Abstract
Significant progress has been made in identification of genes and gene networks involved in key biological processes. Yet, how these genes and networks are coordinated over increasing levels of biological complexity, from cells to tissues to organs, remains unclear. To address complex biological questions, biologists are increasingly using high-throughput tools and systems biology approaches to examine complex biological systems at a global scale. A system is a network of interacting and interdependent components that shape the system’s unique properties. Systems biology studies the organization of system components and their interactions, with the idea that unique properties of that system can be observed only through study of the system as a whole. The application of systems biology approaches to questions in plant biology has been informative. In this review, we give examples of how systems biology is currently being used in Arabidopsis to investigate the transcriptional networks regulating root development, the metabolic response to stress, and the genetic regulation of metabolic variability. From these studies, we are beginning obtain sufficient data to generate more accurate models for system function. Further investigation of plant systems will require data gathering from specific cells and tissues, continued improvement in metabolic technologies, and novel computational methods for data visualization and modeling.
Keywords: Root development, transcriptional networks, metabolism, quantitative traits, high-throughput methods
The shift to systems biology
Advances in molecular biology over the past 30 years have led to one of the most exciting and fruitful times in biological research. This has largely been accomplished through a reductionist approach to answer biological questions. This approach is based on the idea that complex systems are simply a sum of their parts. For example, in molecular genetics, a gene’s function is inferred from the phenotype of the organism when that gene is disrupted. This approach has been highly successful in identifying molecules that play key roles in diverse biological processes; however, it requires a straightforward relationship between the gene and its function. This method is confounded by mutations that result in either undetectable or uninterpretable phenotypes. Further complexity is revealed when gene function depends on context, such as developmental stage or environment [1]. Therefore, a simple summation of parts cannot adequately account for the complexity observed in biological systems [2]. In the past decade, high-throughput technologies coupled with an exponential rise in computing power has generated the next wave of molecular data and a shift from a reductionist to a more holistic or systems biology perspective has taken place.
Systems biology attempts to study complex biological systems by examining all of the components and the relationships between those components in the context of the whole system (for example, an organ). Systems theory puts forth the notion that system complexity can only be understood through study of the whole system, with all of its components and their interactions [3, 4]. This is because systems exhibit emergent properties, which cannot be observed in, or predicted from, examination of isolated components. The first step in a systems approach is to identify all of the system components (e.g. genome, transcripts, proteins, metabolites). The next step is to perturb the system and monitor its response. The resulting data are then integrated into models of system function [5, 6]. New hypotheses generated by these models can be experimentally tested and ultimately, lead to revised models and new testable hypotheses. For example, modeling of experimental data for the Arabidopsis circadian clock revealed that while a single feedback loop is sufficient to generate oscillatory gene expression, a clock comprised of multiple interconnecting loops is required to account for all of the observed clock behaviors. Further experiments based on this modeling identified additional system components, which confirmed the model containing multiple interconnecting loops and added a photoperiod sensing mechanism into the clock [7–10]. Because information concerning the function of the system as a whole is desired, all of the measurements and experimental observations are most informative when conducted on a global scale at high spatial and temporal resolution. Although, many molecular biologists view systems biology as synonymous with high-throughput approaches or the ‘omic’ technologies, such as genomics, transcriptomics, and metabolomics, this is not the case. These high-throughput technologies must be complemented by modeling of the experimental data to truly investigate the molecular mechanisms driving systems-level biological phenomena.
Unicellular organisms were initially the focus of systems biology approaches and therefore, were limited to intracellular systems. However, many fundamental biological processes, such as development, take place in multicellular eukaryotes and therefore require orchestration across multiple cell types. There are several key features of plant growth and development that make plants amenable to systems biology approaches [11]. First, plants have a simple body plan composed of reiterated elements and continuously develop organs through activity of two primary stem cell populations. Plant growth and development is very robust, plants encounter variety of strong perturbations in nature (and in the laboratory) and are able to respond by altering these processes to adapt to external conditions. Plants are also vitally important to our quality of life as a key source of food, shelter, fiber, medicine, and increasingly fuel. Given their significance, it is surprising how little is known about the mechanisms regulating plant growth [12]. Finally, nearly all plant growth and development is postembryonic and continuous, this is in contrast to animal development which is embryonic, often inaccessible, and finite.
Certainly considerable progress has been made in identifying key factors required for plant growth, development, and environmental response. However, it remains largely unknown how these factors are coordinated across the levels of biological organization: from molecules to cells, tissues, organs, whole organisms and populations. A systems level understanding is required if we intend to fully capitalize on plants to address global challenges such as food and fuel shortages. In this review, we highlight recent progress in advancing systems biology approaches in the model plant Arabidopsis to (1) identify transcriptional networks underlying root development, (2) characterize of metabolic responses to environmental stress, and (3) examine of transcriptional and metabolic variation in natural populations. These topics were selected to show the progress in two types of system component identification and the powerful synergy between these two approaches when examined together and at the population level.
Cell-type specific and temporal analyses reveal complex transcriptional networks underlying development
Organogenesis requires coordination of developmental processes in the context of single cells, among cell types or tissues, and in time. The Arabidopsis root provides an elegant model for the study of organ development in a spatiotemporal context. Root growth and development occurs predominately in the radial (spatio) and longitudinal (temporal) axes and is continuous throughout the life of the plant due to regulated division of stem cells (Figure 1) [13, 14]. Newly formed cells will differentiate along the longitudinal axes according to positional cues [15, 16]. By assaying development at the whole-organ level, it is likely that a majority of the cell-type specific information is lost; therefore, one of the main obstacles to understanding organ development is isolation of specific cell types for high-throughput analysis. This has been overcome in the Arabidopsis root where florescence-activated cell sorting has been employed to examine the transcriptional profiles of individual cell types [17–19]. Recently, Brady and colleagues [20] utilized a set of fluorescent protein reporters, which show specific expression in 14 cell types of the developing root. Root cells were isolated and sorted by fluorescence and the transcriptional profile of each cell type was assayed by microarray. Transcriptional profiling of 13 longitudinal sections, representing progression through developmental time, complemented this cell-type specific data set. This high-resolution transcriptional map is the most complete for any organ to date. Cell-type specific analysis identified cell-type specific expression profiles and common transcriptional profiles in distinct cell types. Longitudinal analysis revealed complex temporal regulation of gene expression. Interestingly, many longitudinal expression patterns appeared to fluctuate along developmental time. Coexpression profiling identified a number of transcriptional modules. One module for biosynthesis of a plant hormone was supported by data in the literature; additional predicted modules can be experimentally tested and will provide novel frameworks for genetic regulation of developmental processes.
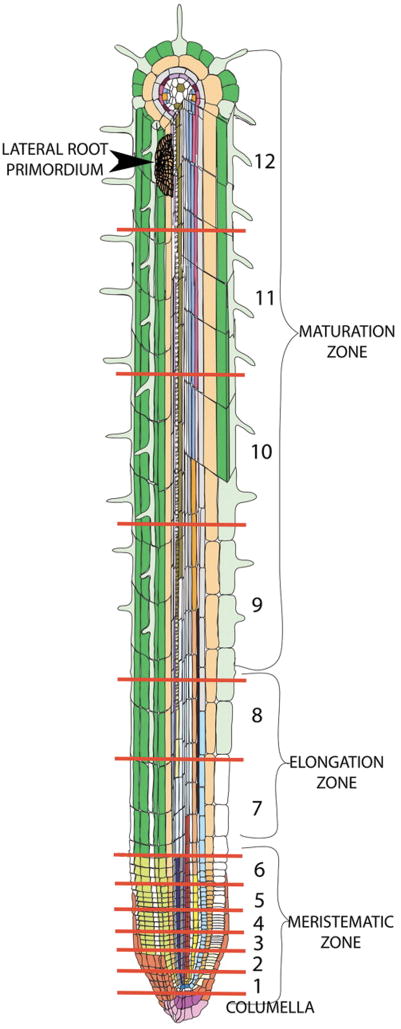
Figure 1.
Diagram of the Arabidopsis root tissues and longitudinal sections used in microarray expression profiles in Brady et al. Colors represent the different developmental stage and cell types examined (14 individual tissues). Thirteen longitudinal samples were analyzed and the red lines indicate their relative positions. Stem cells are located in section 1 and cells further from the stem cells are progressively more differentiated. The developmental zones are bracketed on the right. In the meristematic zone, stem cells undergo asymmetric divisions that recapitulate the stem cells and produce daughter cells, which will divide several more times within this zone. In the elongation zone, division ceases and the cells rapidly expand longitudinally. Finally, in the maturation zone, cells differentiate acquiring their specialized features. Note the radial symmetry particularly in the outer tissues of the root. From Brady, S.M., et al., A high-resolution root spatiotemporal map reveals dominant expression patterns. Science, 2007. 318(5851): p. 801–806. Reprinted with permission from AAAS.
Plant development is plastic and highly responsive to environmental cues; this allows plants to optimize growth depending on changing conditions. To address how root development and environmental response are linked, Dinneny et al. [21] characterized the transcriptional response of five different tissues and developmental stages to salt stress and iron deficiency. These data revealed dramatic cell-type specific transcriptional changes in response to each environmental stress. This suggests that a cell-type specific response to stress is required for the adaptive strategy of the whole organ and begins to reveal how the root partitions functions among cell types. Intriguingly, of the relatively few genes that show transcriptional response to both stresses, those that responded to salt stress in all five tissues were the least likely to be coregulated by iron deficiency. These data are in opposition to the hypothesis that transcriptional responses across multiple cell-types make up a generalized stress response and instead suggest that responses in all cells are stimuli-specific. Additionally, among the genes that respond to salt stress, those with known stress response elements in their promoters were found to be active in multiple cell types. This suggests that distinct, but as yet, uncharacterized cis-elements are responsible for the cell-type specific responses to salt stress. Interestingly, a relatively small number of genes failed to show significant changes in expression during response to either stress. Most of these genes have been previously identified as developmental regulators, suggesting unresponsive genes function as critical determinants of cell fate. Together these two studies make clear the deficiencies of whole organ or organism transcription studies. Because organs are composed of distinct cell-types, transcriptional profiling at a cell-type specific resolution and in response to perturbation is a major step towards achieving a systems level understanding of organ development. The next important step is the modeling of these data to identify transcriptional modules and formulate predictions about module function that can be tested in the plant.
Metabolites provide important insight into a system’s response to perturbation
Systematic transcriptional profiling provides valuable insight into system function; however, for a number of reasons it provides an incomplete picture. First, a large proportion of genes remain functionally uncharacterized. Also, the relationship between transcript abundance and translation or protein activity remains unclear. Finally, the metabolic outputs of genetic networks may be the most indicative of system function and its response to perturbation. Metabolites can be viewed as a measure of enzymatic activity over time and metabolites have important roles in response to perturbation because they feedback into the transcriptional network (Figure 2) [22–24]. Thus, it has become clear that the functional definition of a system must include metabolic composition and, ideally, quantitative metabolic flux in response to perturbation.
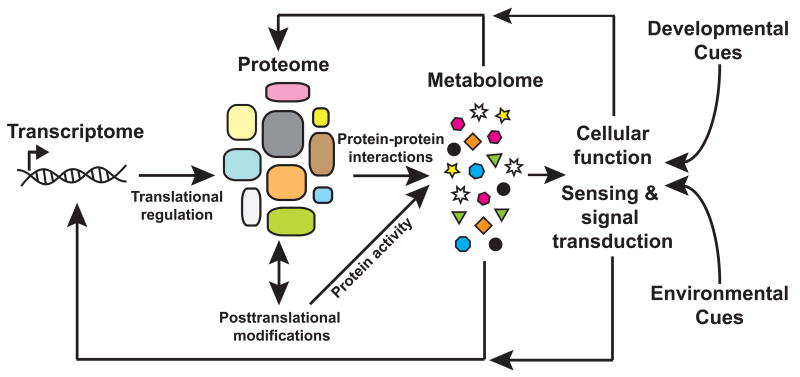
Figure 2.
Schmatic of a network showing interdependence of system components and response to feedback based on developmental or environmental cues. The metabolic profile of a cell or system provides a measure of the output of the genetic network but may also serve as an input. In response to external cues metabolites feed back into networks, ultimately altering behavior of the system. Figure adapted from Weckworth [23].
There are two main types of metabolites: primary and secondary. Primary metabolites, such as lipids and amino acids, function in fundamental cellular processes and are common to all organisms. Secondary metabolites have more specialized functions and are often species-specific. Although secondary metabolites are not explicitly required for survival, they contribute to the overall success of the organism [25]. For example, as sessile organisms, plants cannot escape unfavorable conditions and must employ other adaptations for survival. Diversification of metabolites is thought to be a primary strategy: toxins function in pathogen or herbivore defense, volatiles and pigments function to attract pollinators, other chemicals provide salt or cold tolerance, and a variety of compounds mediate intra- and interplant signaling [26, 27]. These chemicals are also valuable to humans as approximately 12000 distinct alkaloid compounds are predicted to be synthesized in plants and about 2000 have medical applications [28]. Global metabolic profiling allows simultaneous observation of precursors, intermediates, and end products of metabolic pathways. Identification of novel metabolites as well as quantitative analysis of the metabolic flux in a system will provide considerable insight into system function.
Historically, metabolic studies have been targeted at metabolites already predicted to function in a given process; however, to achieve a systems level understanding of that process, unbiased methods are more favorable [29]. Response and acclimation to temperature stress is one of the best examples of system perturbation (stress responses) monitored by unbiased metabolic profiling. Plants exposed to nonlethal temperature extremes acquire a greater tolerance to subsequent extremes [30, 31]. To examine the role of metabolites in temperature acclimation, Kaplan and colleagues metabolically profiled plants exposed to nonlethal high and low temperatures by gas chromatography-mass spectrometry (GC-MS) at three time points [32]. Their study revealed cold treatment had a larger impact on plant metabolism than heat, and found 81 known and 416 unknown mass spectral tags. Comparison of the two data sets showed the majority of heat response metabolites were also present in response to cold, implying a previously unknown relationship between these opposing stresses. In a complementary study, the metabolic profiles of two genotypes with differential tolerance to cold were examined upon exposure to low temperatures. Interestingly, both genotypes showed similar metabolic responses to cold treatment, but in the cold sensitive genotype this response was quantitatively different, with a reduction in metabolite accumulation. Additionally, Cook et al. examined the metabolic profile of plants overexpressing a low-temperature induced transcription factor and showed these plants have a similar metabolic profile as wild type plants after cold treatment [33, 34]. Together these studies reveal extensive changes in metabolic profiles in response to cold stress, a striking similarity between high and low temperature acclimation, and identify metabolites not previously associated with thermal acclimation. Additionally, a genetic mechanism for metabolic changes in response to cold was proposed. While these studies represent significant progress in unbiased metabolic profiling in plants, they reveal the need for systematic examination of metabolites, novel metabolite identification, and metabolic profiling at a higher spatiotemporal resolution before these data can be modeled to reveal novel hypotheses about system function.
Using natural variation to identify the molecular mechanisms underlying multigenic traits
Integration of data comprising multiple system components is a key step in systems biology. Researchers have begun combining results from various types of data sets. For example, Gutierrez and colleagues developed a qualitative multinetwork model for response to carbon and nitrogen in Arabidopsis roots that includes metabolic and gene regulatory components and their interactions [35]. Transcript and metabolite profiling are powerful tools in identification of system components and characterizing the links between these components is important for understanding system function. Recent studies combining these two approaches with natural variation illustrate their additive strength to address complex biological processes at the population level [36]. Natural variation offers an alternative to laboratory-induced mutations in the study of plant function [37]. Natural populations of Arabidopsis vary in diverse morphological and physiological traits (Figure 3). Because reproduction occurs primarily by self-pollination, geographically isolated Arabidopsis populations (accessions) can have genetic and phenotypic differences which are often adaptive to their specific environment. Phenotypic variation among accessions is predicted to be due to genetic polymorphisms that alter gene expression, protein levels, or protein or metabolite activity. Natural variation is typically observed on a phenotypic continuum for a given trait and is described as a quantitative trait [37]. Quantitative traits often result from interactions between multiple genes and environmental factors.
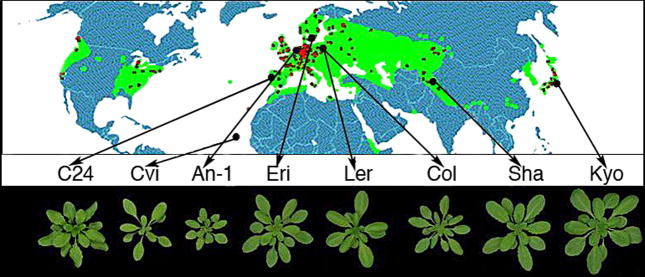
Figure 3.
Natural variation in leaf growth and development in several Arabidopsis accessions collected from distinct geographical locations. Natural variation is often observed as a phenotypic continuum, as opposed to presence or absence of the trait. Here, for example, all accessions have leaves, but the size, shape, and number is variable. The molecular mechanisms regulating quantitative traits such as leaf size and shape can often be applied to improve crop science and plant breeding programs. Figure from the website of Dr. Matthieu Reymond, Max Planck Institute for Plant Breeding Research, used with permission of Dr. Martin Koornneef.
Quantitative trait analyses aim to identify the molecular mechanisms responsible for variation in a given trait [38]; this goal can be complicated by numerous factors. Quantitative traits are genetically mapped to chromosomal regions called quantitative trait loci (QTL). Depending on the size of the mapping population, a QTL may contain hundreds of genes, any of which might be involved in trait variability [39, 40]. Additionally, quantitative traits are often multigenic; therefore, a single trait may be associated with multiple loci, each with positive or negative impacts on the trait. To address these difficulties, populations of recombinant inbred lines (RILs) were developed for many Arabidopsis accessions. RILs are generated by first crossing two accessions and then allowing individual F2 progeny to repeatedly self over many generations, until nearly all loci are homozygous. RILs allow more accurate phenotypic analysis of traits because repeated measurements on each genotype is possible [41]. Characterization of multigenic traits requires dissection of complex interactions, which is the primary goal of systems biology; therefore, use of large populations to examine a system with high-throughput technologies has clear advantages.
Recent unbiased metabolic profiling of Arabidopsis accessions and their associated RILs has revealed extensive genetic control of metabolic variation. An untargeted examination of secondary metabolites among 14 Arabidopsis accessions conducted by liquid chromatography-time of flight-mass spectrometry (LC-QTOF-MS) revealed qualitative and quantitative differences in their metabolic profiles [29, 42]. To address the genetic control of this variability, metabolic profiles of a RIL population generated from the two most biochemically divergent accessions was analyzed [43]. Interestingly, many mass peaks among individual RILs were significantly different from either parent. These results suggest that complex genetic interactions control metabolic output. Furthermore, QTL analysis of about 2000 mass peaks indicated that 75% of these peaks could be assigned to genetic loci. In another study, primary metabolites were examined using gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) in a second pair of accessions and their respective RILs [44]. These accessions were selected because they were previously subjects of targeted metabolic QTL and global gene expression QTL analyses [45–50]. Based on unbiased metabolic profiling, combined with the existing data for this population, the authors identified many metabolic QTLs with phenotypic effects and found that epistatic interactions between multiple loci controlled much of the metabolic variation. Additionally, integration of all the available data allowed modeling of two biochemical networks, one of unknown function and one involving in a central metabolic pathway; both network models included novel metabolites. These studies indicate that metabolic traits are generally less heritable than transcriptional traits, suggesting that metabolic components may be more sensitive to environmental influences and that metabolic traits are regulated by complex interactions between multiple loci. The complex molecular interactions between transcripts, metabolites, and genetic loci revealed by these studies will provide insight into the relationship between plant metabolism and plant growth and development.
Conclusion
Systems biology strives to understand complex biological systems through identification of system components and characterization of their interactions within the context of the whole system [3]. Here we have briefly highlighted three examples of how systems biology approaches in Arabidopsis are forging ahead in the effort to attain a systems level understanding of complex biological processes in plants [5, 51, 52]. Complex transcriptional programs operating in both space and time were found to regulate root development. Identification of coexpressed genes has led to predictions about functional networks that underlie developmental processes and revealed functional relationships between seemingly dissimilar cell types [20]. Network response to perturbation is important for understanding and modeling dynamic networks and it was shown that transcriptional response to abiotic stress in the root is highly dependent on cell identity [21]. At the metabolic level, response to temperature perturbations revealed extensive changes in metabolite profiles [32, 33]. Numerous novel molecules were identified as temperature responsive and surprisingly, a high degree of similarity was found between the metabolic responses to heat and cold. At the organismal level, transcriptional, metabolic, and quantitative genetic data from different Arabidopsis accessions showed that metabolic traits are regulated by complex interactions between many genetic loci [42, 44]. Each of these examples serves as a framework to address additional biological questions using system biology approaches and to begin to elucidate complex system functions.
Despite recent advances in high-throughput technologies, limitations exist. Metabolic techniques lack comprehensive extraction and analysis methods, particularly for low-abundance and novel molecules [23]. The diversity in size, biosynthesis, and structure of metabolites precludes establishment of a single protocol for metabolite extraction and identification. New advances in metabolic technologies will greatly facilitate identification and inclusion of more metabolites into systems studies [24]. Further progress will come from profiling metabolites from distinct cell types [18]. Based on studies of transcriptional profiles in the root, it is expected that individual cells types will have unique metabolic profiles [17, 20, 21]. Although transcriptional profiling in the root has revealed important networks regulating growth and development, plants are comprised of many more cell types than are represented by the root. Transcriptional profiling of all cell types is required to formulate models for plant form and function. Furthermore, data from each system component at a higher spatial and temporal resolution is the key to more precise modeling of underlying networks and system function. Generation of these models requires significant increase in computational and modeling expertise. In order for systems biology to succeed, none of the components can be analyzed in isolation [3]. Large-scale data sets from multiple system components must be integrated to create a holistic model for how a system functions. Finally, achievement of these goals requires collaborative and community efforts between biologists, mathematicians, physicists, statisticians, and computer scientists.
Supplementary Material
Acknowledgments
Work in P.N.B.’s lab on systems biology is funded by grants from the NIH, NSF, and DARPA. We thank Siobhan Brady, Terri Long, Rosangela Sozzani, Anjali Iyer-Pascuzzi and Jayson Punwani for critical reading of the manuscript.
References
- 1.Benfey PN, Mitchell-Olds T. From genotype to phenotype: systems biology meets natural variation. Science. 2008;320(5875):495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Regenmortel MH. Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological and overcome the limitations of reductionism. EMBO Rep. 2004;5(11):1016–1020. doi: 10.1038/sj.embor.7400284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Ideker T. Systems biology 101--what you need to know. Nat Biotechnol. 2004;22(4):473–475. doi: 10.1038/nbt0404-473. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez RA, Shasha DE, Coruzzi GM. Systems biology for the virtual plant. Plant Physiol. 2005;138(2):550–554. doi: 10.1104/pp.104.900150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee SY, Dickerson J, Xu D. Bioinformatics and its applications in plant biology. Annu Rev Plant Biol. 2006;57:335–360. doi: 10.1146/annurev.arplant.56.032604.144103. [DOI] [PubMed] [Google Scholar]
- 7.Locke JCW, Millar AJ, Turner MS. Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol. 2005a;234:383–393. doi: 10.1016/j.jtbi.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Locke JCW, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005b;1:2005.0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke JCW, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeilinger MN, et al. A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady SM, Benfey PN. A systems approach to understanding root development. Canadian Journal of Botany. 2006;84(5):695–701. [Google Scholar]
- 12.Achieving the in Silico Plant. Systems Biology and the Future of Plant Biological Research. Plant Physiol. 2003;132(2):404–409. [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119(1):71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414(6859):98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 15.van den Berg C, et al. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390(6657):287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 16.Benfey PN, Scheres B. Root development. Curr Biol. 2000;10(22):R813–815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- 17.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 18.Birnbaum K, et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2(8):615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- 19.Nawy T, et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17(7):1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 21.Dinneny JR, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320(5878):942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 22.Hall R, et al. Plant metabolomics: the missing link in functional genomics strategies. Plant Cell. 2002;14(7):1437–1440. doi: 10.1105/tpc.140720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weckwerth W. Metabolomics in systems biology. Annu Rev Plant Biol. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- 24.Schauer N, Fernie AR. Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 2006;11(10):508–516. doi: 10.1016/j.tplants.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5(10):439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 26.Gershenzon J. Plant volatiles carry both public and private messages. Proc Natl Acad Sci U S A. 2007;104(13):5257–5258. doi: 10.1073/pnas.0700906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heil M, Silva Bueno JC. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc Natl Acad Sci U S A. 2007;104(13):5467–5472. doi: 10.1073/pnas.0610266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegler J, Facchini PJ. Alkaloid biosynthesis: metabolism and trafficking. Annu Rev Plant Biol. 2008;59:735–769. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- 29.Vorst O, de Vos CHR, Lommen A, Staps RV, Visser RGF, Bino RJ, Hall RD. A non-directed approach to the differential analysis of multiple LC-MS-derived metabolic profiles. Metabolomics. 2005;1(2):169–180. [Google Scholar]
- 30.Guy C. Molecular responses of plants to cold shock and cold acclimation. J Mol Microbiol Biotechnol. 1999;1(2):231–242. [PubMed] [Google Scholar]
- 31.Thomashow MF. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan F, et al. Exploring the Temperature-Stress Metabolome of Arabidopsis. Plant Physiol. 2004;136(4):4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook D, et al. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci U S A. 2004;101(42):15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilmour SJ, et al. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124(4):1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez R, et al. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8(1):R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keurentjes JJ, Koornneef M, Vreugdenhil D. Quantitative genetics in the age of omics. Curr Opin Plant Biol. 2008;11(2):123–128. doi: 10.1016/j.pbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 2000;5(1):22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- 38.de Meaux J, Koornneef M. The cause and consequences of natural variation: the genome era takes off! Curr Opin Plant Biol. 2008;11(2):99–102. doi: 10.1016/j.pbi.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Mauricio R. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat Rev Genet. 2001;2(5):370–381. doi: 10.1038/35072085. [DOI] [PubMed] [Google Scholar]
- 40.Hansen BG, Halkier BA, Kliebenstein DJ. Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends Plant Sci. 2008;13(2):72–77. doi: 10.1016/j.tplants.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Maloof JN. Genomic approaches to analyzing natural variation in Arabidopsis thaliana. Curr Opin Genet Dev. 2003;13(6):576–582. doi: 10.1016/j.gde.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Keurentjes JJ, et al. The genetics of plant metabolism. Nat Genet. 2006;38(7):842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 43.Alonso-Blanco C, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14(2):259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 44.Rowe HC, et al. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20(5):1199–1216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loudet O, et al. Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104(6–7):1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 46.Loudet O, et al. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol. 2003;131(1):345–358. doi: 10.1104/pp.102.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calenge F, et al. Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics. Plant Physiol. 2006;141(4):1630–1643. doi: 10.1104/pp.106.082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kliebenstein DJ, et al. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics. 2006;7:308. doi: 10.1186/1471-2105-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wentzell AM, et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3(9):1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West MA, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175(3):1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan JS, et al. Plant systems biology comes of age. Trends Plant Sci. 2008;13(4):165–171. doi: 10.1016/j.tplants.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Provart NJ, McCourt P. Systems approaches to understanding cell signaling and gene regulation. Curr Opin Plant Biol. 2004;7(5):605–609. doi: 10.1016/j.pbi.2004.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.