburgh phase 2, genotyping was conducted using Illumina Infinium custom arrays according to the manufacturer’s protocols. For both phases 1 and 2, a SNP was deemed to have failed if fewer than 95% of DNA samples generated a genotype at the locus. To ensure quality of genotyping, a series of duplicate samples were genotyped, and cases and controls were genotyped in the same batches in both phases 1 and 2.
Phase 3 genotyping was conducted using either competitive allele-specific PCR KASPar chemistry (KBiosciences) or Taqman (Applied Biosystems), except for Canadian samples, which were genotyped using single-base primer extension chemistry matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) detection (Sequenom). All primers and probes used are available on request. Genotyping quality control was tested using duplicate DNA samples within studies and SNP assays, together with direct sequencing of subsets of samples to confirm genotyping accuracy. For all SNPs, >99.9% concordant results were obtained.
MSI in CRCs was determined using the following methodology: 10 µm sections were cut from formalin-fixed paraffin-embedded tumors, lightly stained with toluidine blue, and regions containing at least 60% tumor microdissected. Tumor DNA was extracted using the QIAamp DNA Mini kit (Qiagen) according to the manufacturer’s instructions and genotyped for the mononucleotide microsatellite loci BAT25 and BAT26, which are highly sensitive markers of MSI32. Samples showing novel alleles at either BAT26 or BAT25 or both markers were assigned as MSI (corresponding to a high level of instability33).
Statistical analysis
Genotype data were used to search for duplicates and closely related individuals among all samples in phases 1 and 2. Identity-by-state values were calculated for each pair of individuals; for any pair with allele sharing >80%, the sample generating the lowest call rate was removed from further analysis.
In London phase 1, genotyped samples were excluded from analyses for the following reasons: carriers of another susceptibility allele (five cases); first-degree relative with CRC (11 controls); duplicated (two cases, seven controls); relatedness (one case, 15 controls). In London phase 2, genotyped samples were excluded from analyses for the following reasons: duplicated (eight cases, two controls); relatedness (two cases, 18 controls); gender discrepancies (13 controls). In Edinburgh phases 1 and 2, genotyped samples were excluded from analyses for the following reasons: identified as of non-European descent (seven in phase 1, three in phase 2); previously unrecognized carriers of known monogenic hereditary CRC syndromes (five DNA MMR gene mutation carriers in phase 2); gender discrepancies between records and genotype (14 in phase 1, 22 in phase 2); hidden relatedness (five in phase 1).
The adequacy of the case-control matching and possibility of differential genotyping of cases and controls was formally evaluated using Q-Q plots of test statistics. The inflation factor λ was calculated by dividing the mean of the lower 90% of the test statistics by the mean of the lower 90% of the expected values from a χ2 distribution with 1 d.f. Deviation of the genotype frequencies in the controls from those expected under Hardy–Weinberg equilibrium was assessed by χ2 test (1 d.f.), or Fisher’s exact test where an expected cell count was <5.
Association between SNP genotype and disease status was primarily assessed using the allelic 1 d.f. test or Fisher’s exact test where an expected cell count was <5. The risks associated with each SNP were estimated by allelic, heterozygous and homozygous ORs using unconditional logistic regression, and associated 95% CIs were calculated in each case.
Joint analysis of data generated from multiple phases was conducted using standard methods for combining raw data based on the Mantel-Haenszel method34. Joint ORs and 95% CIs were calculated assuming a fixed-effects model, and tests of significance of the pooled effect sizes were calculated using a standard normal distribution. Cochran’s Q statistic to test for heterogeneity and the I2 statistic35 to quantify the proportion of the total variation due to heterogeneity were calculated.
We used Haploview software (v3.2) to infer the LD structure of the genome in the regions containing loci associated with disease risk. Patterns of risk for associated SNPs were investigated by logistic regression, coding the SNP genotypes according to additive, dominant and recessive models. Models were then compared by calculating the Akaike information criterion and Akaike weights for each mode of inheritance. Associations by site (colon/rectum), MSI status, family history status (at least one first-degree relative with CRC), gender and age at diagnosis (stratifying into two groups by the median age at diagnosis) were examined by logistic regression in case-only analyses, using all cases from replication phases for whom the clinicopathological variable being tested was available. Results for gender and age at diagnosis were based on all case series apart from London phase 1, VCQ58 and FCCPS; for site, on data from London phase 2, Edinburgh phases 1 and 2, London replication, SEARCH, Canada, DACHS and Kiel; for family history status and MSI status, on London phase 2 and London replication. The combined effect of each pair of loci identified as associated with CRC risk was investigated by logistic regression modeling, and evidence for interactive effects between SNPs assessed by likelihood ratio test assuming an allelic model. The OR and trend test for increasing numbers of deleterious alleles was estimated based on the London and Edinburgh phase 2 data by counting two for a homozygote and one for a heterozygote.
The sibling relative risk attributable to a given SNP was calculated using the formula36,37
where p is the population frequency of the minor allele, q = 1 − p and r1 and r2 are the relative risks (estimated as OR) for heterozygotes and rare homozygotes relative to common homozygotes. Assuming a multiplicative interaction, the proportion of the familial risk attributable to a SNP was calculated as log(λ*)/log(λ0), where λ0 is the overall familial relative risk estimated from epidemiological studies, assumed to be 2.2 (ref. 38). A naïve estimation of the contribution of all of the loci identified to the excess familial risk of CRC under an additive model was calculated using the formula39
We based our estimate of study power on a joint analysis of phases 1 and 2 assuming a multiplicative model for each SNP40. Samples in phase 1 of both GWAs were potentially enriched for genetic susceptibility by virtue of either having familial disease or being diagnosed young. To estimate the power of the meta-analysis, we therefore also computed power estimates accounting for genetic enrichment. For familial cases, the sample size required to detect a common disease susceptibility allele is typically reduced by more than twofold41. Similar statistical considerations apply to early-onset cases. Hence, for the power calculation, we conservatively inflated phase 1 case samples size by a factor of 2. The power to identify epistatic interactions between SNPs was estimated assuming a multiplicative model of gene–gene interaction using the Lubin and Gail approach42,43.
Relationship between SNP genotypes and expression levels
To examine for a relationship between SNP genotype and expression levels of MYC, LOC120376, CDH1, CDH3, BMP4, BMP2 and RHPN2 in lymphocytes, we made use of publicly available expression data generated from analysis of 90 European-derived Epstein-Barr virus–transformed lymphoblastoid cell lines using Sentrix Human-6 Expression BeadChips (Illumina)23,24. Online recovery of data was done using WGAViewer Version 1.25 Software. We compared differences in the distribution of levels of mRNA expression between SNP genotypes using a Wilcoxon-type test for trend44.
URLs
Haploview, http://www.broad.mit.edu/personal/jcbarret/haploview/; QUASAR, http://www.octo-oxford.org.uk/alltrials/trials/q2.html; European Collection of Cell Cultures, http://www.hpacultures.org.uk; WGAViewer, http://www.genome.duke.edu/centers/pg2/downloads/wgaviewer.php; Genetic Lung Cancer Predisposition Study, http://pfsearch.ukcrn.org.uk/StudyDetail.aspx?TopicID=1&StudyID=781, http://www.dh.gov.uk/assetRoot/04/01/45/13/04014513.pdf; NSCCG, http://pfsearch.ukcrn.org.uk/StudyDetail.aspx?TopicID=1&StudyID=1269; 1958 Birth Cohort, http://www.cls.ioe.ac.uk/studies.asp?section=000100020003.
Supplementary Material
ACKNOWLEDGMENTS
Cancer Research UK provided principal funding for this study (A6360). We would like to thank all individuals that participated in this study.
Institute of Cancer Research: Work was supported by the Bobby Moore Cancer Research UK (C1298/A8362). Additional funding was provided by the European Union (CPRB LSHC-CT-2004-503465) and CORE. I.C. was in receipt of a clinical training fellowship from St George’s Hospital Medical School. S.L. was supported by a PhD studentship from Cancer Research UK. We are grateful to colleagues at the UK National Cancer Research Network.
Oxford: Work was supported by CORE and the Bobby Moore Fund. We are grateful to colleagues at UK Clinical Genetics Centres and the UK National Cancer Research Network.
Edinburgh: The work was supported by Cancer Research UK grant numbers C348/A3758 and A8896, C48/A6361 and the Bobby Moore Fund, which supports the work of the Colon Cancer Genetics Group through Cancer Research UK. Additional funding was provided by the Medical Research Council (G0000657-53203), Scottish Executive Chief Scientist’s Office (K/OPR/2/2/D333, CZB/4/449), center grant from CORE as part of the Digestive Cancer Campaign. J.P. was funded by an MRC PhD studentship. We gratefully acknowledge the work of the COGS and SOCCS administrative teams; R. Cetnarskyj and the research nurse teams, all who recruited to the studies; the Wellcome Trust Clinical Research Facility for sample preparation; and all clinicians and pathologists throughout Scotland at collaborating centers. The study used the biological and data resource of Generation Scotland.
Finland: This work was supported by grants from Academy of Finland (Finnish Centre of Excellence Program 2006–2011), the Finnish Cancer Society, the Sigrid Juselius Foundation and the European Commission (9LSHG-CT-2004-512142).
Cambridge: We thank the SEARCH study team and all the participants in the study. P.D.P.P. is a Cancer Research UK Senior Clinical Research Fellow. T.K. is funded by the Foundation Dr Henri Dubois-Ferriere Dinu Lipatti.
Kiel: The study was supported by the German National Genome Research Network (NGFN) through the PopGen biobank (BmBF 01GR0468) and the National Genotyping Platform. Further support was obtained through the MediGrid and Services@MediGrid projects (01AK803G and 01IG07015B). SHIP is part of the Community Medicine Research net (CMR) of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grant number. ZZ9603), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania.
Heidelberg: We wish to thank all participants and the staff of the participating clinics for their contribution to the data collection, B. Kaspereit, K. Smit and U. Eilber in the Division of Cancer Epidemiology, and U. Handte-Daub, S. Toth and B. Collins in the Division of Clinical Epidemiology and Aging Research, German Cancer Research Center for their excellent technical assistance. This study was supported by the German Research Council (Deutsche Forschungsgemeinschaft), grant numbers BR 1704/6-1, BR 1704/6-3 and CH 117/1-1, and by the German Federal Ministry for Education and Research, grant number 01 KH 0404.
Canada: We gratefully acknowledge the contribution of A. Belisle, V. Catudal and R. Fréchette. Cancer Care Ontario, as the host organization to the ARCTIC Genome Project, acknowledges that this project was funded by Genome Canada through the Ontario Genomics Institute, by Génome Québec, the Ministère du Dévelopement Économique et Régional et de la Recherche du Québec and the Ontario Institute for Cancer Research (B.W.Z., T.J.H. and S.G.). Additional funding was provided by the National Cancer Institute of Canada (NCIC) through the Cancer Risk Assessment (CaRE) Program Project Grant. The work was supported through collaboration and cooperative agreements with the Colon Cancer Family Registry and PIs, supported by the National Cancer Institute, National Institutes of Health under RFA CA-95-011, including the Ontario Registry for Studies of Familial Colorectal Cancer (S.G.) (U01 CA076783). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating institutions or investigators in the Colon CFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the Colon CFR. This study made use of genotyping data on the 1958 Birth Cohort. Genotyping data on controls was generated and generously supplied to us by Panagiotis Deloukas of the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk.
Appendix
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
The study was designed and financial support obtained by R.S.H., I.P.M.T., M.G.D. and H.C. The manuscript was drafted by R.S.H. and E.W. with contributions from M.G.D. and I.P.M.T. Statistical analyses were conducted by E.W., with contributions from M.C.D.B., J.-B.C., S.L.S. and A.T. E.W. and A.M.P. performed bioinformatic analyses.
Institute of Cancer Research: Subject recruitment and sample acquisition to NSCCG were undertaken by S.P. Coordination of sample preparation and genotyping was performed by P.B. Sample preparation and genotyping performed by A.M.P., K.S., J.V. and S.L. Histology review was by I.C. Testing of MSI was performed by S.L. and I.C.
Wellcome Trust Centre for Human Genetics: Subject recruitment and sample acquisition were undertaken by E.B., M.G., L.M. and members of the CoRGI Consortium and D.K. Sample preparation was performed by K.H., S.L.S. and E.J. Genotyping was performed and co-ordinated by L.C.-C., K.H., A.S.T., S.L.S., A.W., E.J. and E.D.
Colon Cancer Genetics Group, Edinburgh: Study design and funding: M.G.D., H.C., A.T., S.M.F. and M.E.P. Subject recruitment and sample acquisition was led by M.G.D., H.C., M.E.P., R.C., S.M.F. and members of the SOCCS/COGS study teams. Sample preparation was co-ordinated by S.M.F. and R.A.B. Histology review: M.G.D. and S.M.F. Genotyping was performed and co-ordinated by S.M.F. and M.G.D. A.T. performed statistical analyses. J.G.P. performed bioinformatic analyses. The following authors from the various collaborating groups conceived the local study, undertook assembly of case/control series in their respective regions, collected data and samples, variously undertook genotyping and analysis: I.N., S.T., A.K. and L.A.A. in Finland; T.K. and P.D.P. in Cambridge; S.S., H.V., S.B., C.S., J.T. and J.H. in Kiel; J.C.-C., M.H. and H.B. in Heidelberg; B.W.Z., A.M., T.J.H. and S.G., in Toronto and Montreal. All undertook sample collection and phenotype data collection and collation in the respective centers.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
The full list of authors and affiliations is as follows:
Group 1: Richard S Houlston2, Emily Webb2, Peter Broderick2, Alan M Pittman2, Maria Chiara Di Bernardo2, Steven Lubbe2, Ian Chandler2, Jayaram Vijayakrishnan2, Kate Sullivan2, Steven Penegar2 & Colorectal Cancer Association Study Consortium20
Group 2: Luis Carvajal-Carmona3, Kimberley Howarth3, Emma Jaeger3, Sarah L Spain3, Axel Walther3, Ella Barclay3, Lynn Martin3, Maggie Gorman3, Enric Domingo3, Ana S Teixeira3, CoRGI Consortium20, David Kerr4, Jean-Baptiste Cazier5, Iina Niittymäki6, Sari Tuupanen6, Auli Karhu6, Lauri A Aaltonen6 & Ian P M Tomlinson3
Group 3: Susan M Farrington7, Albert Tenesa7, James G D Prendergast7, Rebecca A Barnetson7, Roseanne Cetnarskyj8, Mary E Porteous8, Paul D P Pharoah9, Thibaud Koessler9, Jochen Hampe10, Stephan Buch10, Clemens Schafmayer11,12, Jurgen Tepel12, Stefan Schreiber11,13, Henry Völzke14, Jenny Chang-Claude15, Michael Hoffmeister16, Hermann Brenner16, Brent W Zanke17, Alexandre Montpetit18, Thomas J Hudson18,19, Steven Gallinger19, International Colorectal Cancer Genetic Association Consortium20, Harry Campbell7 & Malcolm G Dunlop7
2Section of Cancer Genetics, Institute of Cancer Research, Sutton, SM2 5NG, UK. 3Molecular and Population Genetics, Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK. 4Department of Clinical Pharmacology, Oxford University, Old Road Campus Research Building, Oxford, OX2 6HA, UK. 5Bioinformatics and Biostatistics, Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK. 6Department of Medical Genetics, Genome-Scale Biology Research Program, Biomedicum Helsinki, University of Helsinki, Helsinki, Finland. 7Colon Cancer Genetics Group, Institute of Genetics and Molecular Medicine, University of Edinburgh and MRC Human Genetics Unit, Edinburgh EH4 2XU, UK. 8Clinical Genetics, Western General Hospital, Edinburgh EH4 2XU, UK. 9Cancer Research UK Laboratories, Strangeways Research Laboratory Department of Oncology, University of Cambridge, Cambridge, UK. 10Department of General Internal Medicine, University Hospital, Schleswig-Holstein, Campus Kiel, Schittenhelmstraβe 12, 24105 Kiel, Germany. 11POPGEN Biobank, University Hospital Schleswig-Holstein, Campus Kiel, Schittenhelmstrasse 12, 24105 Kiel, Germany. 12Department of General and Thoracic Surgery, University Hospital Schleswig-Holstein, Campus Kiel, Arnold-Heller-Strasse, 24105 Kiel, Germany. 13Institute for Clinical Molecular Biology, University Hospital Schleswig-Holstein, Campus Kiel, Schittenhelmstrasse 12, 24105 Kiel, Germany. 14Institut für Epidemiologie und Sozialmedizin, University Hospital Greifswald, Walther-Rathenau-Strasse 48 Greifswald 17487, Germany. 15Division of Cancer Epidemiology, German Cancer Research Center (DKFZ) and 16Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany. 17The Ottawa Health Research Institute, Division of Epidemiology, 501 Smythe Road, Ottawa, Canada, K1H 8L6. 18The McGill University and Genome Quebec Innovation Centre, 740 Dr Penfield Avenue, Montreal Quebec, Canada H3G 1A4. 19Cancer Care Ontario, 620 University Avenue, Toronto Ontario M5G 1L7 and Ontario Institute for Cancer Research, 101 College Street, Toronto, Canada M5G 2L7. 20A full list of members is provided in the Supplementary Note online.
References
- 1.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin. Cancer Res. 2007;13:356–361. doi: 10.1158/1078-0432.CCR-06-1256. [DOI] [PubMed] [Google Scholar]
- 3.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 4.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 6.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaeger E, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat. Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 8.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 10.Kim JS, et al. Oncogenic β-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–2748. [PubMed] [Google Scholar]
- 11.He XC, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 12.Howe JR, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 13.Zhou XP, et al. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am. J. Hum. Genet. 2001;69:704–711. doi: 10.1086/323703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peck JW, Oberst M, Bouker KB, Bowden E, Burbelo PD. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem. 2002;277:43924–43932. doi: 10.1074/jbc.M203569200. [DOI] [PubMed] [Google Scholar]
- 15.Bellovin DI, et al. Reciprocal regulation of RhoA and RhoC characterizes the EMT and identifies RhoC as a prognostic marker of colon carcinoma. Oncogene. 2006;25:6959–6967. doi: 10.1038/sj.onc.1209682. [DOI] [PubMed] [Google Scholar]
- 16.Chang YW, Marlin JW, Chance TW, Jakobi R. RhoA mediates cyclooxygenase-2 signaling to disrupt the formation of adherens junctions and increase cell motility. Cancer Res. 2006;66:11700–11708. doi: 10.1158/0008-5472.CAN-06-1818. [DOI] [PubMed] [Google Scholar]
- 17.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem. Soc. Trans. 2005;33:672–675. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler JM, et al. Hypermethylation of the promoter region of the E-cadherin gene (CDH1) in sporadic and ulcerative colitis associated colorectal cancer. Gut. 2001;48:367–371. doi: 10.1136/gut.48.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilford P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 20.Richards FM, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum. Mol. Genet. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 21.Salahshor S, et al. A germline E-cadherin mutation in a family with gastric and colon cancer. Int. J. Mol. Med. 2001;8:439–443. doi: 10.3892/ijmm.8.4.439. [DOI] [PubMed] [Google Scholar]
- 22.Li LC, et al. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873–876. [PubMed] [Google Scholar]
- 23.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl. Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penegar S, et al. National study of colorectal cancer genetics. Br. J. Cancer. 2007;97:1305–1309. doi: 10.1038/sj.bjc.6603997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen T, Matakidou A, Consortium G, Houlston R. Identification of low penetrance alleles for lung cancer: The GEnetic Lung CAncer Predisposition Study (GELCAPS) BMC Cancer. 2008;8:244. doi: 10.1186/1471-2407-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith BH, et al. Generation Scotland: the Scottish Family Health Study; a new resource for researching genes and heritability. BMC Med. Genet. 2006;7:74. doi: 10.1186/1471-2350-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int. J. Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. Geneva: World Health Organization; International Classification of Diseases. 1975 Revision. 1977
- 32.Zhou XP, et al. Determination of the replication error phenotype in human tumors without the requirement for matching normal DNA by analysis of mononucleotide repeat microsatellites. Genes Chromosomes Cancer. 1998;21:101–107. doi: 10.1002/(sici)1098-2264(199802)21:2<101::aid-gcc4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Boland CR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 34.Petitti D. Meta-analysis Decision Analysis and Cost-Effectiveness Analysis. Oxford, New York: Oxford; 1994. [Google Scholar]
- 35.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 36.Houlston RS, Ford D. Genetics of coeliac disease. QJM. 1996;89:737–743. doi: 10.1093/qjmed/89.10.737. [DOI] [PubMed] [Google Scholar]
- 37.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 38.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am. J. Gastroenterol. 2001;96:2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 39.Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 40.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 41.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet. Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Closas M, Lubin JH. Power and sample size calculations in case-control studies of gene-environment interactions: comments on different approaches. Am. J. Epidemiol. 1999;149:689–692. doi: 10.1093/oxfordjournals.aje.a009876. [DOI] [PubMed] [Google Scholar]
- 43.Lubin JH, Gail MH. On power and sample size for studying features of the relative odds of disease. Am. J. Epidemiol. 1990;131:552–566. doi: 10.1093/oxfordjournals.aje.a115530. [DOI] [PubMed] [Google Scholar]
- 44.Cuzick J. A Wilcoxon-type test for trend. Stat. Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.