Abstract
Motivation
Mathematically optimal alignments do not always properly align active site residues or well-recognized structural elements. Most near-optimal sequence alignment algorithms display alternative alignment paths, rather than the conventional residue-by-residue pairwise alignment. Typically, these methods do not provide mechanisms for finding effectively the most biologically meaningful alignment in the potentially large set of options.
Results
We have developed Web-based software that displays near optimal or alternative alignments of two protein or DNA sequences as a continuous moving picture. A WWW interface to a C++ program generates near optimal alignments, which are sent to a Java Applet, which displays them in a series of alignment frames. The Applet aligns residues so that consistently aligned regions remain at a fixed position on the display, while variable regions move. The display can be stopped to examine alignment details.
Availability
Available at http://fasta.bioch.virginia.edu/ noptalign. For source code contact the authors at wrp@virginia.edu
Contact
wrp@virginia.edu
INTRODUCTION
Optimal algorithms for global and local alignment of protein and DNA sequences have been available for 30 years (Needleman and Wunsch, 1970; Sellers, 1974; Smith and Waterman, 1981). Given a set of scores for residue identities and replacements, and penalties for beginning and continuing gaps, these algorithms are guaranteed to produce the best possible alignment score. However, while the alignment score is unique, there may be several distinct alternative alignments that produce the optimal score. Optimal alignments that maximize a set of scoring parameters may not be biologically optimal; in some cases, they may not align critical functional residues or structural elements. Such failures of optimal pairwise alignment algorithms are well recognized (Saqi et al., 1998), and are a major motivation for multiple alignment methods.
While optimal alignments sometimes misalign structural or functional features, the errors are usually modest, e.g. a gap that disrupts one end of an alpha helix. In general, we suspect that the most biologically meaningful alignment of two sequences lies close to the algorithmically optimal solution. Various algorithms have been proposed for generating near optimal solutions (Zuker, 1991; Naor and Brutlag, 1994; Waterman and Byers, 1985). These algorithms enumerate some or all sequence alignments within a certain distance from the optimal solution. In addition to generating multiple near optimal solutions, an algorithm might also produce multiple optimal solutions. Often there are a large number of alternatives, and it is difficult to communicate effectively the differences and similarities among alignments within the set of alternative alignments.
Here, we present a tool for finding biologically meaningful alignments among a large set of alternatives. The ability to adjust and configure the display is an important consideration when interacting with decision support systems that provide alternative solutions. When users are presented with a computer-generated ‘optimal’ solution (by some measure), there is a natural tendency to simply accept that solution. This has been called anchoring and, in a different context, automation bias (Kahneman et al., 1982; Parasuraman et al., 1993; Layton et al., 1994). The goal is to allow the user to effectively explore the solution space of a problem and arrive at a solution based not only on the computed solution, but also on user expertise (Vicente, 1999; Guerlain, 2000; Brill et al., 1990).
We have developed a prototype system that generates a set of near-optimal alignments and displays the alignments in a manner that attempts to exploit human pattern recognition capabilities by presenting a ‘moving picture’ of the alignments. This near-optimal alignment program is not a similarity searching program, and does not provide an estimate of the statistical significance of the alignment. Nonetheless, if the original alignment is statistically significant, suboptimal alignments with scores within 90% of optimal will typically be significant as well. In the Systems and Methods section, we describe our implementation of the Zuker (1991) algorithm for generating sets of near-optimal alignments. In the Results section, we describe the software and new algorithms developed for displaying sets of alternative alignments and highlighting the similarities among the alignments. We conclude by presenting different alignment examples that highlight features of the display.
SYSTEMS AND METHODS
The system consists of three separate programs: (1) a Perl CGI script that collects the sequences, annotations and alignment parameters using the BioPerl (Stajich et al., 2002) libraries and then manages the creation of alignments; (2) C++ code that generates the set of near-optimal alignments defined by (1) and (3) the Java Applet that displays the alignments. The alignment generation algorithm (Zuker, 1991) produces a sample of global alignments that meet a score threshold. The CGI script is a straightforward Web form that manages the data input from the user. The display is discussed in the Results section.
Data input and integration
The sequences, alignment parameters and annotations on the sequences are entered using a Web form (http://fasta.bioch.virginia.edu/noptalign). Sequences can be entered by cutting and pasting, or by specifying an NCBI recognized accession or GI number. The researcher then specifies the scoring matrix and gap penalty parameters. Multiple combinations of scoring matrix and gap penalties can be specified to generate a comprehensive set of alignments. The user also specifies the near-optimal neighborhood that the alignment generation algorithm should consider. At present, only global alignments are supported but local alignments can be emulated by specifying subsequences to be aligned.
To identify more easily alternative alignments that are biologically ‘correct’, features can be highlighted on either sequence. These features can be automatically downloaded from the NCBI databases or entered by the investigator.
Alignment generation
Alternative alignments are pre-computed using a C++ program that implements an algorithm described by Zuker (1991). The algorithm has three basic steps. First, a standard dynamic programming technique aligns the two sequences, generating a forward score matrix. We chose an implementation close to that described by Gotoh (1982). Unlike the most space efficient versions of this algorithm, the entire score matrix is maintained in memory. Once the forward score matrix is generated, the process is repeated on the reversed sequences, creating a reverse score matrix.
Next, the forward and reverse score matrices are combined into what we call the Zuker matrix. The value for node i, j in the Zuker matrix is calculated by the score in the forward matrix at point i − 1, j − 1, plus the score in the reverse matrix at m − i + 1, n − j + 1 (where m and n are lengths of the respective sequences), plus the value of the scoring matrix for the residues at locations i and j in the respective sequences. The value at node i, j of the Zuker matrix is the optimal score of a global alignment that is constrained to align residue i in sequence one with residue j in sequence two. For a more detailed discussion of this matrix and the algorithm in general see Zuker (1991) and the RNA folding paper where the technique was originally developed (Zuker, 1989).
During the creation of these two score matrices, a second pair of matrices is created that keeps track of the path used to generate the scores at each node in the respective score matrix. These path information matrices are used to reconstruct the actual alignments when generating a set of alignments. For a given point, i, j , the algorithm constructs the alignment by working forwards and backwards from that point through the information matrices. This process is repeated for every point i, j where the value of that point in the Zuker matrix is greater than the near-optimal neighborhood threshold value specified in the input. The result is a sample of near-optimal alignments of the input sequences. As noted by Zuker, this algorithm only creates a sample of near-optimal alignments; at any given node there is often more than one direction that the alignment can follow and produce an alignment with the optimal or near optimal score, but only one direction is chosen. Once the set of optimal and near-optimal alignments is generated, the alignments are formatted for transmission to a Java Applet that will display the alignment ‘movie’. The actual alignments are encoded using the FASTA -m9c encoding (Pearson and Lipman, 1988). For protein and DNA alignments, matches, insertions and deletions are encoded by ‘=’, ‘+’ and ‘−’ followed by the length of the match, insertion or deletion. Thus, the alignment
PYL-IDGSSHITQS : . ::: .::. PLVEIDG--MLTQT
would be encoded as ‘=3−1=3+2=5’.
The parameter information, sequence data and alignment information is encoded in General Feature Format (GFF2, http://www.sanger.ac.uk/Software/formats/GFF), and transmitted to the Java display applet.
RESULTS
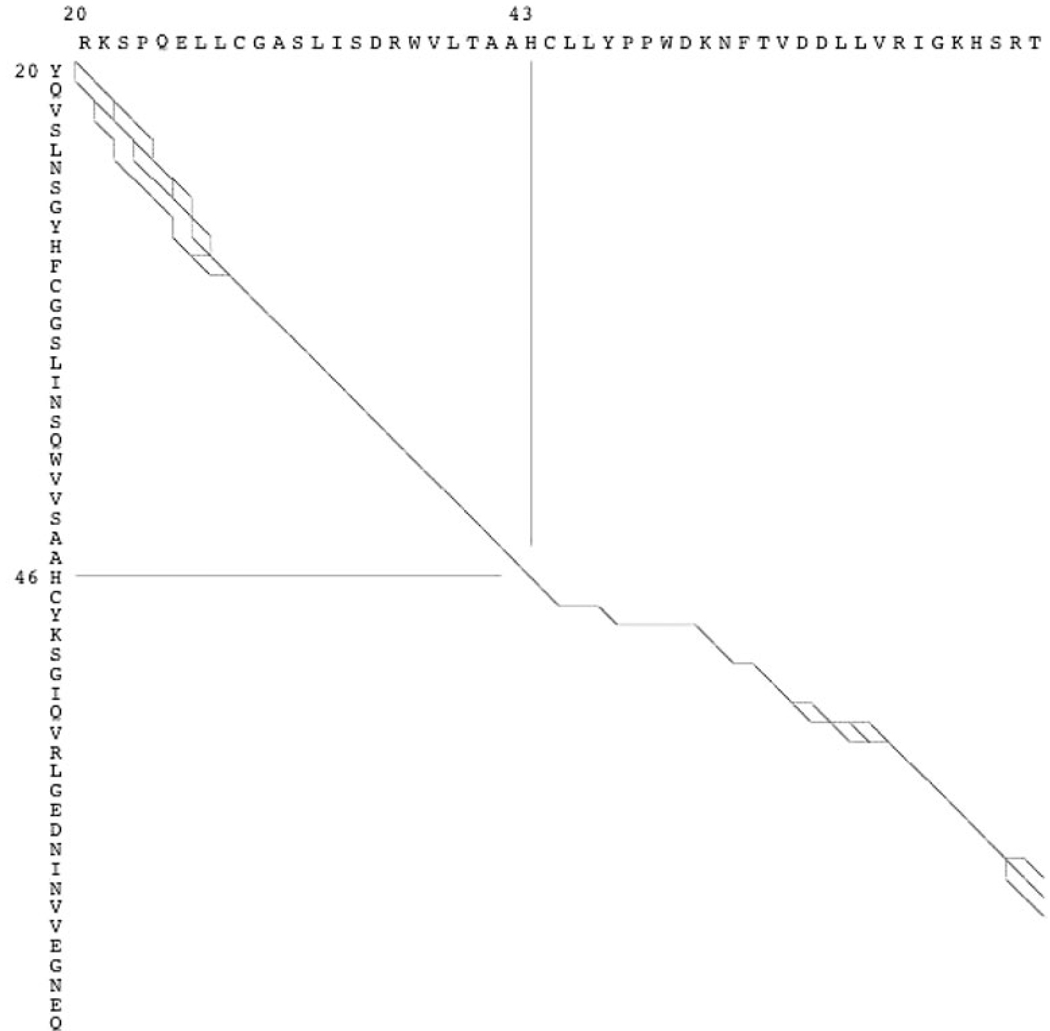
Traditionally, near-optimal alignments are presented together in a single display with a path graph or dot plot (Zuker, 1991; Naor and Brutlag, 1994). This representation effectively high-lights sections of high variability between alignments, but these displays lose information by combining all alignments into one display. Figure 1 provides an example of a partial path graph of a subset of near-optimal alignments of two serine proteases 1TGSZ (NCBI GI: 230350) and 1UVTH (NCBI GI: 2781297). The alignment shows variability at the beginning and end of the alignment by drawing the multiple paths that the alignment could follow. In contrast, in the middle of the alignment there is only one path and hence only one way for the amino acids to align (this is expected as the ‘H’ in the active site is aligned). The primary difficulty with a path graph display is determining which of the alignment paths are more likely. Additionally, path graphs are difficult to use for residue level analysis, because the actual sequences are placed perpendicular and hence distant from one another. Readers of path graphs may have difficulty mentally mapping horizontal, vertical and diagonal edges into insertions and deletions (e.g. with which amino acids in 1UVTH does the 25th amino acid in 1TGSZ align?). Options for annotating a path graph with additional information are also limited.
Fig. 1.
A partial path graph of multiple near-optimal alignments of 1TGSZ (vertical) and 1UVTH (horizontal) generated with the SUBOPT program (Naor and Brutlag, 1994) using the PAM250 matrix and a gap penalty of −3 per residue. While variable regions are easy to spot, reliable regions are harder. It is difficult to determine how two specific amino acids align and annotations are hard to add. The alignment of the active site histidines is indicated.
Traditional text-based alignments, with one sequence placed above the other [as seen in BLAST Altschul et al. (1990) and FASTA (Pearson and Lipman, 1988) output], are ideal for displaying the precise residue-to-residue mapping between the two sequences. However, it is difficult to show alternative text-based alignments and highlight the differences between the alignments. One strategy puts alternate alignments above and below the optimal alignment in some regions. This becomes more difficult when the number of gaps differs among the alignments, as this changes the overall alignment length. As the number of alignments increases, the difficulty increases.
Near-optimal alignment viewer
To address the problems with displaying alternative alignments in a text-based format, we have developed a program to display the alignments sequentially, each for a short period of time, like a movie. Our movie presentation exploits the pattern recognition capabilities of the human eye so that the user will be able to discern changes or areas of stability that occur between alignments as they are displayed.
Steady display
Although it is relatively straightforward to display a large set of alignments as successive frames in a movie, the naïve approach does not make it easy to distinguish constant from variable alignment regions. We seek to highlight the residues that consistently align with one another and distinguish them from those positions that are more variable. To do this, the sections of an alignment that are most consistent should remain steady on the screen while more variable regions should move around. This is not possible with the conventional constant spaced character placement used by BLAST and FASTA.
To address this difficulty, we developed an algorithm for placing pairs of residues (the two residues that align—an edge in a path graph) according to the frequency with which the pairs occur in the set of all alignments and the relative position of that pair within an alignment. The result is a display where residues that consistently align with one another remain stationary in the display, while those that align with many different residues appear to move about.
The first step is to determine the relative position of each aligned pair of residues in relation to the overall length of the alignment. To calculate this, we divide the index (position in the alignment) of the pair by the length of the alignment. Next this relative position is added to all the other relative positions for the given pair, to produce an average position for the pair. Pairs can occur in many different alignments within a set of solutions. The frequency at which each pair occurs with respect to the number of alternative alignments is also calculated.
When we are rendering the text on the screen, we use the cumulative position of given pairs to determine the placement of the particular pair. To do this we access a lookup table to get the position of a pair, which is expressed as a percentage of the length of the alignment. This percentage is multiplied by the width of the display to get the exact position on the screen where the pair will be rendered. The width of the display is determined by the longest alignment.
Recall that this cumulative position is an average value for this pair of residues across all alignments in the near optimal set. Each pair only has one cumulative position and is therefore rendered in the same location on the screen every time. The visual effect of stability is an emergent property of the data. Pairs that appear in a large number of alignments appear stationary on the screen, since they are always rendered in the same location. Residues that are part of pairs that appear infrequently move around, since the different pairs have different cumulative positions.
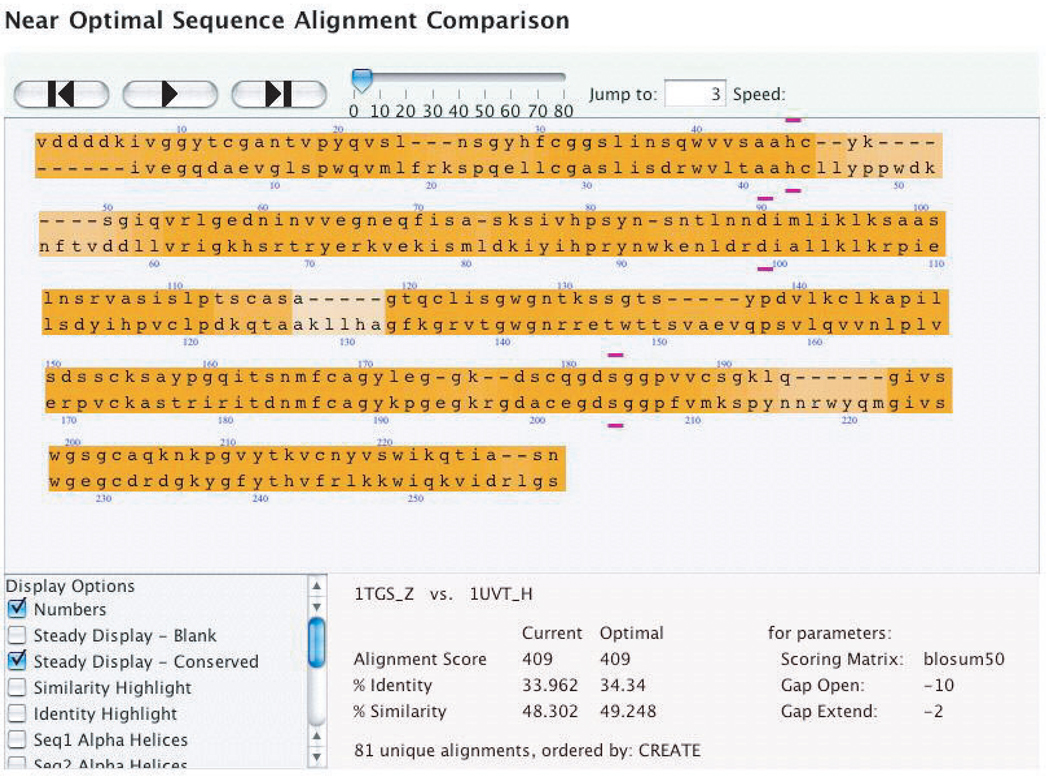
Alone, the Steady Display algorithm provides powerful visual evidence of reliably aligned regions of a set of alignments. However, we can also use the recorded pair-frequency information to color the background of each pair according to the frequency of the pair (Fig 2 and Fig 3A). The most frequent pairs are colored a saturated orange, with the color gradually decreasing in saturation in proportion to the frequency of the pairings. Thus, in Figure 2, the regions around each of the three residues in the serine protease catalytic triad of 1TGSZ and 1UVTH are the most saturated. Figure 3A shows an alternative alignment, where residues 48, 49 (YK) in 1TGSZ are moved to the right, and a second gap is opened after position 54. The least frequent pairings have a white background. In the specific alignment shown, the lightest colors correspond to regions that are not present in 1TGSZ. This coloring provides further visual indication of the consistency of certain sections of alignments.
Fig. 2.
A screen shot of an optimal alignment of 1TGSZ and 1UVTH. Eighty-one alignments within 2% of optimal were calculated. The Steady Display Conserved, numbering and active site options are selected. The three active site residues of the serine protease catalytic triad (46H, 90D, 183S in 1TGSZ; 43H, 99D, 205S in 1UVTH) highlighted with purple bars above or below the residues. The orange highlighting indicates the most conserved regions of the alignment. The darker the orange, the more conserved the region. The alignment is substantially conserved around all three active sites.
Fig. 3.
(A) A subsection of an alternative alignment of 1TGSZ and 1UVTH. This section shows a light region following the active site ‘H’ in the two proteins. While there is variation near the active site, the rest of the alignment is substantially similar to the first one. (B) A subsection of an alignment of 1TGSZ and 1UVTH with Steady Display Blank, 1TGSZ secondary structure information and 1UVTH secondary structure information selected. Near residue 120 in 1UVTH the alpha helix icons appear like X’s, while near residue 150 in 1TGSZ the alpha helix icons that appear like arcs or upside down U’s; when these to align regions, the helix symbols combine to form a loop symbolizing a helix. Likewise, the icons for beta strands come together to form an arrow. (C) A subsection of the 1TGSZ and 1UVTH alignment with the Steady Display Blank, Identity and Similarity options selected.
The display applet can also highlight matching (red) and similar residues (pink) (Fig. 3C). If the user specifies a protein by the GID/accession number then we also have access to the annotation information available in the NCBI database. If any alpha helices or beta strands are specified, we provide an option for those features to be displayed as well. The icons for both the alpha helices and beta strands are designed to combine into a more meaningful icon when both residues in a given pair share the same secondary structure. For alpha helices, an arch beneath the upper sequence combines with an X above the lower sequence to create a small loop that symbolizes a helix (Fig. 3B). Similarly, the icons for beta strands combine to produce an arrow. In addition to these automated features, users can specify their own features to be highlighted on the alignments. All display features may be turned on and off by the user to suit individual preferences.
DISCUSSION
Static images on paper cannot communicate the movement underlying the design of this display. Nonetheless, the figures provide some sense of how the display accomplishes its goals. The ability to communicate the stable regions of the alignment has been accomplished with the Steady Display feature. The large amount of orange coloring in Figure 2 and Figure 3 shows that large portions of the alignment of 1UVTH and 1TGSZ are fairly well conserved. We can also see more variable regions in these figures. For instance, the section of the alignment following the first active site (amino acids 46 and 43) demonstrates how the saturation of the background color varies to communicate variability. In Figure 3A, this section (amino acids 48–55 and 45–62) appears with a white background, indicating that among the 81 alignments this alignment is relatively rare. Figure 2 shows how this same section can have a darker background indicating that the alignment of these residues in Figure 2 is more common than that in Figure 3A. By providing an interactive algorithmic search engine and display mechanism, researchers can conduct more informed alignment comparisons.
This display generation method provides considerable flexibility in attaching annotations to specific alignments. Prototype implementations of conditional highlights and alignment constraints have been developed, although the user interface still remains somewhat cumbersome. We plan to enhance the interface so that different user annotations, such as predicted secondary structure, can be displayed with a choice of icons and colors to facilitate the needs of different researchers.
The general concept of providing a sequenced set of alignment frames for displaying alignment variation may have more general applications. By providing a GFF2 interface to the alignment viewer, it is possible to separate completely the alignment set from the display process. For example, a set of structure-based alignments might be presented, or the results of multiple alignments. In addition, one could imagine that other variable features might be displayed, such as varying DNA-binding sites along a promoter sequence.
ACKNOWLEDGEMENTS
We thank Dr Michael Sierk, Dr Ellen Bass and the reviewers for their helpful comments. We would also like to thank Dr Douglas Brutlag for access to his source code for near-optimal alignment generation. M.E.S. is supported by University of Virginia Biotechnology Training Program (NIH T32 GM08715). This work was supported by a grant from the National Library of Medicine (LM04969).
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brill ED, Flach JM, Hopkins LD, Ranjithan S. MGA: a decision support system for complex incompletely defined problems. IEEE Trans. Sys. Man Cyber. 1990;20:745–757. [Google Scholar]
- Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- Guerlain S. Interactive advisory systems; Proceedings of the Human Performance, Situation Awareness and Automation Conference; Savannah, GA. 2000. pp. 166–171. [Google Scholar]
- Kahneman D, Slovic P, Tversky A. Judgment Under Uncertainty: Heuristics and Biases. Cambridge, UK: Cambridge University Press; 1982. [DOI] [PubMed] [Google Scholar]
- Layton C, Smith PJ, McCoy E. Design of a cooperative problem-solving system for en-route flight planning: an empirical evaluation. Hum. Factors. 1994;36:94–119. [Google Scholar]
- Naor D, Brutlag DL. On near-optimal alignments of biological sequences. J. Comp. Biol. 1994;4:349–366. doi: 10.1089/cmb.1994.1.349. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequences of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Parasuraman R, Molloy R, Singh IL. Performance consequences of automation-induced complacency. Int. J. Aviat. Psychol. 1993;3:1–23. [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence analysis. Proc. Natl Acad. Sci., USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saqi MAS, Russell RB, Sternberg MJE. Misleading local sequence alignments: implications for comparative protein modeling. Protein Eng. 1998;11:627–630. doi: 10.1093/protein/11.8.627. [DOI] [PubMed] [Google Scholar]
- Sellers PH. An algorithm for the distance between two finite sequences. J. Comb. Theory (A) 1974;16:253–258. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilber JGR, Korf I, Lapp H, et al. The BioPerl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente KJ. Cognitive Work Analysis; Lawrence Earlbaum Associates; Mahwah, New Jersey, USA. 1999. [Google Scholar]
- Waterman MS, Byers TH. A dynamic programming algorithm to find all solutions in a neighborhood of the optimum. Math. Biosci. 1985;77:179–188. [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–53. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- Zuker M. Suboptimal sequence alignment in molecular biology: analysis with errors. J. Mol. Biol. 1991;221:403–420. doi: 10.1016/0022-2836(91)80062-y. [DOI] [PubMed] [Google Scholar]