Abstract
The mylk1 gene is a large gene spanning ~250 kb and comprising at least 31 exons. The mylk1 gene encodes at least four protein products: two isoforms of the 220-kDa myosin light chain kinase (MLCK), a 130-kDa MLCK, and telokin. Transcripts encoding these products are derived from four independent promoters within the mylk1 gene. The kinases expressed from the mylk1 gene have been extensively characterized and function to regulate the activity of nonmuscle and smooth muscle myosin II. Activation of these myosin motors by MLCK modulates a variety of contractile processes, including smooth muscle contraction, cell adhesion, migration, and proliferation. Dysregulation of these processes contributes to a number of diseases. The noncatalytic gene product telokin also has been shown to modulate contraction in smooth muscle cells through its ability to inhibit myosin light chain phosphatase. Given the crucial role of the products of the mylk1 gene in regulating numerous contractile processes, it seems intuitive that alterations in the transcriptional activity of the mylk1 gene also will have a significant impact on many physiological and pathological processes. In this review we highlight some of the recent studies that have described the transcriptional regulation of mylk1 gene products in smooth muscle tissues and discuss the implications of these findings for regulation of expression of other smooth muscle-specific genes.
Keywords: myocardin, serum response factor, GATA, gene expression, Hox genes
MYOSIN LIGHT CHAIN KINASES (MLCK) are encoded by at least two genes in mammals, the mylk1 and mylk2 genes (23, 41, 90, 95).1 The mylk2 gene encodes a MLCK isoform that is exclusively expressed in skeletal muscle cells (95). In contrast, the products of the mylk1 gene are widely expressed, being detectable in many, if not all, tissues and cell types, including skeletal and cardiac muscle (19, 27). In this review we focus on the regulation of the mylk1 gene, because there are no published studies on the regulation of the mylk2 gene. Three protein products encoded by the mylk1 gene have been described, and they include two different isoforms of MLCK and the noncatalytic gene product, called telokin (Fig. 1). The two kinases expressed from the mylk1 gene have been extensively characterized and function to regulate the activity of nonmuscle and smooth muscle myosin II. Activation of these myosin motors by MLCK modulates a variety of contractile processes, including smooth muscle contraction, cell adhesion, migration, and proliferation (36, 38), and dysregulation of these processes contributes to a number of diseases. For example, in the vascular system, alterations of these contractile processes directly contribute to the vascular pathologies that occur during the development of atherosclerosis, restenosis, and hypertension (62). Given the crucial role of MLCKs in regulating numerous contractile processes, it seems intuitive that alterations in the transcriptional activity of the mylk1 gene also will have a significant impact on the pathogenesis of these diseases. In this review we highlight some of the recent studies that have investigated the transcriptional regulation of mylk1 gene in smooth muscle tissues and discuss the implications of these findings for regulation of expression of other smooth muscle-specific genes.
Fig. 1.
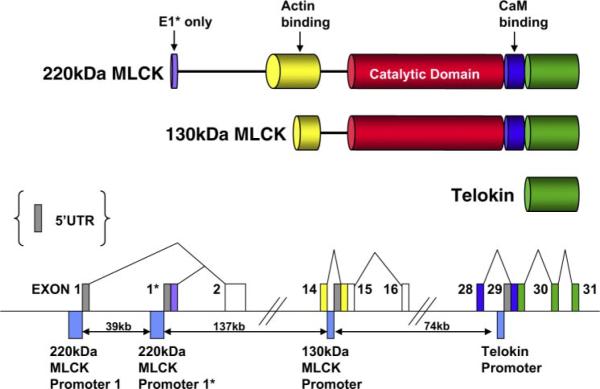
Structure of the mouse mylk1 gene. Top schematic is a representation of the domain structure of protein products of the mylk1 gene. As illustrated, the amino acid sequence of telokin is identical to the COOH terminus of the myosin light chain kinase (MLCK) molecules. Likewise, the amino acid sequence of the 130-kDa MLCK is identical to the common COOH-terminal portion of the 220-kDa MLCK. Bottom schematic is a representation of a portion of the mouse mylk1 gene (not to scale; see Supplemental Data for sequence information).2 Exons are indicated by boxes and are color coded to match the domains in the top schematic. The dark gray boxes represent the unique 5′-untranslated region (UTR) segments of each transcript. Promoter regions are indicated by the blue boxes below the line. Genomic sequence predicts that two 5′ promoters direct the expression of the two isoforms of the 220-kDa MLCK that differ from each other only in the sequence encoded by exon 1* of the 220-kDa MLCK E1*. The protein produced from usage of exon 1* is predicted to be 9 amino acids longer than the protein resulting from usage of exon 1. Two internal promoters direct the expression of transcripts encoding the 130-kDa MLCK and telokin. (For specific sequences, see Supplemental Data).
MLCK FUNCTION
MLCK phosphorylates the 20-kDa myosin regulatory light chain (RLC) of smooth and nonmuscle myosin II in the presence of Ca2+ and calmodulin, which facilitates myosin interaction with actin filaments (20). In most, if not all, cell types, RLC phosphorylation induced by the calcium/calmodulin-dependent activation of MLCK is important for regulating actomyosin-based cytoskeletal functions such as focal adhesion and stress fiber formation, secretion, ion exchange, cytokinesis, neurite growth cone advancement, cell spreading, endothelial and epithelial barrier formation, and cell migration (36). In smooth muscle, phosphorylation of the myosin RLC is also known to be an obligatory step involved in the initiation of contraction. Given the pivotal role of MLCK in regulating numerous functions in nonmuscle cells and for smooth muscle contractile activity, it is likely that changes in the expression of MLCK will have profound effects on the physiological functions of cells. For example, changes in MLCK expression in the vascular system will affect barrier function in endothelial cells and tone in vascular smooth muscle cells, thereby affecting blood pressure. Consistent with this concept, several recent studies have described changes in MLCK expression during development (13, 17, 27) and under various pathological conditions, including vascular injury (21), intestinal inflammation (3, 11, 47, 82), airway hypersensitivity (35, 72), and dedifferentiation of smooth muscle following culture in vitro (48).
Although the 220- and 130-kDa MLCKs have distinct intracellular localizations and cell-specific expression patterns (4), both of the MLCK products of the mylk1 gene have an identical catalytic domain. It remains unclear whether each of the MLCK isoforms have distinct or overlapping functions (Fig. 1). Mice harboring a specific knockout of the 220-kDa MLCK (both isoforms shown in Fig. 1) exhibited no gross morphological abnormalities, were viable and fertile, and exhibited no major cardiovascular defects (61, 79). However, the 220-kDa MLCK knockout mice exhibited decreased susceptibility to lung injury and an impairment of shear stress-induced vasodilatation (61, 79). These data suggest that the 220- and 130-kDa MLCKs exhibit at least some nonredundant functions. As a result of the overlapping structures of the mylk1 gene products, to generate specific knockouts of the 130-kDa MLCK and telokin without altering expression of other products of the mylk1 gene, it is necessary to delete key promoter elements responsible for their expression. This approach was recently used to generate telokin knockout mice (38).
TELOKIN FUNCTION
Telokin is a 17-kDa acidic protein that is expressed exclusively in smooth muscle tissues and cells (29, 34). Telokin is expressed at very high levels in intestinal, urinary, and reproductive tract smooth muscle, at lower levels in vascular smooth muscle, and at undetectable levels in skeletal or cardiac muscle or nonmuscle tissues (10, 18, 29). Recent studies have begun to resolve the physiological function of telokin. Telokin previously has been shown to stabilize myosin filaments in vitro and to inhibit MLCK activity toward intact myosin substrates (37, 59, 68). Telokin also has been shown to activate myosin light chain phosphatase and to be important for cGMP-mediated calcium desensitization of phasic smooth muscle tissues (10, 38, 40, 80, 87). Telokin knockout mice, generated by deleting key regulatory elements from the core of the telokin promoter, exhibit decreased myosin phosphatase activity, resulting in a leftward shift in the calcium-force relationship in visceral but not vascular smooth muscle tissues (38). The higher level of expression of telokin in visceral compared with vascular and smooth muscle cells can thus account for the lower calcium sensitivity of contraction in visceral compared with vascular tissues (10). These findings demonstrate that regulation of the expression of telokin in smooth muscle tissues will thus modulate its contractile properties.
The biochemical and physiological functions of MLCK and telokin in regulation of contractile processes have been studied extensively, and numerous reports have highlighted changes in the activity of the mylk1 gene in pathological processes. The mechanisms responsible for mediating the changes in mylk1 gene expression, however, have only recently begun to be scrutinized.
MYLK1 GENE STRUCTURE
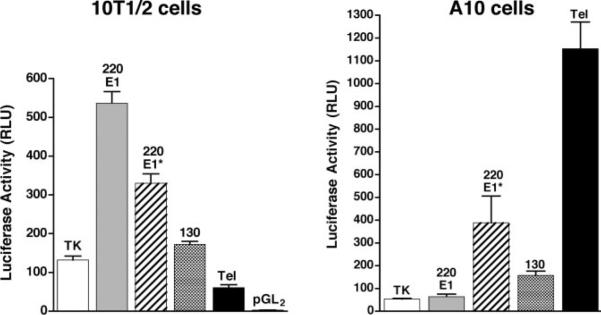
Comparison of MLCK and telokin cDNA sequences, obtained from our laboratory and available databases, with the mouse genome sequence facilitated mapping of the entire mouse mylk1 gene that encodes these proteins (Fig. 1). A similar structure also has been reported based on the sequence of a BAC clone and homology to the human genome (23). The mylk1 gene is a large gene spanning ~250 kb and comprising at least 31 exons, located on mouse chromosome 16. Sequence analysis, together with RNA and protein analysis, suggests that the gene encodes at least four protein products: two isoforms of the 220-kDa MLCK [also known as endothelial (22), long (36), or nonmuscle MLCK (70)], a 130-kDa MLCK [also known as short (36) or smooth muscle MLCK (19)], and telokin [also known as kinase-related protein (85)]. Transcripts encoding these products are derived from four independent promoters within the gene (Figs. 1 and 2). The published mouse 220-kDa MLCK cDNA begins in exon 1*; however, a number of mouse EST clones skip this exon and include a more 5′ exon 1 instead, suggesting that there are two distinct isoforms of the 220-kDa MLCK. The genomic sequences immediately 5′ of exon 1 and exon 1* both exhibit promoter activity, suggesting that the two 220-kDa MLCK isoforms likely arise from alternative promoter usage rather than alternative splicing (Fig. 2). Recently, work in our laboratory (90) showed that intron 14 of the mylk1 gene contains the proximal promoter that drives expression of the 130-kDa MLCK. It also has been shown that telokin mRNA is transcribed from a promoter, located in intron 28 of the mylk1 gene, that interrupts exons encoding the calmodulin-binding domain of the kinase (Fig. 1 and Refs. 12, 18, 31, 94). In addition to the mouse MLCK transcripts produced by the independent promoters, it is likely that additional transcripts are derived by alternative splicing within the mylk1 gene, as described for the human MYLK1 locus (41). Each of the promoters within the mylk1 gene exhibits cell-specific differences in their activity. For example, in 10T1/2 fibroblasts, the 220-kDa MLCK E1 promoter exhibits the highest level of activity, whereas in A10 smooth muscle cells, the telokin promoter has the highest levels of activity (Fig. 2). In the current review, we summarize results from studies aimed at unraveling the transcription regulatory network that controls the activity of the mylk1 gene, and we highlight how these studies have provided important paradigms central to our understanding of the general mechanisms that regulate gene expression in smooth muscle cells.
Fig. 2.
Transcriptional activity of the 4 predicted promoters within the mylk1 gene. Luciferase reporter genes were generated containing genomic fragments (700 and 750 bp, respectively) immediately 5′ of exons E1 (220 E1) or E1* (220E1*) or comprising nucleotides −389 to +115 of the 130-kDa MLCK (130) promoter, −190 to +180 of the telokin (Tel) promoter, or −113 to +20 of the thymidine kinase (TK) promoter. These reporter plasmids or the empty luciferase vector pGL2B were transfected into 10T1/2 embryonic fibroblasts of A10 smooth muscle cells, together with an internal control renilla luciferase plasmid. At 24 h after transfection, lysates were prepared and analyzed for luciferase activity. Results presented are relative light units (RLU) of luciferase activity normalized to the renilla luciferase internal control. Values are means ± SE of 6 samples.
AN AT-RICH ELEMENT AND CARG BOX ARE CRITICAL FOR TELOKIN PROMOTER ACTIVITY
A 370-bp (−190 to +180 bp) mouse telokin promoter has been shown to be sufficient to mediate cell-specific expression of a reporter gene in vitro and to direct smooth muscle-specific expression in vivo in adult transgenic mice (33). Within this promoter, a CArG element that binds to serum response factor (SRF) and an adjacent AT-rich element were found to be essential for promoter activity in vitro (30, 33) and in vivo (38). In this latter study, the AT/CArG core of the endogenous telokin promoter was deleted using a Cre-based knockout strategy. After Cre-mediated recombination, a 30-bp region including the AT/CArG motif was deleted and replaced by a single loxP site. Mice homozygous for the 30-bp deletion within the telokin promoter revealed an almost complete loss of telokin protein and mRNA expression with only minimal effects on MLCK expression (Ref. 38 and Fig. 3). These results unequivocally indicate that in the context of the native mylk1 gene, the AT-CArG region of the telokin promoter is critical for endogenous promoter activity. This study represents one of the first examples in which regulatory elements have been demonstrated to be important for regulating smooth muscle-specific gene expression in their native chromatin context in vivo. Analysis of regulatory elements in their native chromatin context is likely to be particularly important for a complex gene such as the mylk1 gene, in which a single gene contains a number of overlapping transcription units. Although the telokin promoter is only active in smooth muscle cells, the other MLCK promoters are likely to be active in most cell types. Hence, the mylk1 gene is likely to be transcribing through the telokin exons in many cell types in which the telokin promoter itself is inactive. Thus, although the telokin promoter is not active in nonmuscle cells, the body of the telokin gene is likely to be in a relaxed, transcriptionally favorable conformation because of chromatin remodeling induced by the MLCK promoters. A detailed analysis of the chromatin structure throughout the mylk1 locus, in different cell types, is required to understand how chromatin structure regulates expression of the mylk1 transcripts.
Fig. 3.
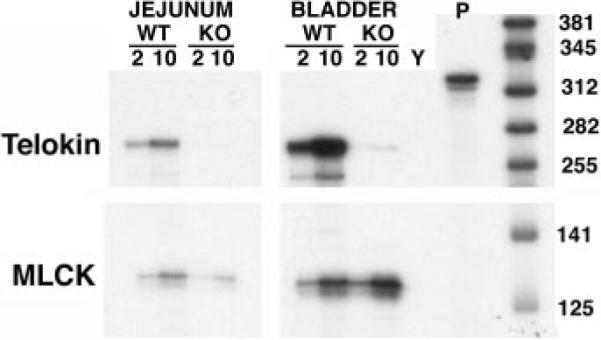
Deletion of the AT-rich region and CArG box from the endogenous telokin promoter specifically abolished telokin expression. An RNase protection assay was performed on RNA isolated from jejunum and bladder of wild-type (WT) and telokin knockout (KO) mice. The probe used (P) protects an ~255-bp fragment of the telokin mRNA and a 125-bp fragment of the MLCK mRNA. The positions of RNA size markers are indicated at right of the blot in base pairs. Either 2 or 10 μg of RNA were used in the assay, as indicated. Y, yeast RNA.
TELOKIN AT/CARG MOTIF ACTS AS A BLADDER SMOOTH MUSCLE CELL-RESTRICTED ENHANCER
Addition of the AT-rich region/CArG box from the telokin promoter (−49 to −94 bp) to the SM22α promoter (−475 to +61 bp) increased the activity of the SM22α promoter in visceral smooth muscle cells (33). This fragment of the SM22α gene normally directs transgene expression exclusively to arterial smooth muscle (33, 39, 42). Of the 10 AT-CArG/SM22α promoter transgenic lines that transmitted and expressed β-galactosidase activity, all exhibited staining in bladder smooth muscle cells in addition to vascular smooth muscle cells (33). Interestingly, this chimeric promoter fragment did not increase transgene expression in intestinal or reproductive tract smooth muscle cells. These data suggest that other elements, outside the −49 to −94-bp region of the telokin promoter, are required for transcription in intestinal and reproductive tract smooth muscle cells (Fig. 4A). These findings also highlight an important paradigm in which a single promoter uses distinct cis-acting regulatory elements to drive expression in different smooth muscle tissues. This raises the exciting possibility that it may be possible to develop reagents that specifically alter expression of a protein, in a single smooth muscle cell type, through targeting the tissue-specific elements.
Fig. 4.
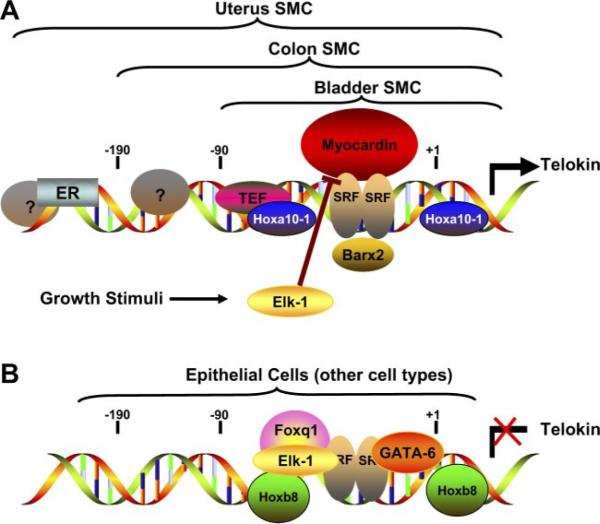
Telokin promoter regulation in smooth muscle and nonmuscle cells. A schematic model is shown of the transcription factors that bind to the telokin promoter in smooth muscle cells (SMC) (A) and in epithelial or nonmuscle cells (B). Analysis of promoter fragments in transgenic mice has suggested that the core of the telokin promoter, which includes a CArG box and adjacent AT-rich region, is required for expression in all smooth muscle tissues (38), and a −90 to +180-bp fragment that includes this region is sufficient for expression in bladder smooth muscle cells (33). Additional unidentified elements between −90 and −190 bp are required for telokin expression in smooth muscle cells in the gastrointestinal tract (33). In most higher mammals, with the exception of rodents, the estrogen receptor (ER) and perhaps other proteins that bind to more distal sequences are required for expression in uterine smooth muscle cells (71). Transcription factors that have been found to bind to the core of the telokin promoter directly [Hoxa10-1, thyrotroph embryonic factor (TEF), and serum response factor (SRF) (16, 30, 97)] or indirectly through their association with SRF [Barx2 and Myocardin (28, 96)] and activate the promoter are shown in A. Transcriptional inhibitors that repress the activity of the telokin promoter in epithelial cells or other nonmuscle cell types are shown in B. The relative contribution of each of the transcription factors to the transcriptional activity of the telokin promoter is likely to be different in smooth muscle cells from distinct tissues and in different nonmuscle cells. GATA-6 may inhibit telokin promoter activity in vascular smooth muscle cells by competing with myocardin for binding to SRF. Similarly, because Elk-1 also can compete with myocardin for binding to SRF, growth factor stimulation of smooth muscle cells may result in activation of Elk-1 and inhibition of telokin expression (98).
SERUM RESPONSE FACTOR PLAYS A CENTRAL ROLE IN REGULATING EXPRESSION OF MYLK1 GENE
Similar to the telokin promoter, the proximal promoter of the 130-kDa MLCK contains a CArG element that has been shown to be critical for transcriptional activity. In addition, a highly conserved CArG element within 130-kDa MLCK intron 1 (intron 15 of the mylk1 gene) has been proposed to be important for promoter activity (Fig. 5) (90). Each of these CArG boxes within the mylk1 gene has been shown to bind to serum response factor (SRF) both in vitro and in vivo in native chromatin (30, 90, 98). SRF and SRF-associated factors play a central role in the expression of many different smooth muscle-specific genes, including smooth muscle myosin heavy chain (smMHC), smooth muscle α- and γ-actin, SM22α, and calponin, in addition to 130-kDa MLCK and telokin (53, 90). SRF is an evolutionarily conserved MADS (MDM1, agamous, deficiens, SRF) domain-containing protein that is required for specification of smooth, cardiac, and skeletal muscle lineages from early mesoderm (2). SRF is a multifunctional protein that binds a highly conserved cis-regulatory element, CC(A/T)6GG, termed a CArG box, found in the promoters of most cardiac and smooth muscle-specific genes. SRF also provides docking surface, within the conserved core MADS domain, for interaction with a wide variety of accessory cofactors. Although SRF expression is greatest in muscle tissues and plays an important role in the differentiation of smooth, cardiac, and skeletal muscle tissues, it is expressed in all tissues, where it is important for the growth factor-mediated induction of immediate-early genes (53). To perform this myriad of functions, SRF associates with various cell-restricted and/or signal-dependent accessory factors that confer coactivator or corepressor activity via ternary complex formation. A number of different SRF-complexes have been shown to regulate the activity of smooth muscle-specific genes. For example, Nkx3.1 and Nkx2.3 are tissue-restricted homeodomain-containing proteins that have been shown to interact with SRF and cooperatively activate the smooth muscle γ-actin promoter (6, 64). Several other homeodomain transcription factors, including Barx1b, Barx2b, and Prx (MHox), have been shown to enhance the DNA binding activity of SRF (26, 28, 57, 91). An architectural transcription factor, HMGI(Y), also has been reported to interact with SRF and activate the SM22α promoter (9). In addition, a trimeric complex of SRF-GATA6-CRP2 has been reported to be required for the induction of smooth muscle differentiation in cells from the proepicardial organ, which give rise to coronary artery smooth muscle cells (7), and a complex of Nkx3.2/SRF/GATA-6 has been shown to cooperatively activate the SM22α promoter (58). However, none of these trimeric complexes has yet been shown to regulate either MLCK or telokin promoters. On the other hand, GATA-6 has been shown to repress the activity of the 130-kDa MLCK and telokin promoters, possibly through disruption of SRF-myocardin complexes (Figs. 4 and 5) (90). The myocardin family of SRF-associated proteins are very powerful activators of many smooth muscle-specific promoters, including both the 130-kDa MLCK and telokin promoters (Figs. 4A and 5A) (8, 14, 15, 43, 45, 84, 90, 92, 93, 96).
Fig. 5.
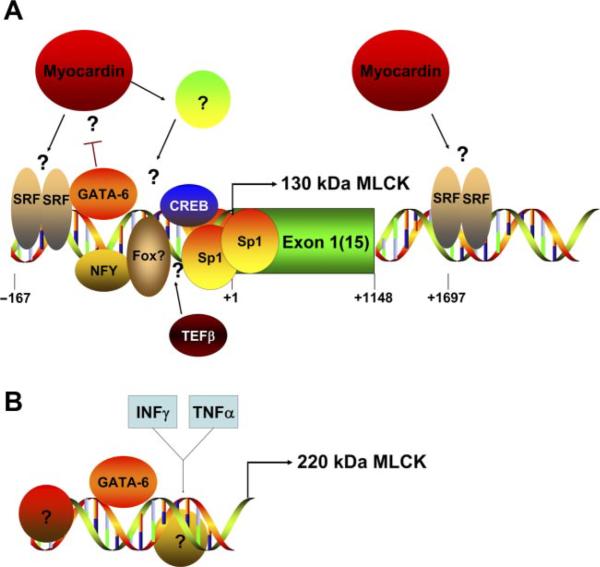
Regulation of the MLCK promoters. In A, the schematic representation depicts the transcriptional regulation of the 130-kDa MLCK promoter (90). Two conserved CArG boxes, one in the proximal promoter and one in the first intron, have been shown to bind SRF in vivo (90). Whether myocardin directly or indirectly regulates the promoter remains to be determined. GATA-6 has been shown to bind to the promoter and inhibit its activity. TEFβ has been shown to increase expression of the 130-kDa MLCK, although its binding site on the promoter has not been defined (97). The presence of the other factors (Fox, Sp1, CREB, NFY) on the promoter is based purely on their consensus binding sites within the promoter sequence and has not yet been verified experimentally. In B, the putative structure of the 220-kDa MLCK promoter(s) is indicated. To date, these promoters have not been characterized, except for the observations that GATA-6 and INF-γ/TNF-α stimulation can increase 220-kDa MLCK expression (47, 82, 90).
MYOCARDIN, AN SRF-DEPENDENT COACTIVATOR, IS IMPORTANT FOR MLCK AND TELOKIN GENE EXPRESSION
Myocardin is a SAP (SAF-A/B, acinus, PIAS) domain-containing protein that was identified by an in silico screen of cardiac-restricted cDNAs (81). Myocardin contains a basic domain, a polyglutamine-rich domain (Q-rich), and a SAP domain in the NH2-terminal portion of the protein and a strong transcriptional activation domain at the COOH terminus (81). Myocardin is restricted to cardiac and smooth muscle lineages in developing mouse embryos. It is an extraordinarily potent activator of CArG box-containing cardiac and smooth muscle promoter-reporter gene constructs, in a manner that is strictly dependent on its association with SRF (81). Overexpression of myocardin in a number of different cell types leads to the induction of many smooth muscle-restricted proteins, including the 130-kDa MLCK and telokin (8, 15, 75, 76, 81, 84, 90, 92, 96). Myocardin null mice die around embryonic day 10.5 due to defects in the placental blood vessels (84). These mice also display no detectable vascular smooth muscle cells in the developing aorta, suggesting that myocardin is required for the development of vascular smooth muscle cells. Unfortunately, because myocardin null embryos die before the normal onset of smooth muscle differentiation in visceral smooth muscle tissues, it has not yet been possible to determine its role in smooth muscle differentiation in these tissues. Intriguingly, it has recently been reported that myocardin null embryonic stem cells can give rise to smooth muscle cells that express smooth muscle-specific markers in vitro, suggesting that not all smooth muscle lineages are dependent on myocardin (65). Myocardin downregulation in cultured smooth muscle cell lines correlates with the decreased expression of smooth muscle-specific contractile proteins (8). These data suggest that myocardin may function not only in the initial steps of smooth muscle cell differentiation but also in the maintenance of differentiation (15, 93). Myocardin specifically activates the promoters of SRF-dependent cardiac- and smooth muscle-restricted genes without affecting other SRF-dependent genes such as c-fos or egr1 (96). It has been proposed that the number of CArG boxes within a gene is critical for myocardin to distinguish between different SRF-dependent genes, with myocardin-dependent genes having two or more CArG boxes and myocardin-independent genes having only a single CArG box (84). Although two CArG boxes are important for the activity of many smooth muscle-specific genes, this mechanism is not sufficient to explain the gene-specific effects of myocardin. For example, some myocardin-independent promoters, such as the Egr1 promoter, contain multiple CArG boxes, and the smooth muscle-specific myocardin-dependent telokin promoter contains only one CArG box (96). In addition, analysis of the smooth muscle α-actin promoter in transgenic mice demonstrated that a promoter fragment containing two CArG boxes was not sufficient to drive smooth muscle cell-specific transgene expression (49).
Myocardin differentially affects the promoters within the mylk1 gene and is able to induce expression of the 130-kDa MLCK and telokin in many cell types without altering expression of the 220-kDa MLCK (90, 96). Although the telokin promoter, which contains only a single CArG box, is activated several hundredfold by myocardin (Fig. 3), the proximal 130-kDa MLCK promoter, which also contains a single CArG box, is only activated two- to threefold (90). In addition, depletion of myocardin in A10 smooth muscle cells by short hairpin RNA significantly decreased telokin expression but did not affect 130-kDa MLCK expression (96). These data could simply reflect a longer half-life of the 130-kDa MLCK protein compared with telokin. Alternatively, it is more likely that telokin is more dependent on myocardin for expression in A10 smooth muscle cells compared with 130-kDa MLCK. In fact, the broad tissue distribution of 130-kDa MLCK compared with telokin would suggest that myocardin-independent pathways must exist for activating this promoter in other cell types (Fig. 5A). Nevertheless, the ability of myocardin to induce 130-kDa MLCK expression in 10T1/2 fibroblasts without significantly activating the promoter in transient reporter assays suggests that the elements required for myocardin to activate this promoter may be located a large distance from the promoter (>6 kb). This arrangement would be unique among myocardin-stimulated genes. Alternatively, the 130-kDa MLCK promoter might be activated indirectly by another myocardin-stimulated transcription factor. Careful analysis of myocardin binding to the endogenous 130-kDa MLCK promoter is required to distinguish between these possibilities.
How myocardin distinguishes smooth muscle-restricted genes from growth factor-responsive SRF-dependent genes remains elusive. Data obtained from analysis of the telokin and c-fos promoters suggest that additional cis elements and trans factors contribute to myocardin's promoter-specific effects (96). For example, binding of Elk-1 adjacent to the CArG box, within the c-fos serum response element, may in part prevent myocardin from activating this promoter (Fig. 4). Although Elk-1 and myocardin have been shown to compete for binding to SRF, the binding of Elk-1 to the c-fos promoter is not sufficient to explain why myocardin does not activate this promoter. In fact, Elk-1 also has been shown to bind to the telokin and SM22α promoters and to partially block myocardin's interaction with SRF on these promoters, leading to downregulation of their activity (83, 98). In addition, a chimeric telokin promoter containing the Elk-1 binding site and CArG box from the c-fos promoter was still strongly activated by myocardin, suggesting that the presence of these elements alone is not sufficient to prevent myocardin from activating the promoter (96). Identification of additional factors that bind to the telokin promoter and cooperate with myocardin is required to help unravel the mechanisms of promoter selectivity by myocardin.
ACTIVATION OF MYLK1 GENE BY THYROTROPH EMBRYONIC FACTOR
In addition to SRF-associated proteins, several other trans factors have been identified that regulate mylk1 transcription, including thyrotroph embryonic factor (TEF) (97), Foxq1 (32), and Hox proteins (16) (Figs. 4 and 5). These proteins bind to the AT-rich region adjacent to the CArG box in the telokin promoter. TEF is a member of the PAR (proline- and acidic-amino acid-rich) subfamily of bZIP transcription factors. Overexpression of TEFβ upregulated endogenous telokin and 130-kDa MLCK expression in A10 smooth muscle cells without affecting expression of other smooth muscle-restricted proteins, such as smooth muscle α-actin or calponin (97). Although overexpression of a dominant negative TEF repressed telokin and 130-kDa MLCK expression, no studies have examined the expression of these proteins following knockdown or knockout of endogenous TEF. The physiological role of TEF in regulating expression of the mylk1 gene in different smooth muscle tissues thus remains to be determined.
REGULATION OF MYLK1 GENE BY HOMEOBOX PROTEINS
In addition to being regulated by the homeodomain containing protein Barx, which can increase SRF DNA binding activity (28), the telokin promoter is directly regulated by the products of clustered homeobox genes (Hox) (16). Homeobox genes encode a family of highly conserved transcription factors that share a conserved 60-amino acid homeodomain, which binds to AT-rich consensus sequences in DNA (67). Homeobox proteins are master regulatory proteins that control basic developmental processes. They are differentially expressed along the anterioposterior (AP) axis and are believed to play an important role in organogenesis and pattern formation during both vertebrate and invertebrate embryogenesis (51). Several studies also have indicated an important role of homeodomain-containing proteins in regulating differentiation, proliferation, and migration in different cell types (52). Homeodomain protein expression often persists in adult tissues, where these proteins are believed to play a role in maintaining cell type specification (54). The spatial pattern of homeobox protein expression makes them ideal candidates for specifying distinct smooth muscle cell phenotypes in different tissues. A number of Hox proteins have been identified in smooth muscle: Hoxa2, a4, a5, and b7 were identified in an adult rat vascular smooth muscle cDNA library (24), and Hoxa10 and b8 in an adult mouse bladder cDNA library (16). In addition, HOXA5, A11, B1, B7, and C9 have been detected in human adult and fetal aortic smooth muscle tissues (54), and HOXA10 in human adult myometrium and endometrium (74). Of the identified Hox proteins, only Hoxa10, b8, and b7 have been shown to have direct effects on the mylk1 gene and other smooth muscle-specific genes. Hoxb7 overexpression promoted differentiation of C3H10T1/2 cells into myofibroblasts and induced expression of SM22α and smooth muscle α-actin (5). However, a comparison of the increased expression of HOXB7 RNA in human atherosclerotic plaques to normal adult smooth muscle tissue suggests that HOXB7 plays a role in either smooth muscle cell proliferation or dedifferentiation under pathological conditions. Within the mylk1 gene, Hoxa10 has been shown to bind directly to multiple sites in the telokin promoter and to specifically regulate telokin promoter activity (16) (Fig. 4A). Hoxa10 is expressed in adult uterine, colonic, and bladder smooth muscle tissues and cells. Several studies have indicated that Hoxa10-1 is important for the process of uterine epithelium cell differentiation and the development of subcellular structures required for implantation (73). In addition, the expression of Hoxa10-1 in the myometrium is regulated during the menstrual cycle, with high levels of estrogen increasing Hoxa10 expression (46). Previously we have shown that telokin expression is also regulated during the menstrual cycle, with high levels of expression being observed during the estrogen-dominant follicular phase and low levels of expression during the progesterone-dominant luteal phase. Telokin expression is also elevated in the uterus during pregnancy (71). A link between Hoxa10-1 and telokin expression in uterine smooth muscle was established by studies in which small interfering RNA-mediated knockdown of Hoxa10-1, in uterine smooth muscle cells, resulted in decreased telokin expression (16). Previous studies also demonstrated that the telokin promoter is directly regulated by estrogen bound to the estrogen receptor (71). Together, these studies suggest that both Hoxa10 and estrogen receptor may synergize with each other to mediate hormone-dependent changes in telokin expression in the uterus (Fig. 4). Because telokin has been shown to play a role in decreasing the calcium sensitivity of contraction through its activation of myosin light chain phosphatase (10, 38), the elevated levels of telokin in the myometrium during pregnancy may contribute to the maintenance of uterine quiescence.
In contrast to Hoxa10, Hoxb8 inhibits the activity of the telokin promoter as well as the promoters of a number of other smooth muscle-specific genes (16). The expression of both Hoxa10 and Hoxb8 mRNA in some of the same smooth muscle cells suggests that the regulation of telokin gene activity is determined by the relative ratio of Hoxa10 and Hoxb8 or that the activity of these proteins is modified posttranscriptionally. In support of this latter possibility, a microRNA has been identified that specifically represses Hoxb8 expression at the posttranscriptional level (88). The physiological significance of Hoxb8 in regulating the expression of smooth muscle-specific genes currently remains unclear. It will be interesting to determine whether increases in the level of Hoxb8 protein expression occur in pathological conditions associated with downregulation of smooth muscle-specific gene expression, as reported previously for HOXB7 (5). Finally, the restricted expression of Hox proteins along the AP axis suggests that other Hox proteins may be playing similar roles in regulating telokin expression in more anterior or posterior tissues. This spatial pattern of expression of the Hox proteins makes these proteins excellent candidates for helping to specify the unique identity of smooth muscle cells in different smooth muscle tissues.
INHIBITORY FACTORS CONTRIBUTE TO CELL-SPECIFIC TRANSCRIPTION OF TELOKIN
The tissue specific pattern of mylk1 expression is likely to be the result of the combined actions of both positive- and negative-acting transcription factors. For example, although telokin is strongly activated by myocardin and myocardin is highly expressed in vascular smooth muscle cells, there is relatively little expression of telokin in these cells (18, 29, 96). This suggests that an inhibitory factor must be attenuating the activity of the telokin promoter in vascular smooth muscle cells. One possible candidate for this inhibitory factor is GATA-6 (89, 90). A number of other proteins also have been reported to inhibit the activity of the telokin promoter, including Hoxb8, Foxq1, and Elk-1 (16, 32, 76, 98) (Fig. 4B). GATA-6 and Elk-1 both appear to act by disrupting the interaction of myocardin and SRF, and the inhibitory activity of both proteins has been proposed to be dependent on the location of their cognate cis-acting elements in the telokin promoter adjacent to the CArG box (89, 98). Interestingly, although GATA-6 also inhibits the 130-kDa MLCK promoter, and expression of the 130-kDa MLCK also can be induced by myocardin, overexpression of Elk-1 had no effect on the levels of the 130-kDa MLCK in A10 smooth muscle cells (98). These data further highlight the differences in the regulation of the 130-kDa MLCK and telokin promoters by myocardin. The mechanisms by which Foxq1 and Hoxb8 inhibit telokin promoter activity have not yet been elucidated. However, the close proximity of the consensus binding sites for Foxq1 and Hoxb8 to the CArG box, the ability of a number of homeodomain proteins to interact with SRF (6, 25, 28, 64, 91), and a recent report describing the interaction of Foxo4 (44) and myocardin make it tempting to speculate that these proteins also may disrupt key SRF activation complexes (Fig. 4B).
REGULATION OF MLCK AND TELOKIN EXPRESSION BY GATA FAMILY MEMBERS IN SMOOTH MUSCLE CELLS
GATA proteins are a family of transcription factors with two zinc fingers that directly bind DNA regulatory elements containing a consensus (A/T)GATA(A/G) motif. To date, six mammalian members of the GATA family have been identified that can be divided, on the basis of sequence and expression similarities, into two subgroups (63). The GATA-1/2/3 subfamily is preferentially expressed in the hematopoietic cells (86), and the GATA-4/5/6 subfamily is expressed within various mesoderm- and endoderm-derived tissues, including the heart, liver, lung, gonads, and small intestine (55). During early murine embryonic development, the expression patterns of GATA-6 and GATA-4 are very similar, with these factors being detected in the precardiac mesoderm, the embryonic heart tube, and the primitive gut. Notably, during development GATA-6 becomes the only member of the family expressed in vascular smooth muscle cells and has been linked to the differentiated phenotype of these cells (56). GATA-6 expression in adult vascular smooth muscle cells is rapidly down-regulated upon mitogen stimulation or vascular injury. Adeno-virus-mediated GATA-6 gene transfer to the vessel wall after balloon injury partially inhibited lesion formation and reversed the downregulation of smMHC, smooth muscle α-actin, calponin, vinculin, and metavinculin expression that is normally associated with injury-induced vascular smooth muscle cell phenotypic modulation (50). In addition to the critical role of GATA-6 in the maintenance of the differentiated phenotype in vascular smooth muscle cells, a recent report has revealed that GATA-6 may act as the predominant GATA factor in the maintenance of endodermal gene expression by TGF-β signaling in gastrulating embryos (1).
Recent studies have shown that the transcripts of the mylk1 gene are differentially regulated by GATA-6 (89, 90). Over-expression of GATA-6, by adenoviral transduction, significantly decreased endogenous telokin and 130-kDa MLCK expression in A10 vascular smooth muscle cells. In contrast, expression of the 220-kDa MLCK, SM22α, and calponin were markedly increased (89, 90). GATA-6 has been shown to bind directly to the telokin and 130-kDa MLCK promoters at consensus binding sites that are closely opposed to the CArG boxes within these promoters (Figs. 4 and 5). In addition to being recruited to promoters through binding to consensus GATA sites, GATA-6 also can be recruited to promoters through its binding to the MADS domain of SRF (60). Because both GATA-6/4 and myocardin interact with same domain of SRF, it is likely that a single SRF molecule is able to interact with either GATA-6/4 or myocardin but not with both molecules at the same time (Fig. 4). Whether GATA-6/4 or myocardin are bound to SRF appears to depend on the specific promoter context. In the case of telokin and 130-kDa MLCK promoters, there is a consensus GATA binding site adjacent to a CArG box, which would greatly facilitate the ability of GATA factors to compete with myocardin for interacting with SRF by increasing the local effective concentration of GATA-6. In contrast, GATA-6 binding to a more distal site in the smMHC promoter can activate the promoter synergistically with myocardin (78). In support of this model, the activation of both the 130-kDa MLCK and telokin promoters by myocardin can be blocked by GATA-6, and overexpression of GATA-6 also attenuates the ability of myocardin to induce expression of the endogenous 130-kDa MLCK and telokin genes (89, 90). This mechanism to explain the GATA-6-mediated repression of the 130-kDa MLCK should be interpreted with caution, because myocardin has not yet been shown to directly activate this promoter or to bind to the promoter in vivo.
Through its interaction with SRF, GATA-6 also has been shown to form complexes with other proteins to regulate expression of a number of smooth muscle-specific genes. For example, an SRF-GATA-6-Nkx3.2 complex cooperatively transactivated α-integrin, SM22α, and caldesmon promoters. This activation also required direct binding of each of these factors to their respective consensus sequences within the promoters (58). It has been reported that cysteine-rich protein 1/2 (CRP1/2) may act as scaffold protein that can stabilize SRF and GATA-6 into a transcriptosome that confers strong activation of smooth muscle-specific promoters (7). However, there have been no reports demonstrating that any of these complexes regulate the activity of the mylk1 gene. Unpublished studies from our laboratory have failed to show any differences in the regulation of the telokin promoter by a GATA-6-SRFCRP2 complex compared with GATA-6 alone. It should be noted, however, that in our hands, adenovirus-mediated over-expression of SRF, GATA-6, and CRP2 together in 10T1/2 cells was not sufficient to induce expression of any of the smooth muscle-specific genes tested.
A number of other transcription cofactors also have been reported to be involved in GATA-induced smooth muscle-specific gene expression. For instance, the transcriptional coactivator p300, together with GATA-6 and NFATc1, form a large complex during the transcription of the smMHC gene. This complex has been shown to play a role in the maintenance of the differentiated phenotype of vascular smooth muscle cells (77, 78). In contrast, the basic helix-loop-helix (bHLH) transcription factor CHF1/Hey2 can suppress the smMHC promoter activity by interacting with GATA-6 through its bHLH domain (69), and LMCD1/Dyxin has been identified to repress GATA-6 function by inhibiting its ability to bind to DNA (66). It will be important to determine whether these cofactors also are involved in GATA-6-mediated regulation of 130-kDa MLCK and telokin expression.
SUMMARY OF MECHANISMS REGULATING TISSUE-SPECIFIC EXPRESSION OF MYLK GENE
Figure 4 summarizes our current model describing the tissue-specific expression of telokin. In this model, high levels of telokin transcription in smooth muscle cells is mediated by a combination of cell- and tissue-restricted factors, acting together with more general transcription factors (Fig. 4A). Among different smooth muscle tissues, unique but overlapping sets of transcription factors are used to direct high levels of telokin expression. For example, in uterine smooth muscle, the estrogen receptor and Hoxa10-1 act together with myocardin to drive high levels of telokin expression. In contrast, in intestinal smooth muscle cells, the estrogen receptor is not required for telokin expression, and an as yet unidentified factor is required in its place (Fig. 4A). In contrast, negative-acting factors such as Foxq1 and Hoxb8, present in nonmuscle cell types, help to prevent telokin transcription in these cells (Fig. 4B). Unlike telokin, the 130-kDa MLCK, although expressed at highest levels in smooth muscle cells, is also expressed in most other cell types. Transcription of the 130-kDa MLCK is thus likely to be regulated by both smooth muscle-specific mechanisms, as well as by more general mechanisms. For example, expression of high levels of the 130-kDa MLCK in smooth muscle cells may result from the activity of myocardin. In nonmuscle cells, which express no detectable myocardin, the activity of more general factors such as Sp1 and cAMP response element binding protein (CREB) may be sufficient to direct the low levels of transcription of the 130-kDa MLCK in these cells (Fig. 5A). Analysis of the expression of the mylk gene in animal models harboring cell- and tissue-specific knockouts of factors shown to regulate the mylk gene is required to further unravel the mechanisms regulating its tissue-specific expression.
SUMMARY AND PERSPECTIVES
A number of studies have provided important insights into the transcriptional regulation of the mylk1 gene, but this is at best a partial view of the regulatory mechanisms. To date, most studies of the promoters within the mylk1 gene have primarily focused on the smooth muscle-specific regulation of telokin promoter, and many unanswered questions remain regarding the regulation of this complex gene. For example, how is the chromatin structure of the mylk1 locus regulated? Do the MLCK promoters regulate the chromatin structure of the entire mylk1 locus, including the telokin promoter? Are there additional promoters within the mylk1 gene? What fragments of the 130-kDa MLCK promoter are required to mimic endogenous 130-kDa MLCK expression in vivo? Where are the promoters of the 220-kDa MLCK isoforms, and how are they regulated? The current studies also have been limited to defining the transcriptional mechanisms responsible for regulation of mylk1 gene expression under normal physiological conditions. An important challenge of future studies will be to elucidate how these regulatory mechanisms are disrupted under pathological conditions. It also will be important to determine how the subsequent changes in expression of the mylk1 gene products contribute to the pathology. By manipulating the regulatory elements that control expression of the mylk1 gene under normal physiological conditions, researchers should be able to design in vivo mouse models with decreased expression of individual mylk1 gene products. Analysis of the physiological and pathological properties of these mice can then be used to define the specific roles of individual mylk1 gene products in vivo.
NOTE ADDED IN PROOF
Since completion of this review, two additional studies have been published that describe the transcriptional regulation of the mylk1 gene. Graham et al. describe the regulation of the human 220-kDa MLCK E1 promoter and its activation by TNFα (Graham WV, Wang F, Clayburgh DR, Cheng JX, Yoon B, Wang Y, Lin A, and Turner JR. TNF induced long myosin light chain kinase transcription is regulated by differentiation-dependent signaling events: Characterization of the human long myosin light chain kinase promoter. J Biol Chem 2006). Han et al. demonstrated that the rat 130-kDa MLCK promoter is also SRF dependent (Han YJ, Hu WY, Chernaya O, Antic N, Gu L, Gupta M, Piano M, and de Lanerolle P. Increased Myosin Light Chain Kinase Expression in Hypertension: Regulation by SRF via an Insertion Mutation in the Promoter. Mol Biol Cell 17:4039–4050, 2006).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Avril Somlyo for providing tissues from telokin knockout mice.
GRANTS This work was supported by National Institutes of Health Grants HL-58571, DK-61130, and DK-65644 (to B. P. Herring) and DK-62810 (P. J. Gallagher). J. Zhou is supported by an American Heart Association postdoctoral fellowship, and O. El-Mounayri is supported by an American Heart Association predoctoral fellowship.
Footnotes
In this review we have used the 220-kDa MLCK and 130-kDa MLCK terminology to described the MLCK products of the mylk gene for simplicity. This terminology is derived from the apparent molecular masses of the mouse MLCK molecules determined by SDS-PAGE. The true molecular masses of these molecules predicted from their cDNA sequences are 213 and 214 kDa for the two forms of the 220-kDa MLCK and 114 kDa for the 130-kDa MLCK.
The online version of this article contains supplemental data.
REFERENCES
- 1.Afouda BA, Ciau-Uitz A, Patient R. GATA4, 5 and 6 mediate TGFβ maintenance of endodermal gene expression in Xenopus embryos. Development. 2005;132:763–774. doi: 10.1242/dev.01647. [DOI] [PubMed] [Google Scholar]
- 2.Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair SA, Kane SV, Clayburgh DR, Turner JR. Epithelial myosin light chain kinase expression and activity are upregulated in inflammatory bowel disease. Lab Invest. 2006;86:191–201. doi: 10.1038/labinvest.3700373. [DOI] [PubMed] [Google Scholar]
- 4.Blue EK, Goeckeler ZM, Jin Y, Hou L, Dixon SA, Herring BP, Wysolmerski RB, Gallagher PJ. 220- and 130-kDa MLCKs have distinct tissue distributions and intracellular localization patterns. Am J Physiol Cell Physiol. 2002;282:C451–C460. doi: 10.1152/ajpcell.00333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostrom K, Tintut Y, Kao SC, Stanford WP, Demer LL. HOXB7 overexpression promotes differentiation of C3H10T1/2 cells to smooth muscle cells. J Cell Biochem. 2000;78:210–221. doi: 10.1002/(sici)1097-4644(20000801)78:2<210::aid-jcb4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle γ-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J Biol Chem. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- 7.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, Marx JG, Moore MS, Beckerle MC, Majesky MW, Schwartz RJ. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 9.Chin MT, Pellacani A, Wang H, Lin SS, Jain MK, Perrella MA, Lee ME. Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y) J Biol Chem. 1998;273:9755–9760. doi: 10.1074/jbc.273.16.9755. [DOI] [PubMed] [Google Scholar]
- 10.Choudhury N, Khromov AS, Somlyo AP, Somlyo AV. Telokin mediates Ca2+-desensitization through activation of myosin phosphatase in phasic and tonic smooth muscle. J Muscle Res Cell Motil. 2004;25:657–665. doi: 10.1007/s10974-004-7807-x. [DOI] [PubMed] [Google Scholar]
- 11.Clayburgh DR, Barrett TA, Tang Y, Meddings JB, Van Eldik LJ, Watterson DM, Clarke LL, Mrsny RJ, Turner JR. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. J Clin Invest. 2005;115:2702–2715. doi: 10.1172/JCI24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collinge M, Matrisian PE, Zimmer WE, Shattack RL, Lukas TJ, Van Eldik LJ, Watterson DM. Structure and expression of a calcium-binding protein gene contained within a calmodulin-regulated protein kinase gene. Mol Cell Biol. 1992;12:2359–2371. doi: 10.1128/mcb.12.5.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalla Libera L, Podhorska-Okolow M, Martin B, Massimino ML, Brugnolo R, Cantini M. Smooth muscle myosin light chain kinase is transiently expressed in skeletal muscle during embryogenesis and muscle regeneration both in vivo and in vitro. J Muscle Res Cell Motil. 1997;18:295–303. doi: 10.1023/a:1018618008483. [DOI] [PubMed] [Google Scholar]
- 14.Du KL, Chen M, Li J, Lepore JJ, Mericko P, Parmacek MS. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279:17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 15.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Mounayri O, Triplett JW, Yates CW, Herring BP. Regulation of smooth muscle-specific gene expression by homeodomain proteins, Hoxa10 and Hoxb8. J Biol Chem. 2005;280:25854–25863. doi: 10.1074/jbc.M501044200. [DOI] [PubMed] [Google Scholar]
- 17.Fisher SA, Ikebe M. Developmental and tissue distribution of expression of nonmuscle and smooth muscle isoforms of myosin light chain kinase. Biochem Biophys Res Commun. 1995;217:696–703. doi: 10.1006/bbrc.1995.2829. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher PJ, Herring BP. The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin. J Biol Chem. 1991;266:23945–23952. [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher PJ, Herring BP, Griffin SA, Stull JT. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991;266:23936–23944. [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- 21.Gallagher PJ, Jin Y, Killough G, Blue EK, Lindner V. Alterations in expression of myosin and myosin light chain kinases in response to vascular injury. Am J Physiol Cell Physiol. 2000;279:C1078–C1087. doi: 10.1152/ajpcell.2000.279.4.C1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia JG, Lazar V, Gilbert-McClain LI, Gallagher PJ, Verin AD. Myosin light chain kinase in endothelium: molecular cloning and regulation. Am J Respir Cell Mol Biol. 1997;16:489–494. doi: 10.1165/ajrcmb.16.5.9160829. [DOI] [PubMed] [Google Scholar]
- 23.Giorgi D, Ferraz C, Mattei MG, Demaille J, Rouquier S. The myosin light chain kinase gene is not duplicated in mouse: partial structure and chromosomal localization of Mylk. Genomics. 2001;75:49–56. doi: 10.1006/geno.2001.6571. [DOI] [PubMed] [Google Scholar]
- 24.Gorski DH, LePage DF, Walsh K. Cloning and sequence analysis of homeobox transcription factor cDNAs with an inosine-containing probe. Biotechniques. 1994;16:856–858. 860–852, 865. [PubMed] [Google Scholar]
- 25.Grueneberg DA, Simon KJ, Brennan K, Gilman M. Sequence-specific targeting of nuclear signal transduction pathways by homeodomain proteins. Mol Cell Biol. 1995;15:3318–3326. doi: 10.1128/mcb.15.6.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hautmann MB, Thompson MM, Swartz EA, Olson EN, Owens GK. Angiotensin II-induced stimulation of smooth muscle α-actin expression by serum response factor and the homeodomain transcription factor MHox. Circ Res. 1997;81:600–610. doi: 10.1161/01.res.81.4.600. [DOI] [PubMed] [Google Scholar]
- 27.Herring BP, Dixon S, Gallagher PJ. Smooth muscle myosin light chain kinase expression in cardiac and skeletal muscle. Am J Physiol Cell Physiol. 2000;279:C1656–C1664. doi: 10.1152/ajpcell.2000.279.5.C1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herring BP, Kriegel AM, Hoggatt AM. Identification of Barx2b, a serum response factor-associated homeodomain protein. J Biol Chem. 2001;276:14482–14489. doi: 10.1074/jbc.M011585200. [DOI] [PubMed] [Google Scholar]
- 29.Herring BP, Lyons GE, Hoggatt AM, Gallagher PJ. Telokin expression is restricted to smooth muscle tissues during mouse development. Am J Physiol Cell Physiol. 2001;280:C12–C21. doi: 10.1152/ajpcell.2001.280.1.C12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herring BP, Smith AF. Telokin expression in A10 smooth muscle cells requires serum response factor. Am J Physiol Cell Physiol. 1997;272:C1394–C1404. doi: 10.1152/ajpcell.1997.272.4.C1394. [DOI] [PubMed] [Google Scholar]
- 31.Herring BP, Smith AF. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am J Physiol Cell Physiol. 1996;270:C1656–C1665. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 32.Hoggatt AM, Kriegel AM, Smith AF, Herring BP. Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J Biol Chem. 2000;275:31162–31170. doi: 10.1074/jbc.M005595200. [DOI] [PubMed] [Google Scholar]
- 33.Hoggatt AM, Simon GM, Herring BP. Cell-specific regulatory modules control expression of genes in vascular and visceral smooth muscle tissues. Circ Res. 2002;91:1151–1159. doi: 10.1161/01.res.0000047508.30800.4f. [DOI] [PubMed] [Google Scholar]
- 34.Ito M, Dabrowska R, Guerriero V, Jr, Hartshorne DJ. Identification in turkey gizzard of an acidic protein related to the C-terminal portion of smooth muscle myosin light chain kinase. J Biol Chem. 1989;264:13971–13974. [PubMed] [Google Scholar]
- 35.Jiang H, Rao K, Liu X, Halayko AJ, Liu G, Stephens NL. Early changes in airway smooth muscle hyperresponsiveness. Can J Physiol Pharmacol. 1994;72:1440–1447. doi: 10.1139/y94-208. [DOI] [PubMed] [Google Scholar]
- 36.Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 37.Katayama E, Scott-Woo G, MI Effect of caldesmon on the assembly of smooth muscle myosin. J Biol Chem. 1995;270:3919–3925. doi: 10.1074/jbc.270.8.3919. [DOI] [PubMed] [Google Scholar]
- 38.Khromov AS, Wang H, Choudhury N, McDuffie M, Herring BP, Nakamoto R, Owens GK, Somlyo AP, Somlyo AV. Smooth muscle of telokin-deficient mice exhibits increased sensitivity to Ca2+ and decreased cGMP-induced relaxation. Proc Natl Acad Sci USA. 2006;103:2440–2445. doi: 10.1073/pnas.0508566103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu S, Miyazaki K, Tuft RA, Ikebe M. Translocation of telokin by cGMP signaling in smooth muscle cells. Am J Physiol Cell Physiol. 2002;283:C752–C761. doi: 10.1152/ajpcell.00501.2001. [DOI] [PubMed] [Google Scholar]
- 41.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Miano JM, Mercer B, Olson EN. Expression of the SM22α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci USA. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu ZP, Wang Z, Yanagisawa H, Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 45.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor cofactors. J Biol Chem. 2004;279:42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 46.Ma L, Benson GV, Lim H, Dey SK, Maas RL. Abdominal B (AbdB) Hoxa genes: regulation in adult uterus by estrogen and progesterone and repression in mullerian duct by the synthetic estrogen diethylstilbestrol (DES) Dev Biol. 1998;197:141–154. doi: 10.1006/dbio.1998.8907. [DOI] [PubMed] [Google Scholar]
- 47.Ma TY, Boivin MA, Ye D, Pedram A, Said HM. Mechanism of TNF-α modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am J Physiol Gastrointest Liver Physiol. 2005;288:G422–G430. doi: 10.1152/ajpgi.00412.2004. [DOI] [PubMed] [Google Scholar]
- 48.Ma X, Wang Y, Stephens NL. Serum deprivation induces a unique hypercontractile phenotype of cultured smooth muscle cells. Am J Physiol Cell Physiol. 1998;274:C1206–C1214. doi: 10.1152/ajpcell.1998.274.5.C1206. [DOI] [PubMed] [Google Scholar]
- 49.Mack CP, Owens GK. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ Res. 1999;84:852–861. doi: 10.1161/01.res.84.7.852. [DOI] [PubMed] [Google Scholar]
- 50.Mano T, Luo Z, Malendowicz SL, Evans T, Walsh K. Reversal of GATA-6 downregulation promotes smooth muscle differentiation and inhibits intimal hyperplasia in balloon-injured rat carotid artery. Circ Res. 1999;84:647–654. doi: 10.1161/01.res.84.6.647. [DOI] [PubMed] [Google Scholar]
- 51.Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 52.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 53.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 54.Miano JM, Firulli AB, Olson EN, Hara P, Giachelli CM, Schwartz SM. Restricted expression of homeobox genes distinguishes fetal from adult human smooth muscle cells. Proc Natl Acad Sci USA. 1996;93:900–905. doi: 10.1073/pnas.93.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 56.Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura M, Nishida W, Mori S, Hiwada K, Hayashi K, Sobue K. Transcriptional activation of β-tropomyosin mediated by serum response factor and a novel Barx homologue, Barx1b, in smooth muscle cells. J Biol Chem. 2001;276:18313–18320. doi: 10.1074/jbc.m101127200. [DOI] [PubMed] [Google Scholar]
- 58.Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
- 59.Numata T, Katoh T, Yazawa M. Functional role of the C-terminal domain of smooth muscle myosin light chain kinase on the phosphorylation of smooth muscle myosin. J Biochem (Tokyo) 2001;129:437–444. doi: 10.1093/oxfordjournals.jbchem.a002875. [DOI] [PubMed] [Google Scholar]
- 60.Oh J, Wang Z, Wang DZ, Lien CL, Xing W, Olson EN. Target gene-specific modulation of myocardin activity by GATA transcription factors. Mol Cell Biol. 2004;24:8519–8528. doi: 10.1128/MCB.24.19.8519-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohlmann P, Tesse A, Loichot C, Ralay Ranaivo H, Roul G, Philippe C, Watterson DM, Haiech J, Andriantsitohaina R. Deletion of MLCK 210 induces subtle changes in vascular reactivity but does not affect cardiac function. Am J Physiol Heart Circ Physiol. 2005;289:H2342–H2349. doi: 10.1152/ajpheart.00511.2004. [DOI] [PubMed] [Google Scholar]
- 62.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 63.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 64.Phiel CJ, Gabbeta V, Parsons LM, Rothblat D, Harvey RP, McHugh KM. Differential binding of an SRF/NK-2/MEF2 transcription factor complex in normal versus neoplastic smooth muscle tissues. J Biol Chem. 2001;276:34637–34650. doi: 10.1074/jbc.M105826200. [DOI] [PubMed] [Google Scholar]
- 65.Pipes GC, Sinha S, Qi X, Zhu CH, Gallardo TD, Shelton J, Creemers EE, Sutherland L, Richardson JA, Garry DJ, Wright WE, Owens GK, Olson EN. Stem cells and their derivatives can bypass the requirement of myocardin for smooth muscle gene expression. Dev Biol. 2005;288:502–513. doi: 10.1016/j.ydbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 66.Rath N, Wang Z, Lu MM, Morrisey EE. LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding. Mol Cell Biol. 2005;25:8864–8873. doi: 10.1128/MCB.25.20.8864-8873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott MP, Tamkun JW, Hartzell GW., III The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 68.Shirinsky VP, Vorotnikov AV, Birukov KG, Nanaev AK, Collinge M, Lukas TJ, Sellers JR, Watterson DM. A kinase-related protein stabilizes unphosphorylated smooth muscle myosin minifilaments in the presence of ATP. J Biol Chem. 1993;268:16578–16583. [PubMed] [Google Scholar]
- 69.Shirvani S, Xiang F, Koibuchi N, Chin MT. CHF1/Hey2 suppresses SM-MHC promoter activity through an interaction with GATA-6. Biochem Biophys Res Commun. 2006;339:151–156. doi: 10.1016/j.bbrc.2005.10.190. [DOI] [PubMed] [Google Scholar]
- 70.Shoemaker MO, Lau W, Shattuck RL, Kwiatkowski AP, Matrisian PE, Guerra-Santos L, Wilson E, Lukas TJ, Van Eldik LJ, Watterson DM. Use of DNA sequence and mutant analyses and antisense oligodeoxynucleotides to examine the molecular basis of nonmuscle myosin light chain kinase autoinhibition, calmodulin recognition, and activity. J Cell Biol. 1990;111:1107–1125. doi: 10.1083/jcb.111.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith AF, Bigsby RM, Word RA, Herring BP. A 310-bp minimal promoter mediates smooth muscle cell-specific expression of telokin. Am J Physiol Cell Physiol. 1998;274:C1188–C1195. doi: 10.1152/ajpcell.1998.274.5.C1188. [DOI] [PubMed] [Google Scholar]
- 72.Stephens NL, Halayko AJ. Airway smooth muscle contractile, regulatory and cytoskeletal protein expression in health and disease. Comp Biochem Physiol B Biochem Mol Biol. 1998;119:415–424. doi: 10.1016/s0305-0491(98)00004-2. [DOI] [PubMed] [Google Scholar]
- 73.Taylor HS. The role of HOX genes in the development and function of the female reproductive tract. Semin Reprod Med. 2000;18:81–89. doi: 10.1055/s-2000-13478. [DOI] [PubMed] [Google Scholar]
- 74.Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 75.Van Tuyn J, Knaan-Shanzer S, van de Watering MJ, de Graaf M, van der Laarse A, Schalij MJ, van der Wall EE, de Vries AA, Atsma DE. Activation of cardiac and smooth muscle-specific genes in primary human cells after forced expression of human myocardin. Cardiovasc Res. 2005;67:245–255. doi: 10.1016/j.cardiores.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 76.Ventura C. Forced myocardin expression primes cardiac and smooth muscle transcription patterning in human mesenchymal stem cells. Cardiovasc Res. 2005;67:182–183. doi: 10.1016/j.cardiores.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 77.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Abe M, Sasayama S. Calcineurin-GATA-6 pathway is involved in smooth muscle-specific transcription. J Cell Biol. 2002;156:983–991. doi: 10.1083/jcb.200106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Sasayama S. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J Biol Chem. 2000;275:25330–25335. doi: 10.1074/jbc.M000828200. [DOI] [PubMed] [Google Scholar]
- 79.Wainwright MS, Rossi J, Schavocky J, Crawford S, Steinhorn D, Velentza AV, Zasadzki M, Shirinsky V, Jia Y, Haiech J, Van Eldik LJ, Watterson DM. Protein kinase involved in lung injury susceptibility: evidence from enzyme isoform genetic knockout and in vivo inhibitor treatment. Proc Natl Acad Sci USA. 2003;100:6233–6238. doi: 10.1073/pnas.1031595100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walker LA, MacDonald JA, Liu X, Nakamoto RK, Haystead TA, Somlyo AV, Somlyo AP. Site-specific phosphorylation and point mutations of telokin modulate its Ca2+-desensitizing effect in smooth muscle. J Biol Chem. 2001;276:24519–24524. doi: 10.1074/jbc.M103560200. [DOI] [PubMed] [Google Scholar]
- 81.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 82.Wang F, Graham WV, Wang Y, Witkowski ED, Schwarz BT, Turner JR. Interferon-gamma and tumor necrosis factor-α synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am J Pathol. 2005;166:409–419. doi: 10.1016/s0002-9440(10)62264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 84.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watterson DM, Collinge M, Lukas TJ, Van Eldik LJ, Birukov KG, Stepanova OV, Shirinsky VP. Multiple gene products are produced from a novel protein kinase transcription region. FEBS Lett. 1995;373:217–220. doi: 10.1016/0014-5793(95)01048-j. [DOI] [PubMed] [Google Scholar]
- 86.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 87.Wu X, Haystead TA, Nakamoto RK, Somlyo AV, Somlyo AP. Acceleration of myosin light chain dephosphorylation and relaxation of smooth muscle by telokin. Synergism with cyclic nucleotide-activated kinase. J Biol Chem. 1998;273:11362–11369. doi: 10.1074/jbc.273.18.11362. [DOI] [PubMed] [Google Scholar]
- 88.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 89.Yin F, Herring BP. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J Biol Chem. 2005;280:4745–4752. doi: 10.1074/jbc.M411585200. [DOI] [PubMed] [Google Scholar]
- 90.Yin F, Hoggatt AM, Zhou J, Herring BP. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am J Physiol Cell Physiol. 2006;290:C1599–C1609. doi: 10.1152/ajpcell.00289.2005. [DOI] [PubMed] [Google Scholar]
- 91.Yoshida T, Hoofnagle MH, Owens GK. Myocardin and Prx1 contribute to angiotensin II-induced expression of smooth muscle alpha-actin. Circ Res. 2004;94:1075–1082. doi: 10.1161/01.RES.0000125622.46280.95. [DOI] [PubMed] [Google Scholar]
- 92.Yoshida T, Kawai-Kowase K, Owens GK. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler Thromb Vasc Biol. 2004;24:1596–1601. doi: 10.1161/01.ATV.0000137190.63214.c5. [DOI] [PubMed] [Google Scholar]
- 93.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 94.Yoshikai S, Ikebe M. Molecular cloning of the chicken gizzard telokin gene and cDNA. Arch Biochem Biophys. 1992;299:242–247. doi: 10.1016/0003-9861(92)90270-7. [DOI] [PubMed] [Google Scholar]
- 95.Zhi G, Ryder JW, Huang J, Ding P, Chen Y, Zhao Y, Kamm KE, Stull JT. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc Natl Acad Sci USA. 2005;102:17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou J, Herring BP. Mechanisms responsible for the promoter-specific effects of myocardin. J Biol Chem. 2005;280:10861–10869. doi: 10.1074/jbc.M411586200. [DOI] [PubMed] [Google Scholar]
- 97.Zhou J, Hoggatt AM, Herring BP. Activation of the smooth muscle-specific telokin gene by thyrotroph embryonic factor (TEF) J Biol Chem. 2004;279:15929–15937. doi: 10.1074/jbc.M313822200. [DOI] [PubMed] [Google Scholar]
- 98.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol Cell Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.