Abstract
Cortical thickness is an important biomarker for image-based studies of the brain. A Diffeomorphic registration based cortical thickness (DiReCT) measure is introduced where a continuous one-to-one correspondence between the gray matter-white matter interface and the estimated gray matter-cerebrospinal fluid interface is given by a Diffeomorphic mapping in the image space. Thickness is then defined in terms of a distance measure between the interfaces of this sheet like structure. This technique also provides a natural way to compute continuous estimates of thickness within buried sulci by preventing opposing gray matter banks from intersecting. In addition, the proposed method incorporates neuroanatomical constraints on thickness values as part of the mapping process. Evaluation of this method is presented on synthetic images. As an application to brain images, a longitudinal study of thickness change in frontotemporal dementia (FTD) spectrum disorder is reported.
Keywords: Cortex, thickness, diffeomorphism, Euclidean distance, deformable models, longitudinal, FTD
1 Introduction
A number of methodologies for cortical thickness measurements have appeared in neuroimaging literature over the past decade (Zeng et al., 1999; Miller et al., 2000; Jones et al., 2000; Fischl and Dale, 2000; Lohmann et al., 2003; Srivastava et al., 2003; Yezzi and Prince, 2003; Thompson et al., 2004; Lerch et al., 2005; Barta et al., 2005; Scott and Thacker, 2005; Hutton et al., 2008). Nonetheless, no gold standard has emerged that one can evaluate a measurement against. One reason is the difficulty of manually measuring thickness values in 3D images, unlike tissue segmentation where voxel-by-voxel manual marking can serve as a gold standard. Thickness measured directly using postmortem brains is also not considered an absolute metric because of possible tissue shrinkage. What complicates matters more is the lack of a consistent definition of cortical thickness. Some methods require explicit point associations between the white matter (WM)/gray matter (GM) surface and the GM/cerebrospinal fluid (CSF) surface. Some of these methods also require explicit construction of surface meshes (Fischl and Dale, 2000; MacDonald et al., 2000), typically using the extracted WM surface as a model that is fit to the GM surface by deformation, thus establishing node-to-node associations. Alternatively, definitions of thickness based on nearest point (Miller et al., 2000; Fischl and Dale, 2000) and distance along the surface normal (Das et al., 2007) do not require explicit point associations. However, regardless of whether a given method requires a priori correspondence maps or not, eventual computation of thickness is always based on some measure of distance between two points. This range of work points to a common definition of thickness that is based on two components: first, a principled point correspondence and, second, a distance measure between the points. Theoretical distinctions between algorithms occur in the definition of correspondence and/or the distance measurement between corresponding points. Practical differences usually occur in the underlying data representation.
Some researchers have argued that methods that require explicit surface extraction may suffer from inaccuracies due to surface generation (Srivastava et al., 2003; Thompson et al., 2005). One popular class of methods that establishes a priori point-to-point correspondence, without an explicit surface representation, is the gamut of PDE based methods (Jones et al., 2000; Yezzi and Prince, 2003; Rocha et al., 2007). Jones et al. (2000) models the cortical mantle as a dielectric and solves Laplace’s equation to compute electric field lines through the cortex in order to establish correspondence. There are still other image-based methods that rely on image operations such as morphological filters (Lohmann et al., 2003), geodesic distance transform (Srivastava et al., 2003), edge detection (Scott and Thacker, 2005) or explicit geometric modeling of the cortical sheet (Barta et al., 2005). The method introduced here is also image-based, but uses Diffeomorphic image registration based point-to-point correspondence.
One important feature of computational methods in neuroanatomy is the ability to include prior knowledge about the anatomy of interest. Such constraints increase reliability and performance in the presence of noise and when image resolution is sub-optimal. von Economo (1929) reported cortical thickness to be between 1.2–4.5 millimeters (mm) from ex vivo measurements, and ≈5 mm is generally reported to be the maximum observed value from in vivo measurements (Fischl and Dale, 2000; Kabani et al., 2001; Hutton et al., 2002). Some surface reconstruction based methods employ proximity constraints on the distance between two surfaces to ensure thickness lies within an anatomic range (Zeng et al., 1999; MacDonald et al., 2000). However, many methods, including Jones et al. (2000), provide no inherent anatomically motivated constraints on the computation of thickness. Note that the method based on solving Laplace’s equation within the cortical sheet provides thickness metrics based on curved paths. Such thickness values cannot be directly related to existing post-mortem knowledge, as provided by von Economo and others. Furthermore, straight-line/Euclidean distances are more easily interpreted in terms of a sheet-like model, as relevant for the cortex. Thus, the majority of thickness definitions use Euclidean distances that are consistent with what might be measured post-mortem. For these clinically relevant reasons – and for the purpose of using clinically motivated priors – we also focus on Euclidean distance as a thickness metric, though our technique applies as easily to curved thickness.
A related anatomical problem in thickness computation is the presence of buried cortex, which refers to the parts of the cortical surface hidden within the deep sulci. Within these sulci, thin strands of CSF are often mislabeled as GM due to partial volume effects, limited resolution and noisy tissue intensity levels. This mislabeling may lead to overestimation of thickness. Note that we use the terms buried cortex and buried sulci to refer to these regions of unresolved CSF, as has been used by other studies of cortical thickness (Kim et al., 2005; Hutton et al., 2008), as opposed to referring to the cortex in deep sulcal regions in general (with CSF separating the sulcal banks resolved or not) that is termed buried rather than cortex visible on the outer surface, as described in Van Essen et al. (1998); Rettmann et al. (2006). Several existing approaches explicitly address this problem: by image enhancement (Jones et al., 2000) to recover CSF, using morphological processing (Lohmann et al., 2003), stochastic modeling (Barta et al., 2005) and explicit image-based labeling of sulcal points (Hutton et al., 2002). The ACE method in Han et al. (2004) uses a level set based framework that corrects for buried sulci using fuzzy CSF membership information. Our own prior work introduced a novel topology preserving segmentation method (Das et al., 2007) that is able to recover some deep sulci. A similar method that does not preserve topology uses the Laplacian to measure thickness on segmentations with digitally recovered sulci (Hutton et al., 2008).
This paper introduces a new method, which we call Diffeomorphic Registration based Cortical Thickness (DiReCT). DiReCT reduces the problems of buried cortex and unconstrained thickness measurement by using a prior-constrained estimate of the distance between the gray/white interface and the gray/cerebrospinal fluid interface. This strategy, in essence, assigns a distance between opposing faces of a thin, sheet-like structure, here, the cortical mantle. Our approach begins with a hypothetical, infinitesimally thin, cortical mantle with ε thickness that lies along the white matter/gray matter interface. The initial model thus has “open” sulci. This thin cortical mantle is then allowed to expand under a one-to-one, differentiable and invertible map (a diffeomorphism) toward the edges of the data-derived gray matter probability. This mapping gives a correspondence field that allows an estimate of the gray/csf tissue interface, and thus thickness. An overview of this process is depicted in Figure 1.
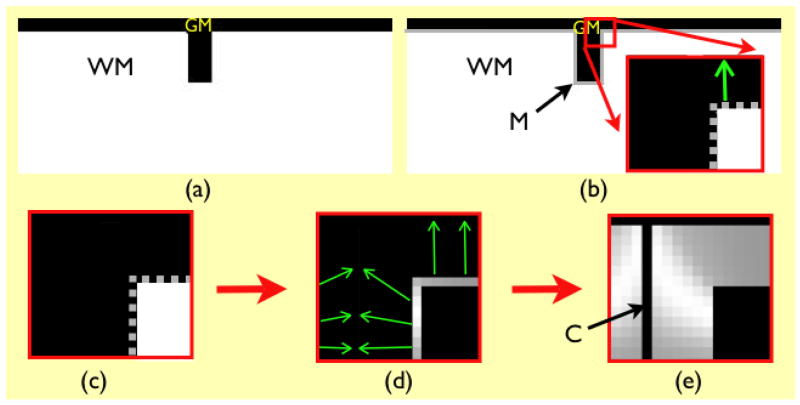
Fig. 1.
Overview of proposed methodology. The original gray matter and white matter images are shown in (a). In (b), M is the initial cortical model, a thin (one voxel thick) sheet, with the inner edge at the gray/white interface, and the outer edge within gray matter. The region enclosed in red is zoomed in to show the initial model (also in (c)), in which M is represented as a gray dotted line. Panel (d) shows how the initial model is deformed to find the estimated gray/csf interface, and establish point-to-point correspondences (green arrows). Gray levels at the gray/white interface denote the distance between corresponding points, which is the measure of thickness. The thickness values are propagated to the GM volume to generate the volumetric thickness map shown in (e), where C denotes the estimated location of gray/csf interface.
The correspondence problem that we solve is made well-posed by the diffeomorphic constraint, which seeks a minimally deforming solution (Dupuis et al., 1998). The topology preservation provided by diffeomorphisms, along with explicit thickness priors, provide shape guidance to this mapping that prevent sulci from unfolding and neighboring banks of gray matter from intersecting. Thus, a maximum a posteriori estimate of the location of buried sulci is gained. Additionally, our thickness measures are guaranteed to stay within a range that is set by user-defined, spatially varying prior constraints. This shape-constrained mapping is able to represent buried sulci as infinitesimally close, opposing banks of gray matter, in the continuous domain, thus yielding sub-voxel resolution. Further, each gray matter bank is connected Diffeomorphically to a unique white matter bank.
In summary, the advantage of our approach, relative to prior work, is that we directly use standard probabilistic segmentation maps – with no further processing – to make a continuous, sub-voxel estimate of cortical thickness, constrained by thickness priors and accurate within buried sulci, within a single algorithmic framework.
The rest of the paper is organized as follows. Section 2.1 introduces various terminologies used. Section 2.2 describes the method for defining correspondences from Diffeomorphic maps and computing cortical thickness. Section 2.3 describes the construction of synthetic images used for evaluation. Methods used for conducting clinical study are discussed in Section 2.4. Section 3.1 presents results of evaluation of our method on synthetic data. Evaluation on a 2D brain slice as well as results of a longitudinal study of atrophy using these methods in a diseased population is presented in Section 3.2. An application of our thickness measurements is presented in the form of a longitudinal study of atrophy in a patient population diagnosed with FTD spectrum disorder. The small number of studies of gray matter atrophy over time in FTD (Avants et al., 2007; Brambati et al., 2007) — compared to a substantial number of similar studies in case of Alzheimer’s Disease (AD) — have either used volumetric measures of atrophy or voxel based measures like the Jacobian. Various issues pertaining to the proposed cortical thickness measure are discussed in Section 4. Section 5 concludes with advantages of the proposed method and future work.
2 Materials and Methods
2.1 Preprocessing and Terminology
Methods for generating a DiReCT map from an MRI volume begin with a set of probabilistic segmentation images, labeling the local mixture of tissue probabilities for each of three tissues, the white matter (WM), gray matter (GM) and the cerebrospinal fluid (CSF), in the brain. A variety of algorithms may be used to derive such images, but we rely on FAST as given in Zhang et al. (2001). Each voxel of the probability images, which are of the same size and spacing, is constrained to sum to 1, such that Pg(x) + Pw(x) + Pc(x) = 1 throughout all x in Ω where Ω is the domain of the image, where Pg(x), Pw(x) and Pc(x) are the GM, WM and CSF probability images respectively. We denote an image containing the sum of any two of these probability images as, in the case of WM and GM, Pwg = Pw + Pg (see Figure 2). Finally, the gray/white interface (GWI) is defined as voxels within Pg where Pg(x) >= 0.5 and at least one neighboring voxel within the 26-connected neighborhood of x has Pw >= 0.5. Note that no explicit surface mesh is extracted from the WM segmentation, thus avoiding complexities of surface generation. Instead, voxels representing the GWI are identified in the image domain.
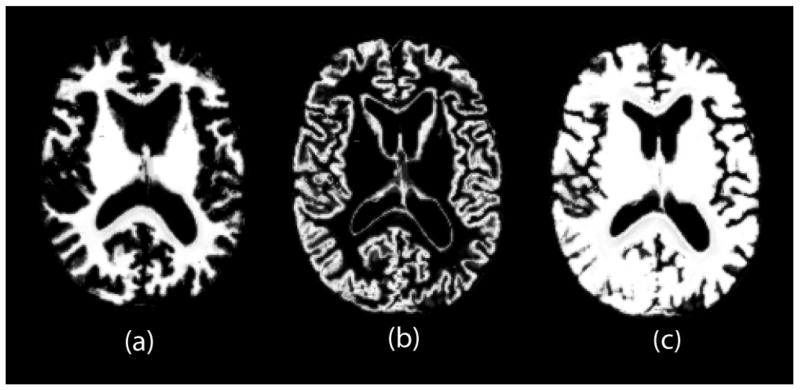
Fig. 2.
Examples of probability map image. WM probability map image in (a) is added to GM probability map in (b) to create combined WM and GM probability map image in (c). Correspondence is found by registering images in (a) and (c).
Our goal is, for every point along the GWI, to find a corresponding GM point as given by Pwg that closely estimates the likely edge of the gray matter cortex. As argued in MacDonald et al. (2000), the cortical GM surface should have the same topology and a coarsely similar shape as the GWI. Note that this GM surface edge is not necessarily bounded, within the segmentation probability images, by neighboring CSF values with high probability, due to partial voluming and the problem of buried cortex. We denote the correspondence map as φ(x, t): Ω × [0, 1] → Ω and specify that φ(x, t) is a diffeomorphism, or a differentiable map with a differentiable inverse. Note that, while we are interested in mapping the GWI to a likely location of the cortical surface, we embed the mapping of the GWI within a volumetric diffeomorphism. We choose the Diffeomorphic transformation space because curves are guaranteed to remain curves and surfaces are guaranteed to remain surfaces under a diffeomorphic mapping. Thus, diffeomorphisms provide a natural shape constraint on the deformation of the GWI towards the cortical surface, which we augment with a thickness constraint. We detail, below, our approach for computing this mapping.
2.2 Thickness Computation
Define the velocity field, v(x, t) : Ω × t ∈ [0, 1] → ℝd, where t is the “time” parameter, d is dimension, and v is a regularized vector field. We restrict the velocity field v to exist in the space of continuous and square-integrable functions with bounded derivatives by choosing a linear operator, L, which measures the energy of v, integrated over time. In practice, we choose L = ∇2 + ℐ (where ℐ is the identity operator) which has a Green’s kernel that is closely approximated by Gaussian smoothing.
As shown in Dupuis et al. (1998), this regularity guarantees that a diffeomorphism can be generated from v by integrating an ordinary differential equation (ODE) (Arnold, 1991),
and thus
| (1) |
The fundamental theorem of ODEs guarantees that this diffeomorphism is unique and varies continuously with initial conditions, φ(x, 0). Here, φ(x, 0) = x for all computations. Standard numerical integration techniques, such as Runge-Kutta methods, may be used to perform the integration. A diffeomorphic map may be applied to deform an image, I, through I(φ(x)).
Given this transformation space, our goal is to generate a time one mapping, φ(x, 1), that estimates correspondence between the GWI and the outer edge of the cortical sheet, with given thickness constraints. Let M(x) be the moving image that represents the initial model of the gray matter sheet, derived from Pw(x), that is to be deformed under this mapping. Thus, we minimize the following variational energy, which captures both the notion that the velocity field should be smooth and the mapping should be a diffeomorphism, with constrained Euclidean displacement T(x, t),
| (2) |
The model image M(x) can be taken as the probabilistic WM segmentation image Pw(x), or a binary white matter segmentation image derived by thresholding Pw(x) at 0.5. In the latter case, one would have non-zero moving image gradient only at the GWI. Our experiments, using FAST (Zhang et al., 2001) as the segmentation method, have shown that the resultant mapping of the GWI, and hence the thickness measures, are nearly identical in either case. In all experiments reported here, we have used M(x) = Pw(x). M (φ(x, 0)) represents the GWI as described in section 2.1. Then, M(φ(x, 1)) represents the other side of the cortical sheet, the estimated gray/CSF interface, in the same way. The function τ: Ω → ℝ+ provides a prior on the particle x’s Euclidean distance, from its origin, as it transports along the diffeomorphic path. Note that, given a Diffeomorphic correspondence, thickness T(x, t) at a GWI point x is defined as the Euclidean length of the displacement that φ(x, t) imposes on x: T(x, t) = |φ(x, t) − x|. The inverse mapping φ−1(x, 1) is easily computed in the Diffeomorphic space by integrating the negative velocity field, as in Avants et al. (2006). Note that because φ−1((φ(x, 1), 1) = ℐ (identity map), we have an invertible correspondence map between the GM and GWI surface points.
Most critically, since we solve for a Diffeomorphic (differentiable, one-to-one and onto) map, the topology of the moving image (the WM segmentation in this case) cannot change. This is consistent with our treatment of the WM segmentation as the space in which we transport our GWI model toward the GM segmentation. This is one way our model preserves the shape of the resolved sulcus in the WM segmentation, thus recovering extremely narrow sulci that might have been mislabeled as GM. In addition to preserving topology, our optimization scheme seeks a minimally deforming solution (first term in equation 2) for a given level of image similarity between the deformed image and the target image (the data term in registration, second term in equation 2). Consider possible solutions to the registration problem for a closed sulcus such as that shown in Figure 1(a). A solution that maps the deepest points in the sulcus in the moving image (white/gray interface points) towards the “gyral” region (Figure 3(a), we’ll call it solution A) provides a comparable level of image similarity as one that maps these points towards the center of the sulcus (Figure 3(b), we’ll call it solution B). This is because, solution B maps the gray/white interface points from opposing banks of sulci to points that are infinitesimally close to each other in the continuous domain – thus the deformed image matches the target image as closely as it does for solution A. In other words, the deformed image “engulfs” the entire closed sulcus equally well in both cases in the continuous domain. In the discrete domain, however, we can see the single-pixel thick resolved sulcus for solution B. In fact, a range of solutions in between these two solutions exist, that resolve a progressively shallower sulcus; all of these solutions have similar image matching quality. Solution B, however, is the one that imposes the least amount of total deformation, and is hence the preferred solution, providing an estimate of a likely gray/CSF boundary. Finally, an explicit thickness prior in our optimization limits the maximum amount of deformation a white/gray interface point can undergo – which may also prevent solutions such as solution A which entails larger deformations. Thus, our shape-constrained mapping prefers to generate solutions in the class of panel (b) as opposed to panel (a) in terms of the classic buried sulcus recovery problem. These solutions are still far from perfect, particularly when CSF is not visible. At the same time, they represent a principled approach in terms of estimating thickness in the absence of perfect segmentation.
Fig. 3.
(a) and (b) show two Diffeomorphic measurements of thickness that fit the model in Figure 1 to the gray matter. Both measurements minimize the image difference term with approximately the same level of quality, but with two very different levels of deformation. Model (a) uses no explicit regularization of the deformation, and yields a more deforming thickness solution with similarity value of 242 and a first derivative based deformation value of 1.74. Model (b), on the other hand, uses Diffeomorphic regularization and produces a continuous thickness solution with similarity value of 250 and a deformation value of 0.93. The solution in (a) requires 1.86 times the deformation of the solution in (b) but gains only 3 percent improvement in similarity, when similarity is measured in the continuous domain, implying “infinitesimal” proximity of the neighboring gray matter banks. Solution (a) is also much more computationally expensive to generate and greatly violates expected thickness values. Neither solution is perfect, because the true solution is unknown within the closed sulcus – but both represent optimal solutions given the respective models.
2.2.1 Optimization Method
In the absence of a prior constraint on the thickness, one may optimize equation 2 by gradient descent on the Euler-Lagrange equations given by Beg et al. (2005) or, relatedly, a greedy approach as described in Avants et al. (2006). Optimization of the energy with the prior constraint can be performed with only a slight modification to use a constrained integration of the ODE that respects the thickness constraint τ. We detail this optimization, assuming a standard integration technique, below, and n steps in the temporal discretization of the velocity field. We take τ(x) and the probability images as inputs to the method and only T(x, 1) is the desired output.
Set v(x, t) = 0, ∀ x ∈ Ω, t∈ [0, 1] and λ the gradient descent parameter. Set t = 0, φ(x, 0) = x, , where m ≥ n is the no. of time steps used in the the scheme for integrating the ODE in equation 1.
Define a temporary gradient field u(x, t) = 0, ∀ x ∈ Ω, t ∈ [0, 1].
-
Compute, for all ti, the values of φ(x, ti), u(x, ti) and T (x, ti), as follows,
-
Integrate φ by φ(x, t + Δt) = φ(x, t) + Δt v(φ(x, t), t) over m steps.
Only update φ if T(x, t) < τ(x) at each step.
-
If T(x, ti+1) < τ (x) then compute the similarity gradient,
(3) If T(φ(x, ti), ti) >= τ(x) then u(x, ti+1) = 0.
-
Set v(x, ti) ← K ⊗ (v(x, ti) + λu(x, ti)) where K is the Green’s kernel for the linear operator, L and ⊗ represents convolution. We estimate K with a Gaussian kernel of standard deviation 1.5mm3, in a 1mm3 image space.
Go to 2 until E (equation 2) can no longer be reduced.
Propagate the value of T(x) defined at the GWI to the full volume of the gray matter to generate the thickness map. This process is described below.
Thus, we perform a greedy minimization of E within the space of diffeomorphisms while maintaining T(x) < τ(x) explicitly.
2.2.2 Thickness Propagation
The map φ(x, t) defines the deformation path a GWI point follows through the GM mantle as a function of time t, reaching its corresponding GM surface point at t = 1. Let Tsurf (x) be the deformation norm image which has the value of measured thickness at every GWI point, and zero elsewhere. In order to propagate the thickness values Tsurf (x) volumetrically, we sample φ(x, t) at time intervals of t in [0, 1] and successively apply the transformations φ(x, t) such that, . This corresponds to transporting the thin thickness valued surface through the same velocity field that generates φ(x, t) and labeling each point in space with the passing value. In theory, this mapping would provide a dense field of thickness values throughout the domain traversed by Tsurf (φ(x, t)), as each point would be passed only once. However, due to discretization, care must be taken in how this integration is performed. We detail the discretized algorithm as,
Define IGW I as the binarized GWI and Tsurf as the thickness values as-signed to IGW I. Define Ihit and Itotal as zero-valued images. Ihit represents the integrated partial volume contribution of the deformed IGW I over time and Itotal represents the associated integrated partial volume of the deformed Tsurf image.
-
Set i = 0. For t = 0 to t = 1 in increments of Δt ∋ t0 = 0, ti+1 = ti + Δt, compute and also .
For all x ∈ Ω, set Tvol(x) = Itotal/Ihit if Ihit(x) > 0, otherwise Tvol(x) = 0.
This method iteratively accumulates the propagating level sets over time in the same image space. The thickness values on the WM surface thus propagate along the deformation path specified by φ(x, t) into the GM volume, generating the dense volumetric thickness map image Tvol, which we refer to as the DiReCT map.
2.3 Evaluation with 3D Phantom
We tested our registration based thickness measurements on a set of 3D phantom images at different image resolutions and noise levels. The gray matter mantle in these images is defined either as a spherical shell or one with cortex-like undulations. Similar phantoms have been used by Srivastava et al. (2003) in 2D. Mathematically, the phantom is given by a family of surfaces S : R2 ↦ R3 parameterized by θ and φ, the azimuth and elevation angles respectively – centered at c :
| (4) |
where R(γ, α) represents the radial distance from the center c of the volume enclosed by the surface S. R determines the shape of the surface. We consider different forms of the function f(γ, α, θ, φ) for different shapes of phantoms that were studied.
2.3.1 Sphere
In the simplest case, f(·) = 0, R = r and S reduces to a sphere of radius r centered at c. If SW (θ, φ; R = rw) and SG(θ, φ; R = rg) are nominally defined as the WM and GM surfaces respectively (rg > rw), the cortical gray matter would be a spherical shell of radius rg−rw, which also represents the theoretical cortical thickness value at every WM surface point on SW.
2.3.2 Ribbon
Next we consider a surface with ribbon-like undulations mimicking the cortex. This is realized as
| (5) |
where γ controls the magnitude and α controls the frequency of the undulations along the surface. Again, we construct the cortical mantle by defining the WM and GM surfaces as SW (θ, φ; R = rw + fr(γ, α, φ)) and SG(θ, φ; R =rg + fr(γ, α, φ))(rg > rw) respectively. An example of such a phantom is shown in Figure 4(a).
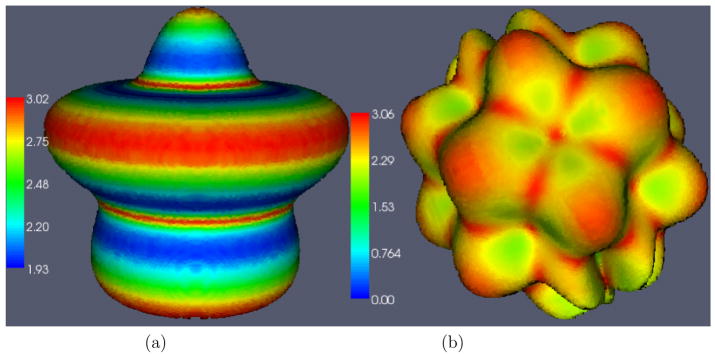
Fig. 4.
(a) Ribbon shaped phantom. (b) Ribbon with sulcus phantom. Color represents measured thickness values on the surface.
2.3.3 Ribbon with Sulcus
The undulations in the phantom above are only along the φ (elevation) axis as can be seen from Figure 4(a). Any cross-section of the surface parallel to the z axis is a circle, since fr is not a function of θ, the azimuth angle. Now we introduce a third phantom shape with undulations along both θ and φ where
| (6) |
In this case, the phantom has bigger variations in curvature along the surface, resulting in deep sulcus-like structures in some areas. Cortical mantle is constructed the same way as described in section 2.3.2 with SW (θ, φ; R =rw + fs(γ, α, θ, φ)) and SG(θ, φ; R = rg + fs(γ, α, θ, ∀))(rg > rw). An example of such a phantom is shown in Figure 4(b).
2.3.4 Resolution and Noise Levels
For each phantom shape described above, we created phantoms with the same values of rw = 7 mm, rg = 10 mm, γ = 2, α = 5, but with different spatial resolution and noise levels. Binary WM and GM masks are generated by spanning the parameter spaces as φ = −π(Δ φ)π, θ = 0(Δ θ)π and letting r = 0(Δr)rw and r = rw(Δr)rg respectively and discretizing at the appropriate resolution. Voxels thus labeled as WM and GM are then set to intensity values obtained from mean WM and GM intensity values in sample brain images. The phantom image I obtained in this manner is then corrupted by noise as Inoisy = Gμ ⊗ (I + νσ) where νσ is an additive Gaussian noise with zero mean and variance σ and Gμ is a Gaussian smoothing kernel with spread μ. Table 1 summarizes the parameters used. Three noise levels are nominally taken as none, low and high with increasing values of both μ and σ. To measure thickness, the WM and GM probability maps are generated by applying the FAST (Zhang et al., 2001) segmentation algorithm to the noisy phantoms.
Table 1.
Voxel sizes in mm and corresponding image sizes at each resolution level.
| Resolution | Voxel Size | Image Size |
|---|---|---|
| Low | 1.0 | 30 × 30× 30 |
| Medium | 0.5 | 60 × 60 × 60 |
| High | 0.1 | 250 × 250 × 250 |
2.4 Application to Real Brain Images
We applied the methodology to a longitudinal study of gray matter thickness changes in a patient cohort diagnosed with frontotemporal dementia. Previous neuroimaging studies on FTD patient populations have identified locally focused atrophy (Chan et al., 2001) and these observations have been validated through correlation of annualized gray matter loss with cognitive decline (Avants et al., 2005) and in patients with autopsy-confirmed disease (Grossman et al., 2007; Murray et al., 2007).
2.4.1 Patient Population
The dataset contains twenty elderly individuals in total, diagnosed with FTD spectrum disorders, imaged at two time points each. The patients were right-handed, high school-educated (education = 16.0 ± 2.7 years) native English-speakers. The age at the first assessment was 61.8 ± 6.8 years with a mean MMSE (mini-mental state examination) score of 21.4±7.3. The second assessment was conducted after an average of 12.4±3.3 months, ranging from 7 to 21 months, with a mean MMSE score of 18.1 ±9.4. Patients were identified clinically in the Department of Neurology at the University of Pennsylvania School of Medicine. Initial clinical diagnosis was established by an experienced neurologist (author MG) using a modification of published criteria (Price et al., 2001). Subsequently, at least two trained reviewers of a consensus committee confirmed the presence of specific diagnostic criteria based on an independent review of the semi-structured history, mental status examination and neurological examination. Rare disagreements were resolved through discussion.
2.4.2 Imaging
All images were acquired from a Siemens Trio 3.0 T MRI scanner. Each study began with a rapid sagittal T1-weighted scan to determine patient position. A T1 structural scan was then acquired using the MP-RAGE sequence with TR (repetition time) = 1620 ms, TE (echo time) = 3 s, slice thickness = 1 mm, in-plane resolution = 0.9766 × 0.9766 mm and field of view (FOV) = 256 × 256 × 192. The same protocol was used at baseline (younger) and followup (older) time points.
2.4.3 Preprocessing and Segmentation
Groupwise morphometric studies are conventionally performed in a normalized coordinate system. We derive a template for this purpose from an expanded dataset (n=117) of FTD patients combined with healthy age-matched controls using symmetric Diffeomorphic normalization (SyN) methods (Avants and Gee, 2004). The template is one that is specific for the scanner, age and the MR protocol. This represents the average shape of the aged brain more accurately than a generic template and aids normalization accuracy. For each subject, we first find the mapping between the baseline (earlier time) image and the followup (later time) image. We then find the mapping between the baseline image and the template. We then compose these two maps to obtain the mapping between the followup image and template (Avants et al., 2005). In each case, the inverse maps were also generated. Once we derive thickness maps for each subject at both time points, we can warp them into the template coordinate system using these maps and perform statistical analysis in the template space. Generating maps between baseline image and template as well as followup image and template this way eliminates inconsistencies that may occur in the conventional approach which directly registers both the baseline and followup images separately to the template.
The semiautomated ITK-SNAP segmentation method (Yushkevich et al., 2006) is used to label the brain in the whole head template. Individual brain parenchyma are then extracted by mapping the template brain mask to each brain volume using the inverse maps obtained by SyN. Fuzzy segmentations for gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) are obtained for each patient scan using the FAST algorithm (Zhang et al., 2001). However, spatially varying three-tissue priors using tissue probability maps from the template brain are used to initialize the segmentation procedure.
2.4.4 Longitudinal Comparison
We use the SyN map between the respective subject space at earlier (or later) time and the template space to map the individual DiReCT maps to the template space. The thickness value for the later time is then annualized by obtaining a linearly interpolated estimate of thickness at every voxel one year after baseline scan from the two values at earlier and later time. The assumption about linear progression of atrophy over time may not be valid in general as progression is likely to vary with disease stage. However, with only two time points available and in the absence of any other known temporal model, it is the only well-constrained assumption (Avants et al., 2005) that may be made. Once we have transported the DiReCT maps in the template space (Figure 9(d)–(e)), it is possible to perform groupwise longitudinal analysis of thickness changes. In order to study mean thickness change in Brodmann regions, we registered a template brain with Brodmann area labels to our local template.
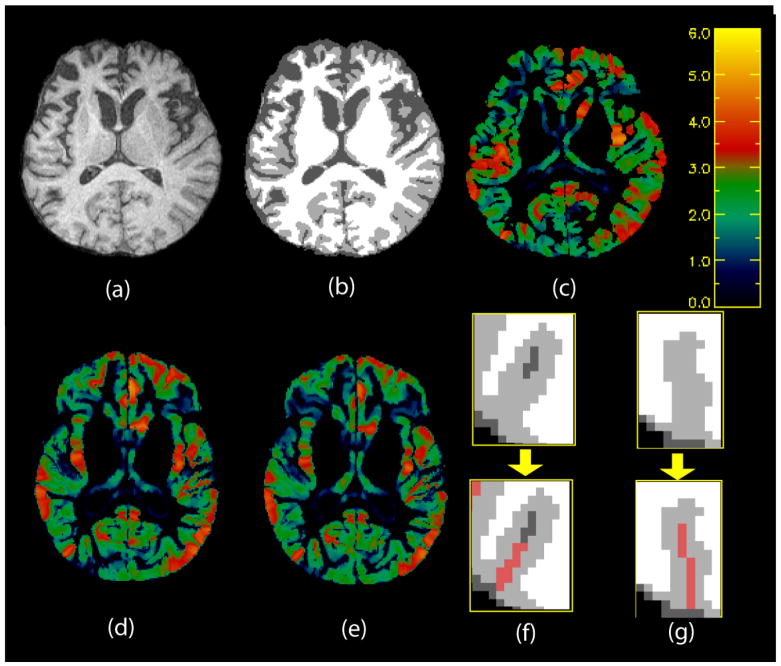
Fig. 9.
Top: (a) Axial slice of skull-stripped brain. (b) Segmentation and (c) thickness map of slice in (a). Colorbar is in mm. Yellow is higher, blue is lower. Bottom Left: DiReCT maps in the template space for the same slice at (d) earlier and (e) later time points. Bottom Right: Examples of recovery of deep sulci in (f) and (g). Upper panels show original segmentation: black = background, white = WM, gray = GM, dark gray = CSF. Lower panels show recovered CSF in red.
3 Results
Results of application of our thickness measurement algorithm in both phantom images and real brain images are presented below. For the phantom images derived from equation 4, we evaluate the correspondences produced by our method and discuss how the thickness measurements depend on image resolution and varying noise levels. We examine various properties of the method on a high resolution 2D slice of the brain. Finally, we describe results of a longitudinal study of thickness changes in a clinical population.
3.1 3D Phantoms
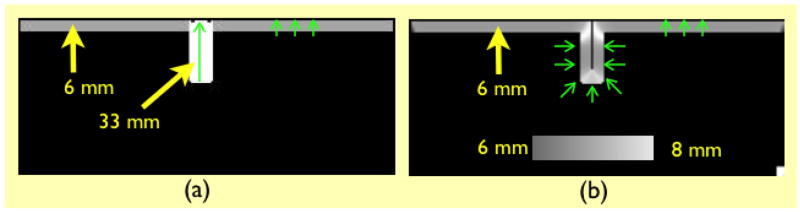
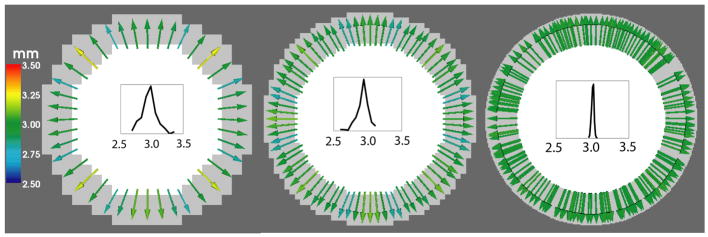
Since we measure thickness as the norm of the deformation field that maps a WM surface point to its corresponding GM surface point, a natural way to evaluate our method is to compare the correspondence map produced with some theoretically prescribed correspondence map. In the simplest case of the 3D phantoms used, the sphere, the corresponding point for any point on the WM surface would be the closest point on the GM surface, which is also the point on the GM surface along the direction of the WM surface normal, as well as the point on the GM surface in the radial direction. The thickness value is expected to be the same everywhere, and is given by T = rg − rw = 3 mm. Figure 5 shows the correspondence map produced by our algorithm.
Fig. 5.
Deformation fields showing correspondences found by Diffeomorphic mapping in a spherical phantom with no noise. A 2D slice is shown here with the solid colors representing WM (white) and GM (gray). Resolution increases from left to right. Color represents norm of the deformation field which is the estimated thickness (in mm). Histogram of measured thickness values are shown in each case.
Notice that the estimated thickness varies roughly between 2.75 – 3.25 mm because of partial volume effects at the low resolution. As the resolution increases, the effects of partial volume diminish and the estimated thickness approaches the theoretical value of 3 mm everywhere. This can be seen from the color of the arrows representing deformation field as well as from the gradual sharpening of the distribution of thickness values as resolution increases.
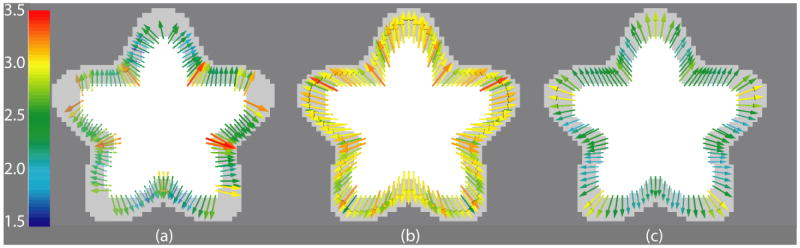
In case of the Ribbon image, as shown in Figure 6, there is no obvious choice for what kind of correspondence map one should use for measuring thickness. Correspondence based on surface normal direction may lead to inconsistencies at valleys with high curvature (Yezzi and Prince, 2003). Correspondence based on the closest GM surface point (Figure 6(a)) is another option. This can be easily defined on arbitrary surfaces once the surfaces have been extracted, but it doesn’t guarantee a smoothly varying deformation field for arbitrary shape, nor does it ensure one-to-one mapping. In case of geometric objects like the ribbon, one could use the radial direction to define correspondence (Figure 6(b)). However, this definition will not work for surfaces with arbitrary shapes. Registration based correspondence (Figure 6(c)), on the other hand, establishes one-to-one mapping and is applicable to arbitrarily shaped objects. It also generates smoothly varying deformation fields. In addition, as we shall see next, it can recover some deep sulci lost due to segmentation errors.
Fig. 6.
Deformation fields showing correspondences defined in different ways in a ribbon shaped phantom with no noise. A 2D slice is shown here with the gray levels representing WM (light gray) and GM (gray). Color represents norm of the deformation field which is the estimated thickness. (a) Closest point-based correspondence. Notice that some correspondence lines do not seem to map to GM boundary points since many of them point to boundary locations at a different slice in 3D. (b) Radial direction based correspondence – by construction, deformation norms are tightly clustered around 3 mm –the variation around the theoretical value is only due to discretization. (c) Correspondence based on Diffeomorphic mapping – this captures the visible variation in thickness along the surface.
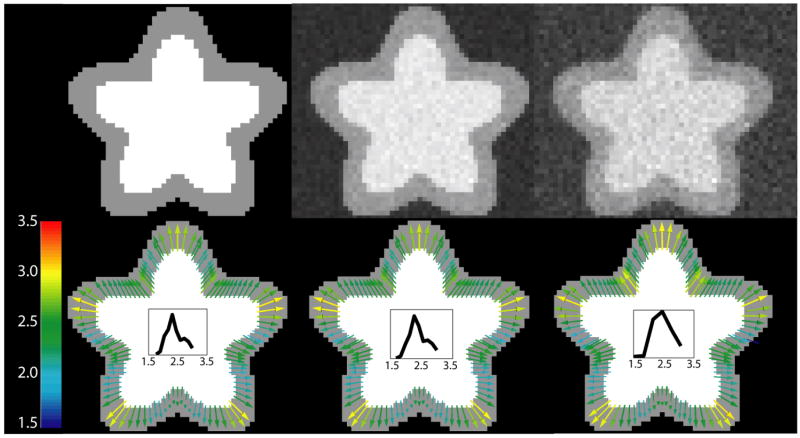
Figure 7 shows a slice through the Ribbon phantom at different noise levels and how it affects the correspondences found. As one would expect, with increasing noise, the histogram of the distribution of thickness widens. Yet, the correspondence map remains fairly robust across different noise levels. Measured thickness, as given by the color of correspondence arrows, varies in few locations. Note that the differences across noise levels may not necessarily reflect the level of reliability of the thickness measurement per se, but may mostly arise from the variability of the segmentation.
Fig. 7.
Left: No noise, Middle: Low noise, and Right: High noise – the same slice at different levels of noise and at the same spatial resolution. Top row shows slices through 3D phantom with noise added. Bottom row shows deformation fields extracted from phantoms with corresponding noise levels as well as histogram of thickness distribution in each case.
Importantly, even though the 3D phantoms described in this section are constructed using an analytical equation (Equation 4), only for certain simple choices of the thickness definition one could obtain an analytical ground truth for the thickness value, such as a definition based on nearest point. In fact, ground truth thickness values for a phantom of a given shape could vary substantially depending on how thickness is defined, as demonstrated in Figure 6. For Diffeomorphic correspondence based definition of thickness, no analytical ground truth is available for a direct evaluation. Nonetheless, it is informative to examine how a given correspondence model (and hence the spatial distribution of measured thickness) captures an intuitive notion of thickness for a thin ribbon-like object (Figure 6). It is also useful to evaluate how a method performs under limited image resolution (Figure 5), as is often the case in practice, and in the presence of noise (Figure 7). Hence, these are the aspects of evaluation we considered using synthetic images, rather than direct comparison with mathematical ground truth. Recovery of buried sulci is another important aspect of evaluation, and is considered in detail in the following subsection.
3.2 Results on Brain Images
The behavior of the proposed method under different noise conditions and spatial resolution has been described with phantom images in section 3.1. Next, different properties of the method are illustrated with the help of a 2D high resolution image of a brain slice in Figure 8. Segmentations in (a), (c), (e) and (i) are drawn such that progressively thicker cortex and narrower sulci are visible. This way, we can examine the solutions obtained by DiReCT under slightly different GM segmentations. We begin with the segmentation in panel (a), where all the sulci are fully resolved (we also separate the gyral crowns by CSF even though they touch in the original image). Panel (b) shows the DiReCT correspondence map of (a). The smoothly varying deformation field, and consequently the smoothly varying thickness values are able to capture the variation of thickness visible in the segmentation of the cortical sheet.
Fig. 8.
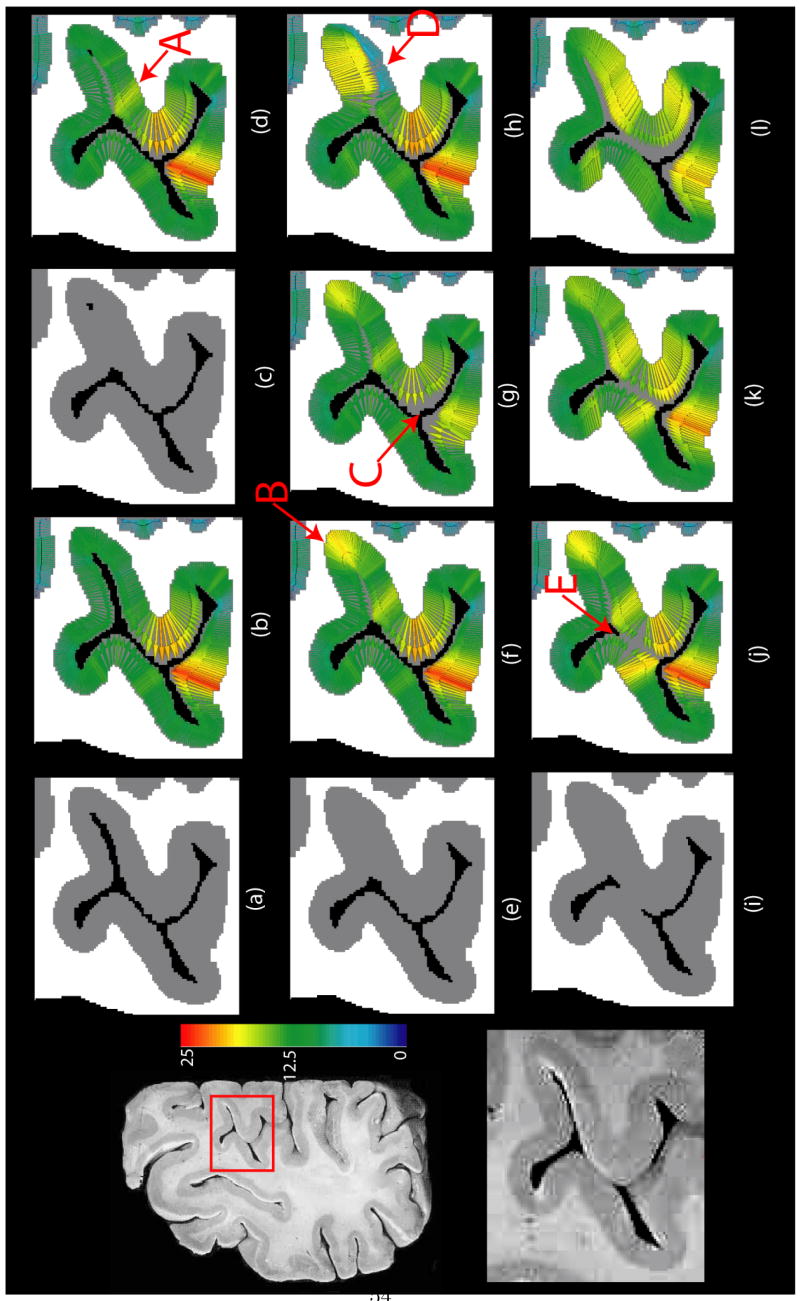
Results of running the proposed algorithm on a 2D high resolution brain slice. Left column: original slice (top) and the zoomed in section of cortex in inset (bottom). (a) Segmentation of the portion of the cortex in inset with all sulci resolved. (b) DiReCT correspondence from segmentation in (a). (c) Segmentation with only a few pixels of CSF resolved in the fundus. (d) DiReCT correspondence from segmentation in (c). (e) Segmentation with closed sulcus. (f) DiReCT correspondence from segmentation in (e) with a high prior making measurements unconstrained. (g) DiReCT correspondence from segmentation in (e) with a prior = 18. (h) DiReCT correspondence from segmentation in (e) with spatially varying prior, with a prior 6 on the lower bank of the closed sulcus and unconstrained elsewhere as in (f). (i) Segmentation with closed sulcus as in (e) and gyri touching each other in the middle. (j) DiReCT correspondence from segmentation in (i) with σ ≈ one voxel length. (k) DiReCT correspondence from segmentation in (i) with σ double that of (j). (l) DiReCT correspondence from segmentation in (i) with σ double that of (k). All quantities are measured in voxel units. See text for discussion.
3.2.1 Recovery of Buried Sulcus
In Figure 8(c), GM in a sulcus gets thicker and no CSF is visible except a few voxels in the fundus. The DiReCT map in (d) is able to resolve this sulcus accurately, and the yellowish color in the region marked A reflects the thicker gray matter there. In (e), the sulcus is completely closed. The correspondence map in (f) is still able to resolve a narrow sulcus. However, since there is no information available about the depth of the sulcus, as was provided by the few CSF voxels in (c), the method produces a thicker fundus, marked B.
3.2.2 Effect of Thickness Prior
Panels (e)–(h) in Figure 8 demonstrate the effect of the thickness prior τ. Sulcal CSF can remain unresolved in the segmentation, as in (e), for various reasons. For a sulcus narrower than a voxel, partial volume effect is often responsible for the CSF therein being mislabeled as GM. In this scenario, setting the prior to a high value compared to the thickness of the sulcal GM such that it essentially doesn’t constrain the measurements there would lead the diffeomorphic mapping to (correctly) resolve an infinitesimally thin sulcus. This is the case in (f), where the prior is set to a high value. Both sulcal banks have similar thickness in this case, which is the most unbiased solution. However, other factors, including segmentation errors, may label even a wider sulcus completely as GM. In this case, an unconstrained measurement would, erroneously, give rise to banks of GM on both sides that are too thick. Also, the two sulcal banks may have asymmetric thickness. If such prior information is known, the proposed method provides the flexibility to use it to take us one step closer to what is neuroanatomically plausible. This is demonstrated in (g) and (h). In (g), a slightly lower thickness prior than the thickest gyral regions (marked C) is used as a prior, leading to a solution with a thinner gyrus, with shape guidance provided by Diffeomorphic mapping. In (h), on the other hand, a spatially varying prior is used, with a prior of about one-fourth of the total thickness of two sulcal banks used for the sulcal bank marked D. This leads to a smooth solution with asymmetric thickness on both sides of the sulcus. In summary, thickness prior may be rendered ineffective by setting it to a very high value, yielding unconstrained thickness measurements, or it can be used in conjunction with Diffeomorphic mapping to get better estimates – if the user so desires and if reliable prior information is available. Using an arbitrary threshold as prior in thickness measurement on populations may bias group analysis. Therefore, we advocate using known, possibly spatially varying, neuroanatomical prior information only to the extent that this information is available. Indeed, when segmentation is perfect, the prior is unnecessary, and can be ignored.
3.2.3 Effect of Smoothing Kernel
Panels (i)–(l) in Figure 8 demonstrate the effect of the width σ of the smoothing kernel K in step 4 of the optimization method in section 2.2.1. In (i), in addition to the closed sulcus in (e), two gyral crowns touch each other. This leads to a solution in (j) that is different than (f), most notably in gyral region marked E, yet the two gyri are separated correctly. As the size of the smoothing kernel is doubled in (k) and quadrupled in (l), we get a smoother and smoother solution, but the GM/CSF boundaries do not match well, even though thickness prior is set to the same high value for (j)–(l). σ is set to 1 voxel width for (b)–(j). As expected, a larger kernel focuses more on larger scale features and is less sensitive to finer scale variations in cortical shape. In general, setting it of the order of one voxel width ensures a sufficiently smooth solution as well as good image similarity.
Next, we describe results of our clinical study.
3.2.4 Clinical Study
We construct DiReCT maps for a longitudinal dataset of elderly individuals diagnosed with FTD spectrum disorders at two different time points. τ is set to 5 mm for results reported here. Figure 9 shows examples of DiReCT maps. Note the recovery of deep sulci as demonstrated in (f) and (g). In Figure 9(f) top panel, the original segmentation is able to recover only some CSF in the sulcus. The rest of the sulcus is resolved by our algorithm as shown in Figure 9(f) bottom panel. In Figure 9(g), the sulcus is completely closed. In this case, an infinitesimally narrow sulcus is recovered. These examples show the ability of our shape-constrained Diffeomorphic mapping to recover buried sulci in 3D brain images.
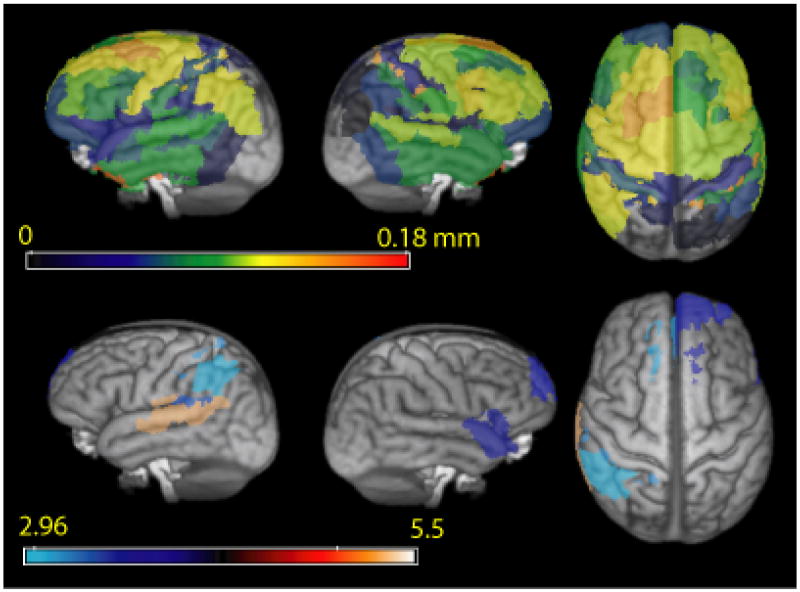
Mean thickness change in Brodmann areas is calculated for every subject. Pairwise t-tests for longitudinal thickness reduction are performed across all subjects in the template space. Figure 10 shows areas of statistically significant change at a false discovery rate (FDR) of 5% to correct for multiple comparisons overlaid on the rendered SyN template. Table 3 lists most significant Brodmann areas. Mean thickness changes are also shown in Figure 10.
Fig. 10.
Top: Mean annual decrease in thickness in different Brodmann regions. Colormap corresponds to annual thickness change in mm. Bottom: FDR corrected (p < 0.05) regions with significant reduction in thickness in 20 patients with FTD spectrum disorder. Colormap corresponds to pairwise t-statistic.
Table 3.
Areas of significant thickness reduction across subjects with FDR corrected p < 0.05. Many of these areas, including area 22 and area 40 are implicated in language comprehension.
| # | Hemisphere | Brodmann Area | T value | Thickness Reduction in mm/year |
|---|---|---|---|---|
| 1 | L | 22 | 5.30 | 0.08 |
| 2 | R | 38 | 3.77 | 0.08 |
| 3 | L | 42 | 3.34 | 0.06 |
| 4 | R | 32 | 3.64 | 0.08 |
| 5 | L | 1 | 3.13 | 0.12 |
| 6 | R | 10 | 3.58 | 0.10 |
| 7 | L | 32 | 3.05 | 0.08 |
| 8 | L | 40 | 2.96 | 0.11 |
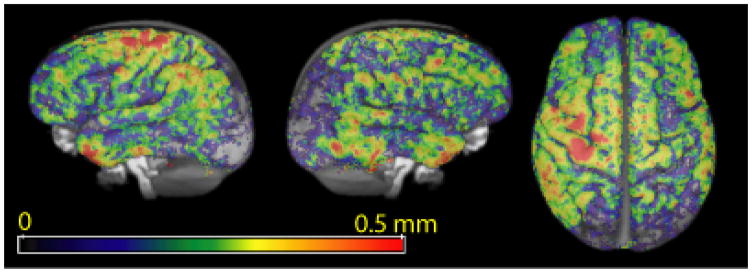
Areas of significant atrophy include large parts of the temporal lobe, as well as some areas of parietal and frontal lobe. Jacobian-based voxelwise measures of atrophy on the same dataset revealed many of the same atrophied regions (Avants et al., 2007). The agreement of these independently gained results help support the robustness of these disease effects. These same areas have been shown to be atrophied compared to healthy age-matched controls in cross-sectional studies (Grossman et al., 2004). In addition, in Table 2, we compare the mean cortical thickness in different lobes of the brain in the baseline scans of our clinical population with published values (Du et al., 2007) in an FTD population of similar age group (61.7 ± 7.5 years as opposed to 61.8 ± 6.8 years for our dataset). The most significant atrophied regions (Table 3) show annual mean thickness reduction of more than 0.10 mm. In contrast, average reduction in the occipital lobe is 0.033 mm (not significant). Cortical thinning due to normal aging is usually reported to be far less (Salat et al., 2004; Preul et al., 2006) – less than 0.10 mm per decade – than the rate of decrease in thickness we found, suggesting atrophy related to neurodegeneration. Even though such corroborative evidence does not conclusively prove that the thickness changes reported here are entirely due to progression of disease, it is also very unlikely that such high rates of cortical thinning in areas known to be affected by the disease could be explained by other factors like normal aging. A voxelwise map of thickness reduction is shown in Figure 11.
Table 2.
Table showing average thickness over different lobes as measured in baseline scans of FTD population.
| # | Hemisphere | Lobe | Average thickness at baseline (mm) | Reported measurements |
|---|---|---|---|---|
| 1 | L | Frontal | 2.26 ± 0.58 | 1.9 ±0.4 (FTD, Du et al. (2007)) |
| 2 | R | Frontal | 2.21 ± 0.55 | N/A |
| 3 | L | Parietal | 1.81 ±0.44 | 1.9 ±0.2 (FTD, Du et al. (2007)) |
| 4 | R | Parietal | 1.81± 0.45 | N/A |
| 5 | L | Temporal | 2.33 ±0.56 | 2.3 ±0.4 (FTD, Du et al. (2007)) |
| 6 | R | Temporal | 2.23 ±0.55 | N/A |
| 7 | L | Occipital | 1.76 ±0.43 | 1.3 – 2.75 (Control, (von Economo, 1929)) |
| 8 | R | Occipital | 1.82 ±0.45 | N/A |
Fig. 11.
Voxelwise average annual thickness reduction map across subjects.
These areas showing significant change over time are directly relevant to the patients participating in this study. Correlations of imaging studies with independent measures of clinical functioning and pathology validate the importance of these areas in FTD spectrum disorders. The atrophied areas in the left frontal and temporal regions are implicated in primary progressive aphasia, one presentation of these conditions; and the atrophied areas in the right hemisphere are limited to the disorder of social comportment and personality that is another presentation of these diseases. Parietal lobe atrophy may be associated with the worsening apraxia and short-term memory disorder of corticobasal degeneration (Murray et al., 2007).
4 Discussion
In this section, we discuss some pertinent issues in the context of cortical thickness measurements as they relate to the proposed methodology.
4.1 How to evaluate cortical thickness measurements ?
We alluded to the lack of a gold standard for cortical thickness measurements in section 1. As we have demonstrated in section 3.1, common thickness measurements may be thought of as a distance measure between a pair of points; depending on how the pair of points is defined, measured thickness can vary greatly. Thus, it is not obvious how to compare clinical studies where thickness measurements are based upon different definitions of thickness. On the other hand, it is challenging to prove the validity of any single thickness definition. Labeling three dimensional brain images – a point operation – is an extremely challenging task for a human. Labeling points on surfaces that are likely to occur in different slices and different orientations – a line operation – is even more challenging, thus making it difficult to generate reliable ground truth thickness measures. This is one reason why we, and other researchers, have not made a quantitative comparison of our evaluation results to a ground truth measurement. Rather, we focused on discussing the variability of our measurements as a function of relevant imaging parameters such as image resolution and noise levels, as well as qualitative visual assessment of the derived correspondence maps. Also, we have demonstrated that our measurements are sensitive enough to be used in a clinical study.
Furthermore, any thickness measure is only as good as the quality of tissue segmentation it is based upon. This makes it difficult to assess the reliability of thickness measures in the presence of noise. While some studies have examined the reliability as a function of image acquisition parameters (Haidar and Soul, 2006; Sato et al., 2003; Han et al., 2006), over repeated scans (Sowell et al., 2004) and as a function of the particular distance measure used to define thickness (Lerch and Evans, 2005; Haidar and Soul, 2006) – no systematic study of the variability of thickness measurements as a function of the tissue segmentation algorithm used has been reported. The issue of recovering deep sulci is also a pertinent one in this context – since it is often segmentation errors coupled with partial volume effects that lead to buried sulci. We have taken a model-based approach to minimize the effects of segmentation errors. As long as the WM segmentation is locally neuroanatomically correct, our constrained Diffeomorphic mapping ensures that the derived GM correspondence preserves the initial anatomical model, in effect often correcting for erroneous GM over-segmentation. It can be noted here that methods that define thickness as distance between corresponding nodes on WM and GM surface meshes (MacDonald et al., 2000), although still dependent on the quality of segmentation, can be evaluated directly in the presence of noise by taking an initial surface reconstruction as the ground truth, corrupting it, and recovering it again (Kabani et al., 2001). The reliability of thickness measurements as a function of segmentation method used deserves to be studied further.
4.2 Recovery of deep sulci
The problem of detecting buried sulci to improve thickness measurements has been addressed explicitly by many researchers (Hutton et al., 2002; Jones et al., 2000; Scott and Thacker, 2005; Lohmann et al., 2003; Barta et al., 2005; Han et al., 2004), often in the form of a pre or post-processing step. On the other hand, labeling cortical sulci directly from MRI volumes (Tu et al., 2007), or from surface models (Sandor and Leahy, 1997) is a well-studied problem in itself. It is hard to say whether any particular approach will produce the result that is closest to “reality” in the context of a given thickness measurement method, including ours. Nonetheless, the proposed solution in this paper is a natural consequence of our constrained Diffeomorphic mapping, and thus directly built into the definition and optimization of our point correspondence. We deal with thickness constraints directly in our optimization, as opposed to having to deal with the problem of thickness over-estimation separately. Once we find a topology preserving, thickness constrained fit of the GWI to the GM surface, the buried sulci are automatically recovered.
Many areas in the cortex are known to have asymmetric thickness on opposing sulcal banks. If spatially varying thickness prior information about such asymmetry is available, the proposed method will recover asymmetric thickness even if no CSF is visible. Also, note that whenever even some amount of CSF (reflected in lowered gray matter probability) is visible in the segmentation, this will guide the registration process (For example, in Figure 9 (f)). However, if no thickness prior information is available and no CSF is visible, our method will still be able to recover an infinitesimally narrow sulcus (For example, in Figure 9 (g)), but produce a symmetric thickness map on either sulcal bank, which is the most unbiased solution under those circumstances. We note that the estimated gray/CSF interface between opposing sulcal banks will be influenced by the specific cortical geometry, in particular the curvature of the fundus. In a “tighter” sulcus (which would usually imply greater curvature at the deepest part of the sulcus), the opposing banks will meet more quickly. Thus – in the absence of a prior – the estimated thickness at the depth of the sulcus will be lower than if the sulcus was wider. This implies the following relationship for sulci with zero resolved CSF: the narrower the sulcus, the lower the estimated thickness at the fundus will be. We believe that this is a natural relationship. However, it is only a “best estimate” as determined by our methodology.
We do not claim that ours is the best approach to recover deep sulci per se, but it is a principled one consistent with how we define point associations for measuring thickness. At the same time, our method naturally builds in neuroanatomic constraints on thickness, with the intention of providing a more plausible and accurate estimate that agrees with what is known about the in vivo brain and the folded, ribbon-like structure of gray matter hidden in buried sulci.
4.3 Groupwise morphometric study
For clinical applications, a statistical framework for carrying out groupwise morphometric studies on measured thickness values is required. When measurements are made on explicitly constructed cortical surface meshes (Lerch et al., 2005; Chung et al., 2005; Fjell et al., 2006), or a hybrid approach where measurements are made in image volumes but then transferred to surface meshes by high dimensional mapping (Thompson et al., 2005) – surface based statistics can be used. Surface based smoothing is often used to increase the signal-to-noise ratio. Similar smoothing schemes can be used in the image domain as well (Avants and Gee, 2003), as was done in Das et al. (2007). In the longitudinal population study presented in section 3, because of the inherent smoothness of the deformation fields, and hence in the DiReCT maps, explicit smoothing was not necessary. Although image registration-based normalization as we have used here for group study poses an inherent limitation to accuracy, since we look at mean region-based quantities – statistics based in the template volume should be adequate. Moreover, for a longitudinal comparison, intra-subject registration error is minimal – even voxelwise statistics would be much more accurate than if it were a cross-sectional study. We note that even though volumetric thickness maps were used in this study, it is obviously possible to use surface based statistics in the SyN template – either on the WM surface in the image domain or by extracting the WM surface mesh on each individual. In that case, one would not need to propagate the thickness measurements to the GM volume. In the clinical study presented here, we use propagation along the dense and Diffeomorphic deformation path to ensure that all GM voxels are assigned a thickness. This is also useful in visualization.
5 Conclusion
In this paper, we introduced a principled approach to finding correspondence between WM and GM surfaces using shape-constrained Diffeomorphic mapping. We evaluated this methodology with 3D phantom data at different spatial resolutions and noise levels. We constructed examples to show the merit of buried sulcus recovery, the use of thickness priors and the effect of the size of smoothing kernel in a 2D brain slice data. Finally, as an application of the methodology to real brain images, we present a groupwise longitudinal study of thickness changes in a patient population.
The advantages of DiReCT in comparison to other approaches include:
The DiReCT algorithm uses a volumetric, Diffeomorphic correspondence model between GWI and the gray/CSF interface to reduce the need for separate preprocessing and mitigate the problem of buried sulci. This technique provides a new level of control over the deformation implied by the correspondence driving the thickness measurement. At the same time, self-intersections of gray matter banks – which cause difficulties and add computational complexity in approaches that are based on deforming meshes (Fischl and Dale, 2000; MacDonald et al., 2000) – are automatically prevented.
DiReCT incorporates a natural geometric definition of thickness that enables clinically motivated Euclidean thickness priors to interact with the optimization of the correspondence. Methods with curved measures of distance (Yezzi and Prince, 2003; Jones et al., 2000) cannot directly use existing post-mortem knowledge. At the same time, if reliable thickness priors are not available, DiReCT provides the flexibility of generating unconstrained measurements as well.
DiReCT encodes thickness measures within the volumetric domain thus enabling voxel-wise statistics either through image-based region of interest analyses – which we have demonstrated in this paper in one of the few longitudinal studies of thickness change in FTD – or through VBM like studies (Hutton et al., 2008).
In summary, the proposed method integrates many advantages of various existing approaches. DiReCT incorporates the model-based, deformation-controlled and regularized approach of methods that use surface deformation, yet works in the volumetric image space. DiReCT also provides an anatomically motivated and prior-constrained, subvoxel measurement of thickness along with an estimate of buried sulci, within the same algorithmic framework. Finally, we showed that DiReCT produces clinically meaningful measurements in a study of cortical thickness within Brodmann areas defined in an FTD cohort.
Our future work will continue to investigate the effect of our new thickness constraints on clinical studies, will incorporate the estimate of GM probability in the thickness estimation step and will contrast surface-based population studies with the volumetric approach used here. We intend to extend the proposed method to use spherical topology representations which are required for inflation and flattening, as in Das et al. (2007). We will also investigate the feasibility of labeling protocols for generating gold standard thickness measurements.
Acknowledgments
This work was partially supported by NIH grants EB006266, NS045839, DA022807, DA14129 and HD046159.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold VI. Ordinary differential equations. Springer-Verlag; Berlin: 1991. [Google Scholar]
- Avants B, Anderson C, Grossman M, Gee J. Spatiotemporal normalization for longitudinal analysis of gray matter atrophy in frontotemporal dementia. 2007 doi: 10.1007/978-3-540-75759-7_37. MICCAI 2007, Lecture Notes in Computer Science 4792, 303–310. URL http://dx.doi.org/10.1007/978-3-540-75759-7_37. [DOI] [PubMed]
- Avants B, Gee J. The shape operator for differential analysis of images. Inf Process Med Imaging. 2003;18:101–113. doi: 10.1007/978-3-540-45087-0_9. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. Neuroimage. 2004;23(Suppl 1):S139–S150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Avants B, Grossman M, Gee JC. The correlation of cognitive decline with frontotemporal dementia induced annualized gray matter loss using Diffeomorphic morphometry. Alzheimer Dis Assoc Disord. 2005;19(Suppl 1):S25–S28. doi: 10.1097/01.wad.0000183083.14939.82. [DOI] [PubMed] [Google Scholar]
- Avants BB, Schoenemann PT, Gee JC. Lagrangian frame diffeomorphic image registration: Morphometric comparison of human and chimpanzee cortex. Med Image Anal. 2006;10(3):397–412. doi: 10.1016/j.media.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Barta P, Miller MI, Qiu A. A stochastic model for studying the laminar structure of cortex from MRI. IEEE Trans Med Imag. 2005;24(6):728–742. doi: 10.1109/TMI.2005.846861. [DOI] [PubMed] [Google Scholar]
- Beg F, Miller M, Trouv’e A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comp Vision. 2005;61:139–157. [Google Scholar]
- Brambati SM, Renda NC, Rankin KP, Rosen HJ, Seeley WW, Ashburner J, Weiner MW, Miller BL, Gorno-Tempini ML. A tensor based morphometry study of longitudinal gray matter contraction in FTD. Neuroimage. 2007;35(3):998–1003. doi: 10.1016/j.neuroimage.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, Fox NC, Jenkins R, Scahill RI, Crum WR, Rossor MN. Rates of global and regional cerebral atrophy in AD and frontotemporal dementia. Neurology. 2001;57(10):1756–1763. doi: 10.1212/wnl.57.10.1756. [DOI] [PubMed] [Google Scholar]
- Chung MK, Robbins SM, Dalton KM, Davidson RJ, Alexander AL, Evans AC. Cortical thickness analysis in autism with heat kernel smoothing. Neuroimage. 2005;25(4):1256–1265. doi: 10.1016/j.neuroimage.2004.12.052. [DOI] [PubMed] [Google Scholar]
- Das S, Avants B, Grossman M, Gee J. Measuring cortical thickness using an image domain local surface model and topology preserving segmentation. IEEE 11th International Conference on Computer Vision. 2007;2007:1–8. [Google Scholar]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, Miller BL, Weiner MW. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain. 2007;130(4):1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis P, Grenander U, Miller MI. Variational problems on flows of diffeomorphisms for image matching. Q Appl Math. 1998;56(3):587–600. [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Reinvang I, Lundervold A, Salat D, Quinn BT, Fischl B, Dale AM. Selective increase of cortical thickness in high-performing elderly–structural indices of optimal cognitive aging. Neuroimage. 2006;29(3):984–994. doi: 10.1016/j.neuroimage.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Grossman M, Libon D, Forman M, Massimo L, Wood E, Moore P, Anderson C, Farmer J, Chatterjee A, Clark C, Branch Coslett H, Hurtig HM-Y, Lee V, Trojanowski J. Distinct antemortem profiles in patients with pathologically defined frontotemporal dementia. Archives of Neurology. 2007;64(11):1601–1609. doi: 10.1001/archneur.64.11.1601. [DOI] [PubMed] [Google Scholar]
- Grossman M, McMillan C, Moore P, Ding L, Glosser G, Work M, Gee J. What’s in a name: voxel-based morphometric analyses of MRI and naming difficulty in Alzheimer’s disease, frontotemporal dementia and corticobasal degeneration. Brain. 2004;127(3):628–649. doi: 10.1093/brain/awh075. [DOI] [PubMed] [Google Scholar]
- Haidar H, Soul JS. Measurement of cortical thickness in 3D brain MRI data: Validation of the Laplacian method. Journal of Neuroimaging. 2006;16:146–153. doi: 10.1111/j.1552-6569.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, Maguire P, Rosas D, Makris N, Dale A, Dickerson B, Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32(1):180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Han X, Pham D, Tosun D, Rettmann M, Xu C, Prince J. CRUISE: Cortical reconstruction using implicit surface evolution. Neuroimage. 2004;23(3):997–1012. doi: 10.1016/j.neuroimage.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Hutton C, De Vita E, Ashburner J, Deichmann R, Turner R. Voxel-based cortical thickness measurements in MRI. NeuroImage. 2008;40(4):1701–1710. doi: 10.1016/j.neuroimage.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton C, De Vita E, Turner R. Sulcal segmentation for cortical thickness measurements. 2002 MICCAI 2002, Lecture Notes in Computer Science 2488, 443–450. URL http://dx.doi.org/10.1007/3-540-45786-0_55.
- Jones SE, Buchbinder BR, Aharon I. Three-dimensional mapping of cortical thickness using Laplace’s Equation. Human Brain Mapping. 2000;11(1):12–32. doi: 10.1002/1097-0193(200009)11:1<12::AID-HBM20>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, Le Goualher G, MacDonald D, Evans AC. Measurement of cortical thickness using an automated 3-D algorithm: a validation study. Neuroimage. 2001;13(2):375–380. doi: 10.1006/nimg.2000.0652. [DOI] [PubMed] [Google Scholar]
- Kim JS, Singh V, Lee JK, Lerch J, Ad-Dab’bagh Y, MacDonald D, Lee JM, Kim SI, Evans AC. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. Neuroimage. 2005;27(1):210–221. doi: 10.1016/j.neuroimage.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005;24(1):163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. Focal decline of cortical thickness in Alzheimer’s disease identified by computational neuroanatomy. Cereb Cortex. 2005;15(7):995–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- Lohmann G, Preul C, Hund-Georgiadis M. Morphology-based cortical thickness estimation. Inf Process Med Imaging. 2003;18:89–100. doi: 10.1007/978-3-540-45087-0_8. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Kabani N, Avis D, Evans AC. Automated 3-D extraction of inner and outer surfaces of cerebral cortex from MRI. Neuroimage. 2000;12(3):340–356. doi: 10.1006/nimg.1999.0534. [DOI] [PubMed] [Google Scholar]
- Miller MI, Massie AB, Ratnanather JT, Botteron KN, Csernansky JG. Bayesian construction of geometrically based cortical thickness metrics. Neuroimage. 2000;12(6):676–687. doi: 10.1006/nimg.2000.0666. [DOI] [PubMed] [Google Scholar]
- Murray R, Neumann M, Forman MS, Farmer J, Massimo L, Rice A, Miller BL, Johnson JK, Clark CM, Hurtig HI, Gorno-Tempini ML, Lee VMY, Trojanowski JQ, Grossman M. Cognitive and motor assessment in autopsy-proven corticobasal degeneration. Neurology. 2007;68(16):1274–1283. doi: 10.1212/01.wnl.0000259519.78480.c3. [DOI] [PubMed] [Google Scholar]
- Preul C, Hund-Georgiadis M, Forstmann BU, Lohmann G. Characterization of cortical thickness and ventricular width in normal aging: a morphometric study at 3 Tesla. J Magn Reson Imaging. 2006;24(3):513–519. doi: 10.1002/jmri.20665. [DOI] [PubMed] [Google Scholar]
- Price C, Davis K, Moore P, Campea S, Grossman M. Clinical diagnosis of frontotemporal dementia (FTD) Neurology. 2001;56(8 Suppl 3):A176. [Google Scholar]
- Rettmann ME, Kraut MA, Prince JL, Resnick SM. Cross-sectional and longitudinal analyses of anatomical sulcal changes associated with aging. Cereb Cortex. 2006;16(11):1584–1594. doi: 10.1093/cercor/bhj095. [DOI] [PubMed] [Google Scholar]
- Rocha KR, Yezzi AJ, Prince JL. A hybrid Eulerian-Lagrangian approach for thickness, correspondence, and gridding of annular tissues. IEEE Transactions on Image Processing. 2007;16(3):636–648. doi: 10.1109/tip.2007.891072. [DOI] [PubMed] [Google Scholar]
- Salat D, Buckner R, Snyder A, Greve D, Desikan R, Busa E, Morris J, Dale A, Fischl B. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14(7):721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Sandor S, Leahy R. Surface-based labeling of cortical anatomy using a deformable atlas. IEEE Trans Med Imag. 1997;16(1):41–54. doi: 10.1109/42.552054. [DOI] [PubMed] [Google Scholar]
- Sato Y, Tanaka H, Nishii T, Nakanishi K, Sugano N, Kubota T, Nakamura H, Yoshikawa H, Ochi T, Tamura S. Limits on the accuracy of 3-D thickness measurement in magnetic resonance images–effects of voxel anisotropy. IEEE Trans Med Imag. 2003;22(9):1076–1088. doi: 10.1109/TMI.2003.816955. [DOI] [PubMed] [Google Scholar]
- Scott MLJ, Thacker NA. Robust tissue boundary detection for cerebral cortical thickness estimation. MICCAI 2005, Lecture Notes in Computer Science. 2005;3750:878–885. doi: 10.1007/11566489_108. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. J Neurosci. 2004;24(38):8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Maes F, Vandermeulen D, Dupont P, Paesschen WV, Suetens P. An automated 3D algorithm for neocortical thickness measurement. 2003 MICCAI 2003, Lecture Notes in Computer Science 2879, 488–495. URL http://www.springerlink.com/content/gn3w3pnwvufq8uf0.
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang Y, van Erp TGM, Cannon TD, Toga AW. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage. 2004;23(Suppl 1):S2–S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Eckert MA, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Reiss AL. Abnormal cortical complexity and thickness profiles mapped in Williams syndrome. J Neurosci. 2005;25(16):4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Zheng S, Yuille A, Reiss A, Dutton R, Lee A, Galaburda A, Dinov I, Thompson P, Toga A. Automated extraction of the cortical sulci based on a supervised learning approach. IEEE Trans Med Imag. 2007;26(4):541–552. doi: 10.1109/TMI.2007.892506. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Joshi S, Miller MI. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. Proc Natl Acad Sci USA. 1998;95(3):788–795. doi: 10.1073/pnas.95.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Economo C. The cytoarchitectonics of the human cerebral cortex. H. Milford Oxford University Press; 1929. [Google Scholar]
- Yezzi AJ, Prince JL. An Eulerian PDE approach for computing tissue thickness. IEEE Trans Med Imag. 2003;22(10):1332–1339. doi: 10.1109/TMI.2003.817775. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zeng X, Staib LH, Schultz RT, Duncan JS. Segmentation and measurement of the cortex from 3-D MR images using coupled-surfaces propagation. IEEE Trans Med Imag. 1999;18(10):927–937. doi: 10.1109/42.811276. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]