Abstract
Photoactive analogues of farnesyl diphosphate (FPP) are useful probes in studies of enzymes that employ this molecule as a substrate. Here, we describe the preparation and properties of two new FPP analogues that contain diazotrifluoropropionyl photophores linked to geranyl diphosphate via amide or ester linkages. The amide-linked analogue (3) was synthesized in 32P-labeled form from geraniol in 7 steps. Experiments with Saccharomyces cerevisiae protein farnesyltransferase (ScPFTase) showed that 3 is an alternative substrate for the enzyme. Photolysis experiments with [32P]3 demonstrate that this compound labels the β-subunits of both farnesyl- and geranylgeranyltransferase (types 1 and 2). However, the amide-linked probe 3 undergoes a rearrangement to a photochemically unreactive isomeric triazolone upon long term storage making it inconvenient to use. To address this stability issue, the ester-linked analogue 4 was prepared in 6 steps from geraniol. Computational analysis and X-ray crystallographic studies suggest that 4 binds to PFTase in a similar fashion as FPP. Compound 4 is also an alternative substrate for PFTase and a 32P-labeled form selectively photocrosslinks the β-subunit of ScPFTase as well as E. coli farnesyldiphosphate synthase and a germacrene-producing sesquiterpene synthase from Nostoc sp. strain PCC7120 (a cyanobacterial source). Finally, nearly exclusive labeling of ScPFTase in crude E. coli extract was observed, suggesting that [32P]4 manifests significant selectivity and should hence be useful for identifying novel FPP utilizing enzymes in crude protein preparations.
Keywords: protein prenylation, prenyltransferase, photoaffinity labeling, DATFP, farnesyl diphosphate, sesquiterpene synthase, germacrene synthase.
INTRODUCTION
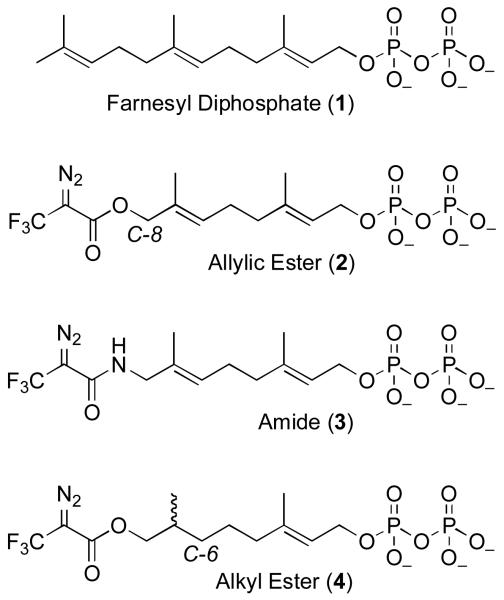
Farnesyl diphosphate (FPP, 1) is an important metabolite in the biosynthesis of a variety of molecules including sesquiterpenes (1, 2) and the side chains of a number of cofactors;(3) FPP also serves as the source of prenyl groups that are appended to proteins.(4) Photoactive analogues of FPP have been useful in identifying proteins and enzymes that use this molecule as a substrate. Hence, a variety of photocrosslinking groups have been used to prepare FPP analogues for use in such experiments including diazotrifluropropionates (DATFP) (5-12), benzophenones (13-22) and aryl azides.(23, 24) While DATFP-based probes do suffer from the disadvantage that they require activation at lower wavelengths than benzophenone- and aryl azide-containing reagents, they also have the advantage that they are intrinsically smaller than most other types of photoactive moieties; in particular, a DATFP group is an excellent mimic of a single isoprene unit. For that reason, Baba and Allen synthesized compound 2 and initially employed it in studies with FPP synthase.(6-8) In subsequent years, this compound has been used extensively by a number of researchers in experiments with protein prenyltransferases.(5, 9, 11, 25)
One feature of compound 2 that makes it suboptimal for photoaffinity labeling experiments is the ester linkage that connects the photoactive DATFP moiety with the isoprenoid skeleton. Because of the allylic nature of this ester, cleavage between the two parts of the molecule can occur by nucleophillic addition to either the ester carbonyl or the allylic position (C-8). This causes problems in the synthesis but more critically, it can result in loss of the radioactive label (usually present at C-1) in subsequent crosslinking experiments. To circumvent this problem, we decided to explore the synthesis and use of DATFP-containing analogues that incorporate more stable linkages between the crosslinking group and isoprenoid. In this paper we first describe the preparation and use of an amide-linked DATFP analogue (3); the design of that compound is based on earlier work with amide-linked benzophenone-containing probes.(19) While this new compound functions as an alternative substrate of Saccharomyces cerevisiae protein farnesyltransferase (ScPFTase) and selectively labels the β-subunit of all three known protein prenyltransferases, its diazo-amide moiety undergoes a rearrangement to a photochemically inert triazoline over time in aqueous solution. This property limits the utility of this probe. Next, we describe the synthesis of 4, an analogue based on a 6,7 dihydrogeranyl framework that replaces the allylic ester present in 2 with an alkyl ester; that design is based on earlier work with azide-containing alternative substrates for ScPFTase.(26, 27) Computational analysis and X-ray crystallographic studies suggest that 4 binds to PFTase in a similar fashion as FPP. Compound 4 is also an alternative substrate for ScPFTase and labels a number of FPP-utilizing enzymes including protein farnesyltransferase, farnesyl diphosphate synthase and a sesquiterpene cyclase. Of particular significance, photolysis of crude E. coli extracts expressing ScPFTase, in the presence of 4, results in labeling of the enzyme suggesting that this probe should be useful for identifying other FPP-utilizing enzymes present in complex protein mixtures.
MATERIALS AND METHODS
General
All trans-geraniol, dimethylallyl alcohol, t-butyl hydroperoxide, H2SeO3, salicylic acid, hydrazine monohydrate, toluene sulfonic acid, and trichloroacetonitrile were obtained from Aldrich. All synthetic reactions were performed at room temperature in open air, with magnetic stirring unless noted otherwise. TLC analysis was performed on pre-coated (250 μm) silica gel 60 F-254 plates plates from Merck, and the plates were visualized by UV light at 254 nm or KMnO4 staining. Flash chromatography silica gel (60-120 mesh) was obtained from Mallickrodt Baker Inc., Paris, KY, USA. Dowex 50W-X8 resin and silver staining kits were obtained from Bio-Rad, Sep-Pak columns were purchased from Waters, and Amplify was from Amersham Life Science. [32P]H3PO4 (specific activity 8500-9120 Ci/mmol) was purchased from DuPont NEN. CH2Cl2 and CH3CN were dried with an M. Braun solvent purification system. Deuterated solvents were used as obtained from Cambridge Isotope Laboratories Inc. 1H-NMR spectra were obtained at 300 MHz and 13C-NMR at 75 MHz. All NMR spectra were acquired on Varian instruments at 25°C. Chemical shifts are reported in ppm with J values reported in Hz. IR readings were taken using NaCl polished plates. HPLC was performed on a Beckman 127/166 instrument equipped with a Linear Instruments LC305 fluorescence detector. Phosphorimaging analysis was performed with a Molecular Dynamics 445 SI Phosphorimager. N-dansyl-GCVIA was synthesized by Dr. Dan Mullen in the Department of Chemistry, University of Minnesota. ScPFTase (21), HsPGGTase I, EcFPPSase (28) and NoSTSase (29) were purified as previously described. RnPGGTase II was a generous gift from Dr. Miguel Seabra (Imperial College, London).
(E,E)-8-N-(2-Diazo-3,3,3-trifluoropropionamido)-3,7-dimethyl-2,6-octadienyl-1-diphosphate (3)
Alcohol 9 (3.1 mg, 10 μmol) was reacted with anhydrous H3PO4 (2.0 mg, 20 μmol, prepared by lyophilization over P2O5) in CH3CN (255 μL) containing 25% (v/v) CCl3CN and triethylamine (40 μmol) for 2 h at rt under N2. The volatile components were then evaporated, and the resulting residue was partially purified by reversed-phase chromatography on a Sep-Pak C18 cartridge with an NH4HCO3/CH3CN step gradient from 0-100% CH3CN; pure product eluted at 25% CH3CN. The fractions containing the desired pyrophosphate product were concentrated and redissolved in 25 mM NH4HCO3, and the final concentration was determined by UV absorbance (236 nm, extinction coefficient = 14,000 cm−1M−1).(30) Diphosphate 3 was obtained in 8% yield, and the purity was determined to be 93% diphosphate, 7% monophosphate by reversed-phase HPLC analysis. 1H-NMR (300 MHz, D2O, pH 8.0): δ = 1.53 (s, 3H), 1.56 (s, 3H), 2.00-2.05 (m, 4H), 3.65 (s, 2H), 4.13 (t, 2H, J = 6.0), 5.26-5.30 (m, 2H); HR-FAB-MS calcd for C13H10N3O8P2F3Na [M+Na]+488.0576, found 488.0609
(E,E)-[α,β(n)32P]-8-N-(2-Diazo-3,3,3-trifluoropropionamido)-3,7-dimethyl-2,6-octadienyl-1-diphosphate ([32P]3)
Alcohol 9 (2.0 mg, 6.3 μmol) was reacted with anhydrous [32P]H3PO4 (1.2 mg, 13 μmol) prepared as previously described in CH3CN (200 μL) containing 20% (v/v) CCl3CN and triethylamine (2.5 mg, 26 μmol) for 2 h.(31) The volatile components were then evaporated, and the resulting residue was purified using a reversed-phase Sep-Pak C18 cartridge with an NH4HCO3/CH3CN step gradient. The product ([32P]3) eluted from the Sep Pak cartridge in fractions containing 20% and 25% CH3CN. After evaporation and redissolving in 25 mM NH4HCO3, [32P]3 (specific activity: 1.25 Ci/mmol) was obtained, giving a 7% yield. The radiochemical purity was determined to be 40%, as assessed by thin layer chromatography in isopropanol/NH4OH/H2O (6:3:1, v/v/v), followed by phosphorimaging analysis.
(E)-8-O-(2-Diazo-3,3,3-trifluoropropanoyloxy)-3,7-dimethyl-2-octenyl-1-diphosphate (4)
Anhydrous phosphoric acid was prepared by lyophilization of a solution of 1% H3PO4 (v/v) (3.8 mL, 0.64 mmol). To that residue was added CH3CN (1.6 mL), and Et3N (180 μL, 1.3 mmol) followed by compound 17 (100 mg, 0.32 mmol), dissolved in 3.2 mL of a solution of CCl3CN in CH3CN (20%, v/v). The reaction was allowed to proceed in the dark under an N2 atmosphere with magnetic stirring for 2 h. The solvent was then removed by rotory evaporation and the residue was redissolved in 10 mL of 25 mM NH4HCO3. The product was purified by reversed-phase HPLC with buffer A (25 mM NH4OH) and buffer B (CH3CN) over 7 injections, using a linear gradient from 0-35% buffer B over 35 min. Samples (2.0 mL) were injected into a 5 mL loop, onto a Phenomex Luna C18 semi-prepartive column (250 × 10 mm). Monitoring at a wavelength of 236 nm, the desired product eluted at 22-26% solvent B. The product-containing fractions were lyophilized to yield 23 mg (16%) of a white powder. 1H-NMR (300 MHz, D2O) δ 0.77 (d, J = 6.6 Hz, 3H), 1.05-1.40 (m, 6H), 1.55 (s, 3H), 1.63-1.80 (m, 1H), 1.90 (t, J = 7.5 Hz, 2H), 3.99 (t, J = 6.9 Hz, 2H), 4.14 (t, J = 6.3 Hz, 2H), 5.31 (t, J = 6.0 Hz, 1H); 31P-NMR (121 MHz, CDCl3) −5.93 (d, J = 22.0 Hz, 1P), −0.91 (d, J = 22.0 Hz, 1P); HR-MS (ESI-TOF) calcd for C13H20F3N2O9P2[M-H]− 467.0596, found 467.0594.
(E)-[α,β(n)32P]-8-O-(2-Diazo-3,3,3-trifluoropropanoyloxy)-3,7-dimethyl-2-octeny-1-diphosphate ([32P]4)
Radiolabeled [32P]4 was synthesized using a procedure similar to that described for unlabeled 4. Briefly, a solution of 1% H3PO4(v/v) (110 μL, 20 μmol) was prepared in a 10 mL round bottom flask and lyophilized overnight as previously described. Compound 17 (3.1 mg, 10 μmol) was dissolved in 50 μL CH3CN and added to the reaction flask containing the anhydrous H3PO4. To this solution, a mixture of CH3CN/CCl3CN 20% (v/v) (200 μL) was added, followed by Et3N (5.6 μL). The reaction was stirred in the dark under an Ar atmosphere for 2 h followed by evaporation under a stream of N2 (g) to yield a yellow residue. The resulting material was then dissolved in 25 mM NH4HCO3 (5 mL) and applied to a reversed-phase Sep-Pak cartridge equilibrated in 25 mM NH4HCO3. The column was eluted with a step gradient of CH3CN (10% steps, 4 × 1.0 mL fractions per step) and the composition of the fractions was determined by TLC (i-PrOH/NH4OH/H2O, 6:3:1 v/v/v) followed by phosphorimaging analysis. After evaporation of the product-containing fractions (30% CH3CN) and redissolving in 25 mM NH4HCO3, [32P]4 (specific activity: 275 Ci/mol) was obtained. The radiochemical purity was determined to be 24%, as assessed by TLC (i-PrOH/NH4OH/H2O, 6:3:1, v/v/v), followed by phosphorimaging analysis; Impurities that were present include the corresponding monophosphate (36%), the triphosphate (11%) and inorganic phosphate (9%). The remaining 20% is likely a dimer formed by condensation of two monophosphates. The concentration of [32P]4 was determined by UV spectroscopy using the extinction coefficient (ε236 = 14,000 cm−1M−1) previously reported for the diazoester fragment.(30)
(E,E)-8-N-(2-Diazo-3,3,3-trifluoropropionamido)-3,7-dimethyl-1-tetrahydropyranyl-2,6-octadiene (8)
The amine 6 was prepared by reacting phthalimide 5 (300 mg, 0.78 mmol) with NH2NH2•H2O (62 mg, 1.2 mmol) in EtOH (10 mL) at rt for 16 h. The reaction was filtered and used directly in the next step of the synthesis without further purification. Amide 8 was prepared by reacting crude 6 (110 mg, 0.43 mmol) with 7 (100 mg, 0.58 mmol) in dry pyridine (0.5 mL) at 0 °C. The reaction was stirred in the dark for 30 min under N2, followed by an additional 2 h at rt. It was then concentrated in vacuo, redissolved in EtOAc, and washed sequentially with 0.10 N HCl, 1.0 M NaHCO3, and aqueous saturated NaCl. The organic layer was dried over MgSO4, filtered, concentrated under reduced pressure, and the crude product was purified by flash chromatography on silica (toluene/EtOAc, 4:1, v/v), giving 8 in 18% yield (30 mg, 77 μmol). Rf = 0.44 (silica gel, toluene/EtOAc, 4:1, v/v); 1H-NMR (500 MHz, CDCl3) δ 1.52-1.81 (m, 6H), 1.62 (s, 3H), 1.67 (s, 3H), 2.05-2.09 (m, 2H), 2.13-2.18 (m, 2H), 3.49-3.53 (m, 1H), 3.85-3.91 (m, 1H), 3.88 (d, 2 H, J = 8.5), 3.98-4.04 (m, 1H), 4.21-4.27 (m, 1H), 4.61-4.63 (m, 1H), 5.28-5.38 (m, 2H); 13C-NMR (75.4 MHz, CDCl3, DEPT) δ 14.38, 16.39 (primary C); 19.62, 25.49, 25.99, 30.71, 39.08, 47.45, 62.31, 63.64 (secondary C); 97.92, 121.03, 126.95 (tertiary C); 122.22, 125.79, 131.20, 139.56, 158.94 (quaternary C); 19F-NMR (282.2 MHz, CDCl3) δ −55.77.
(E,E)-8-N-(2-Diazo-3,3,3-trifluoropropionamido)-3,7-dimethyl-2,6-octadien-1-ol (9)
Alcohol 9 was prepared by reacting protected DATFP ester (8, 50 mg, 0.13 mmol) with TsOH (1.3 mg, 6.9 μmol) in MeOH (13 mL) in the dark at rt for 20 h. The reaction mixture was then concentrated under reduced pressure and purified by flash chromatography on silica (toluene/EtOAc 4:1, v/v) giving the pure product in 67% yield (26 mg, 85 μmol). Rf = 0.15 (silica gel, toluene/EtOAc, 4:1, v/v); 1H-NMR (300 MHz, CDCl3) δ 1.62 (s, 3H), 1.66 (s, 3H), 2.04-2.09 (m, 2H), 2.13-2.18 (m, 2H), 3.87 (d, 2H, J = 5.7), 4.14 (d, 2H, J = 6.9), 5.25-5.30 (m, 1H), 5.36-5.42 (m, 1H), 5.45 (broad s, 1H); 13C-NMR (75.4 MHz, CDCl3, DEPT): δ 14.38, 16.12 (primary C); 25.74, 38.94, 47.41, 59.31 (secondary C); 124.01, 126.50 (tertiary C); 122.22, 125.79, 131.19, 138.78, 159.03 (quaternary C); 19F-NMR (282.2 MHz, CDCl3) δ −55.77; HR-FAB-MS calcd for C13H18N3O2F3Na [M+Na]+ 328.1249, found 328.1259.
2,2′-((2E,6E)-2,6-dimethylocta-2,6-diene-1,8-diyl)diisoindoline-1,3-dione (10)
Compound 5 (150 mg, 0.40 mmol) and PPTS (98 mg, 0.4 mmol) were dissolved in EtOH (4.8 ml) and stirred at 55 °C for 4 h. The solvent was removed in vacuo, redissolved in Et2O (5 mL) and washed with half-saturated brine to remove the catalyst. The ether solution was dried over Na2SO4 and evaporated to yield 83 mg of the desired deprotected alcohol in (70% yield). Rf = 0.30 (silica gel, hexane/EtOAC, 5:2, v/v); 1H NMR (300 MHz, CDCl3): δ 1.60 (s, 3H), 1.63 (s, 3H), 2.04-2.08 (m, 2H), 2.11-2.19 (m, 2H), 4.10 (d, 2H, J = 6.3), 4.17 (s, 2H), 5.24-5.35 (m, 2H), 7.68-7.75 (m, 2H), 7.82-7.87 (m, 2H); HR-ESI-MS calcd for C18H21NaNO3 [M + Na]+ 322.1419, found 322.1397. The above deprotected alcohol (80 mg, 0.27 mmol) was combined with phthalimide (45 mg, 0.27 mmol) and PPh3 (68 mg, 0.27 mmol) in 2.2 mL THF. To the stirred solution was added DEAD (6 μl, 0.27 mmol) in 0.6 mL THF dropwise over 30 min. The reaction mixture was stirred for 18 h at rt followed by a second addition of PPh3 (42 mg, 0.15 mmol) and DEAD (3.0 μL, 0.15 mmol). The reaction was stirred overnight at rt and quenched with hexane (5.0 ml) to precipitate the reaction byproducts. The filtrate was evaporated and the residue purified by flash chromatography (silica gel, hexane/EtOAC, 5:2, v/v) to yield compound 10 (63 mg, 54%) as a white solid. Crystallization of the product was achieved by adding 5.5 mL CH3OH and heating in an oil bath with swirling until the solid was completely dissolved. The flask was removed from the heat and left undisturbed, to cool to rt. Crystals formed over a period of 4 h. Rf = 0.50, silica gel, hexane/EtOAC, 5:2, v/v; 1H NMR (300 MHz, CDCl3): δ 1.59 (s, 6H), 1.98-2.03 (m, 2H), 2.07-2.12 (2H), 4.16 (s, 2H), 4.26 (d, 2H, J = 7.2), 5.24 (t, 1H, J = 6.3), 5.32 (t, 1H, J = 6.3), 7.69-7.33 (m, 4H), 7.83-7.87 (m, 4H); HR-ESI-MS calcd for C26H24NaN2O4 [M + Na]+ 451.1634, found 451.1648.
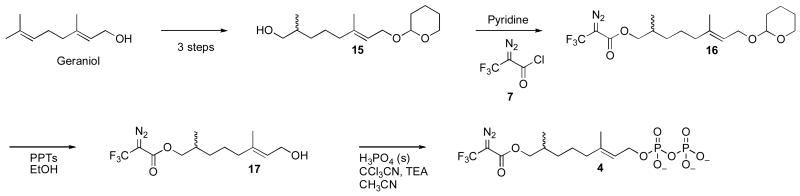
(E)-8-O-(2-Diazo-3,3,3-trifluoropropanoyloxy)-3,7-dimethyl-1-O-tetrahydropyranyl-2-octene (16)
Pyridine (3 mL) was added to alcohol 15 (100 mg, 0.39 mmol) and the reaction flask was sealed with a rubber septum. 2-diazo-3,3,3-trifluoropropanoyl chloride (DATFP-Cl, 7, 85 mg, 0.49 mmol) was then added and the mixture was cooled to 0 °C and allowed to react with stirring, in the dark, for 30 min followed by an additional 2 h at rt. The solvent (pyridine) was removed in vacuo and the resulting residue dissolved in Et2O and washed sequentially with 0.10 M HCl, 1.0 M NaHCO3, and aqueous saturated NaCl. The organic layer was dried over MgSO4, filtered, concentrated under reduced pressure, and the resulting crude product was purified by flash chromatography (hexanes: EtOAc, 6:1) affording compound 16 in 88% yield. 1H-NMR (300 MHz, CDCl3) δ 0.95 (d, 2H, J = 6.6 Hz), 1.12-1.64 (m, 12H), 1.73 (s, 3H), 1.80 (m, 2H), 2.03 (t, 2H, J = 7.5 Hz), 3.52 (m, 1H), 3.91 (m, 1H), 4.05 (m, 2H), 4.16 (dd, 1H, J1 = 5.7 Hz, J2 = 10 Hz), 4.24 (dd, 1H, J1 = 6.6 Hz, J2 = 12 Hz), 4.64 (t, 1H, J = 4.2 Hz), 5.37 (t, 1H, J = 6.9 Hz); 13C-NMR (75 MHz, CDCl3) δ 16.33, 16.73, 19.69, 24.77, 25.55, 30.77, 32.58, 32.76, 39.69, 62.38, 63.71, 70.80, 97.97, 120.88, 121.73, 124.54, 140.04, 161.10; 19F-NMR (282 MHz, CDCl3) δ −58.02 (s, 3F); FT-IR (neat) 1320 (vs), 1352 (vs), 1394 (s), 1732 (vs), 2136 (vs), 2344 (w), 2361 (w), 2869 (s), 2939 (vs) cm−1; HR-MS (ESI-TOF) calcd for C18H27F3N2NaO4 [M+Na]+ 483.2441, found 483.2444.
(E)-8-O-(2-Diazo-3,3,3-trifluoropropanoyloxy)-3,7-dimethyl-2-octen-1-ol (17)
Alcohol 16 (135 mg, 0.34 mmol) was dissolved in 10 mL of absolute EtOH. After adding PPTS (12 mg, 50 μmol), the flask was sealed with a rubber septum and stirred in the dark for 6 h at 60 °C. The product was purified by flash chromatography (hexanes: EtOAc, 5:1, v/v) and obtained in 90% yield. 1H-NMR (300 MHz, CDCl3) δ 0.95 (d, 3H, J = 6.6 Hz), 1.10-1.56 (m, 6H), 1.67 (s, 3H), 1.81-1.84 (m, 1H), 2.01 (t, 2H, J = 7.8 Hz), 4.05 (dd, 1H, J1 = 6.6 Hz, J2 = 11 Hz), 4.15 (m, 3H), 5.41 (t, 1H, J = 6.9 Hz); 13C-NMR (75 MHz, CDCl3) δ 15.83, 16.37, 24.45, 32.26, 32.36, 39.29, 59.04, 70.52, 120.72, 123.43, 124.29, 139.12, 160.97; 19F-NMR (282 MHz) δ −58.01 (s, 3F); FT-IR (neat) 1319 (vs), 1351 (vs), 1396 (vs), 1701 (vs), 2135 (vs), 2857 (s), 2925 (vs), 2962 (vs), 3371 (b) cm−1; HR-MS (ESI-TOF) calcd for C13H19F3N2NaO3 [M+Na]+ 331.1240, found 331.1240.
1-(2-Diazo-3,3,3-trifluoropropionyloxy)-3-methyl-2-butene (18)
Dimethylallyl alcohol (100 mg, 1.16 mmol) was dissolved in freshly distilled pyridine (0.5 mL) and stirred until homogenous. A solution of DATFP-Cl (7, 239 mg, 1.39 mmol) in pyridine (1.0 mL) was slowly added, and the reaction allowed to proceed at 0 °C in the dark for 2.5 h. The pyridine solvent was removed in vacuo and the product was redissolved in Et2O. The organic layer was then washed sequentially with 0.1 M HCl, 1 M NaHCO3, and brine, dried over Na2SO4 and concentrated. Purification was accomplished by flash chromatography (hexanes: EtOAc, 3:1) producing 258 mg (64%) of a pale, yellow oil. 1H-NMR (300 MHz, CDCl3) δ 1.73 (s, 3H), 1.77 (s, 3H), 3.74 (d, 2H, J = 7.2 Hz), 5.35 (tq, 1H, J1 = 1.5 Hz, 8.7 Hz); 13C-NMR (75 MHz, CDCl3) δ 18.04, 25.77, 62.82, 117.76, 121.00, 124.57, 136.14, 160.92; 19F-NMR (282 MHz, CDCl3) δ −58.26 (s, 3F); FT-IR (neat) 1313 (vs), 1355 (vs), 1384 (vs), 1448 (m), 1723 (vs), 2137 (vs), 2341 (w), 2360 (w), 2309 (m), 2977 (m) cm−1.
1-(2-Diazo-3,3,3-trifluoropropionyloxy)-3-methyl-3-butene (19)
Compound 19 was prepared as described for 18 starting from isopentenyl alcohol in 66% yield. 1H-NMR (300 MHz, CDCl3) δ 1.76 (s, 3H), 2.39 (t, 2H, J = 6.9 Hz), 4.37 (t, 2H, J= 6.9 Hz), 4.74 (s, 1H), 4.83 (s, 1H); 13C-NMR (75 MHz, CDCl3) δ 22.33, 36.74, 64.11, 112.87, 120.92, 124.50, 141.01, 160.85; 19F-NMR (282 MHz, CDCl3) δ −58.20 (s, 3F); FT-IR (neat) 1329 (vs), 1344 (vs), 1392 (vs), 1456 (m), 1721 (vs), 2136 (vs), 2256 (m), 2359 (w), 2973 (m), 3081 (m), 3434 (b) cm−1.
Stability studies of 18 and 19
Compound 18 (11 mg, 0.05 mmol) was added to an 8 inch thin-walled NMR tube and dissolved by the addition of 0.7 mL of mildly acidic solution (CD3CN/D2O 10%/TFA-d 0.1%, v/v/v); a separate NMR tube containing 18 (11 mg, 0.05 mmol) dissolved in a mildly basic solution (CD3CN/D2O 10%/ND4OD 0.1%, v/v/v) was also prepared. The samples were then heated directly in the NMR tubes in a 100°C water bath for 4 h followed by 1H-NMR analysis. Experiments with 19 in acidic or basic solutions were prepared and analyzed in a similar manner.
Enzymatic Substrate and Inhibition Studies Monitored by Fluorescence Spectroscopy
The photoactive isoprenoids 3 and 4 were studied as possible substrates for ScPFTase by monitoring the increase in fluorescence of a peptide substrate as it becomes prenylated as previously described.(32, 33) For experiments with the yeast enzyme, the assay conditions used were 50 mM Tris·HCl (pH 7.0), 10 mM MgCl2, 10 μM ZnCl2, 5.0 mM DTT, 0.040% (w/v) n-dodecyl-β-D-maltoside, 2.0 μM N-dansyl-GCVIA, isoprenoid diphosphate (FPP, 3 or 4, 10 μM) and ScPFTase (24-92 nM) in a final volume of 500 μL. For experiments with the mammalian enzyme, similar conditions were employed except that N-dansyl-GCVLS (2.4 μM) was used as a substrate. Enzymatic reactions were initiated by the addition of PFTase and monitored for 300 – 3000 sec (340 nm excitation, 505 emission) to determine the rate. For IC50 determinations, fluorescence assays were performed in triplicate as described above except that reactions contained 2.0 μM FPP along with 3 or 4 at varying concentrations. The IC50 values were determined from a direct plot of rate versus inhibitor concentration.
Enzymatic Substrate Studies Monitored by HPLC
To confirm the enzymatic modification of N-dansyl-GCVIA by 3 and 4, larger scale reactions (10-15 ml) were performed using the conditions reported for the fluorescence assays described above except that the detergent was omitted (detergent is usually included to solubilize the product, not for enzyme activity). In each case, the reaction was initiated with enzyme and incubated at 30 °C for 6-24 h. The reaction mixture was then applied to a Sep Pak cartridge equilibrated in solvent C (5% CH3CN, 95% H2O, 0.1% TFA, v/v/v); product was eluted with solvent B (100% CH3CN, 0.1% TFA, v/v). The fluorescent fractions were concentrated, redissolved in 50% solvent A (100% H2O, 0.1% TFA, v/v) and 50% solvent B, and further purified by reversed-phase HPLC using a Varian Microsorb C18 analytical column (250 × 4.6 mm). Elution was performed using a linear gradient from 100% solvent A to 100% solvent B (solvents described above) over 40 min with a flow rate of 1 mL/min. The product was collected, lyophilized and analyzed by MS. In cases where the reaction kinetics were monitored by HPLC, aliquots of 500 μL were removed at varying time intervals from large scale reactions, flash frozen in N2 (l), and stored at −20°C prior to analysis. The samples were then thawed and analyzed by reversed-phase HPLC as described above.
Photolabeling of prenyltransferases with [32P]DATFP-Amide ([32P]3)
For experiments with ScPFTase, purified enzyme (4 μg) was incubated in 50 mM HEPES (pH 7.5), 1.0 mM DTT, 5.0 mM MgCl2, 10 μM ZnCl2, 15 μM [32P]3 (dissolved in 25 mM NH4HCO3), and 30 μM FPP (where appropriate, dissolved in 25 mM NH4HCO3) or the equivalent volume of 25 mM NH4HCO3 in a final volume of 75 μL. Samples were irradiated at 254 nm at 4 °C for 3 min in quartz test tubes (10 × 45 mm) using a UV Rayonet mini-reactor equipped with 4 RPR-2540 Å lamps and a circulating platform. The samples were heated to 70 °C for 5 min in loading buffer (4% SDS, 12% glycerol (w/v), 50 mM Tris, 2% mercaptoethanol (v/v), 0.01% Serva blue G, pH 6.8) and analyzed by SDS-PAGE with a 10% Tris-tricine gel. The gel was fixed by slow shaking in isopropanol/H2O/acetic acid (25:65:10, v/v/v) for 20 min. It was then exposed to a phosphorimaging screen for 3 hours, followed by silver staining. Experiments with HsPGGTase, were performed as described above except that geranylgeranyl diphosphate (30 μM final concentration) was used instead of FPP to demonstrate substrate protection. Experiments with RnPGGTase II were also performed as described above except that ZnCl2 was omitted from the reaction buffer. Geranylgeranyl diphosphate (30 μM final concentration) was used, instead of FPP, to show substrate protection. I all cases, enzyme concentrations were determined via the method of Bradford using BSA as a standard.(34)
Photolabeling of enzymes with [32P]DATFP-Dihydroester ([32P]4)
Samples of [32P]4 (5 μM) were photolyzed in 52 mM Tris HCl, pH 7, 5.8 mM DTT, 12 mM MgCl2, 12 mM ZnCl2, and either 25 μM FPP dissolved in 25 mM NH4HCO3 or the equivalent volume of 25 mM NH4HCO3 (where appropriate) in a final volume of 100 μL. In experiments employing purified enzymes (ScPFTase, EcFPPSase and NoSTSase), the final concentrations of the proteins were 0.05 mg/mL; for experiments with crude E. coli lysate containing ScPFTase, a final protein concentration of 0.81 mg/mL was employed. Enzyme concentrations were determined via the method of Bradford using BSA as a standard.(34) Samples were irradiated for 5 minutes using the apparatus described above. After photolysis, 50 μL of loading buffer (see above) was added to each sample before heating to 60 °C for 20 minutes. Analysis was accomplished via electrophoresis using a 12% Tris-glycine SDS-PAGE gel and subsequent staining with Sypro Orange followed by phosphorimaging to visualize the radiolabeled bands.
Docking of 4 in the active sites of RnPFTase
For modeling compound 4 in the active site of RnPFTase (pdb file 1JCR)(35), docking was performed using Glide (Schrodinger, version 5.5). A standard precision docking parameter was set and 10,000 ligand poses per docking were run. Five resulting conformations with the lowest docking score for each enantiomer (R-4 and S-4) were chosen. Each one of those was then subjected to QM-MM using QSite (Schrodinger, version 5.5). The three conformations with the overall lowest energy were chosen for display for each enantiomer.
X-Ray crystallography studies of RnPFTase
Crystals of the RnPFTase:4 complex were prepared by soaking 4 into preformed crystals using methods previously described.(36) X-ray diffraction data for the PFTase:4 complex was collected on a Rigaku FRE rotating anode generator equipped with VariMax HR optics and a Raxis-IV++ image plate detector. The detector set at 150 mm, data were collected in 200 contiguous 0.30° oscillation images each exposed for 5 min. The data extend to 2.05 Å resolution and have a Rmerge of 6.1% with a 3.8-fold multiplicity. The structure was refined using autoBUSTER (Global Phasing Limited) to an Rfactor of 17.7% and an Rfree of 21.4%.
Electrostatic potential calculations of 1 and 4
Structures of 1 and 4 were generated in Gaussian View and the geometries were optimized at the b3lyp/6-31G(d) level using Gaussian 03 (M.J. Frisch, G.W. Trucks, H.B. Schlegel et al., Gaussian 03, Revision E.01, Gaussian, Inc., Wallingford, CT, 2004). The extended conformation of each molecule was produced by constraining the distance between remote atoms. A map of electrostatic potential and a map of the electron density were generated for each molecule. To generate the molecular electrostatic potential (MEP), the electrostatic potential was plotted on an isodensity surface (MO = 0.02 and density = 0.02) using Gaussian View. The minimum and maximum potentials were set to −0.05 and 0.2, respectively. Calculations of total polar surface area and LogP were performed using ChemBioDraw Ultra (v. 11.0.1).
RESULTS AND DISCUSSION
Synthesis of DATFP-Amide 3
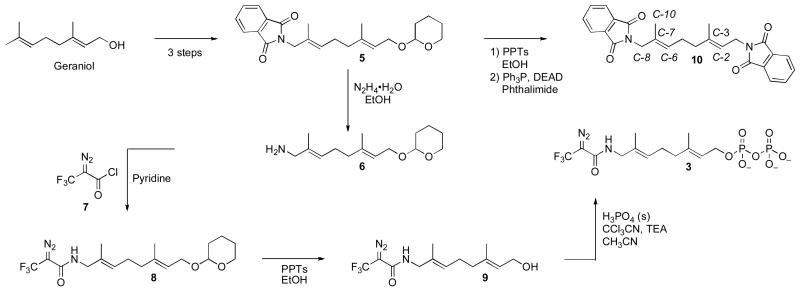
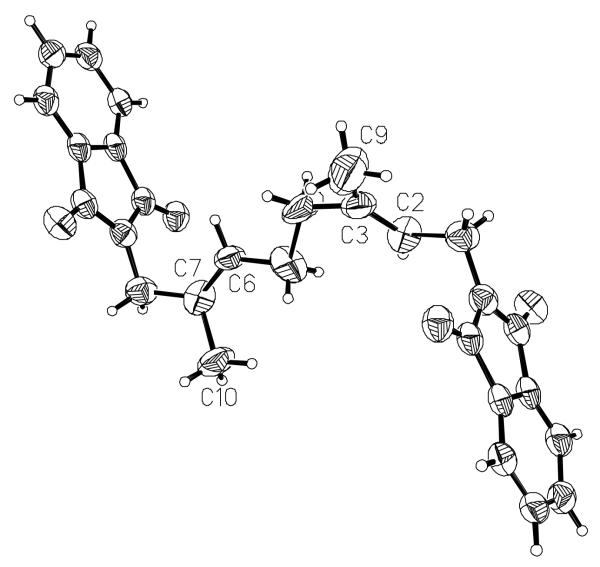
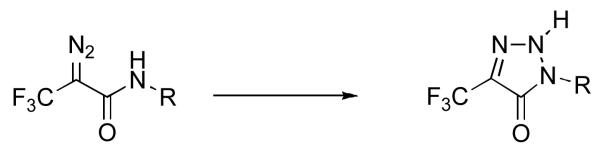
The amide-containing analogue 3 was prepared using the route shown in Scheme 1. Amine 6 was prepared as previously described (19) and acylated with DATFP-Cl (30) (7) in pyridine to produce the protected intermediate 8. That compound was subsequently deprotected and phosphorylated using the method of Cramer (37, 38) to yield a mixture mono and diphosphates. Pure diphosphate 3 was obtained by reversed-phase HPLC and characterized by 1H-NMR, MS and UV/vis spectrophotometry. While the regioselectivity of geraniol hydroxylation with SeO2 has been established in other cases,(39) we elected to confirm this in the THP-protected version by converting intermediate 5 to the corresponding bis- phthalimide derivative 10. That transformation was accomplished by THP removal followed by subsequent Mitsunobu reaction (40, 41) to yield 10 whose structure was then confirmed via x-ray crystallography. The E stereochemistry of the 6,7 alkene is apparent in the structure of this derivative shown in Figure 2. For photolabeling experiments, radiolabeled 3 was prepared via phosphorylation of alcohol 9 with [32P]−H3PO4 as previously described.(42) Solutions of 3 in aqueous NH4HCO3 are stable for several weeks when stored at 4°C. However, storage for longer periods of time results in a change in the UV spectrum from λmax = 238 nm to λmax = 250 nm with no change in the mass spectrum of the compound; these observations are indicative of isomerization of the diazo amide to the corresponding triazolone (see Figure 3).(43, 44) Thus, for maximal crosslinking efficiency compound 3 must be used within one month of its synthesis.
Scheme 1.
Synthesis of amide-linked photoaffinity analog 3.
Figure 2.
Structure of compound 10 determined by X-ray crystallography to establish the E stereochemistry of the C-6 alkene.
Figure 3.
Rearrangement of DATFP-amides to triazolones.
Enzymatic Studies with PFTase using DATFP-Amide 3
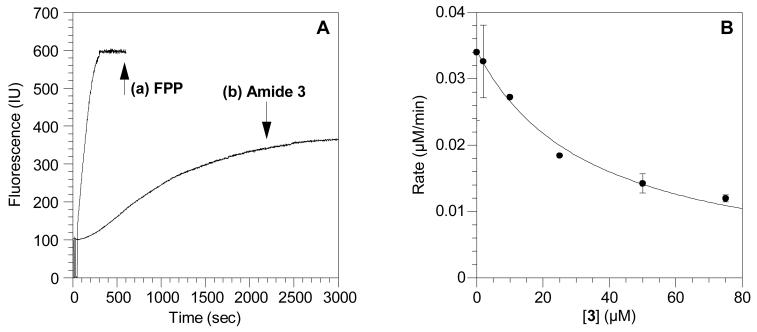
Initially, a continuous fluorescence assay was used to determine whether or not the new photoaffinity analog 3 is a substrate for ScPFTase. Incubation of the substrate peptide N-dansyl-GCVIA with FPP resulted in a rapid increase in dansyl group fluorescence (see Figure 4A, trace a) due to the production of the farnesylated product 11 (see Scheme 2). Replacing the natural substrate, FPP, with 10 μM amide 3 in the assay resulted in a similar time-dependent increase in fluorescence, suggesting that the analog is a substrate for the enzyme (see Figure 4A, trace b); interestingly the fluorescence plateaued at a lower value than that obtained with FPP suggesting that the prenylated product 13 is less fluorescent than 11. However, importantly, the fluorescence increased at a much slower rate with 10 μM 3 than with 10 μM FPP and much longer reaction times (3000 s) were required for significant conversion to product compared with 200 s for complete conversion to the farnesylated peptide product. Also, a significantly higher concentration of enzyme (92 nM) was used, compared with 2.5 nM normally added for assays with FPP as the substrate. Based on those numbers, we estimate that 3 functions some 250-fold times slower than FPP as a substrate for ScPFTase. In order to verify that 3 is a substrate for the enzyme, the product from a large-scale enzyme reaction (15 mL) was purified by reversed-phase HPLC and analyzed by MS. The FAB-MS spectrum showed a [M+H]+ peak of 982.5 and a [M+Na]+ peak at 1004.6, corresponding to the expected peptide product (13).
Figure 4.
Evaluation of amide 3 as an alternative substrate and inhibitor of ScPFTase. (A) Reaction between N-dansyl-GCVIA and FPP or amide 3 catalyzed by ScPFTase monitored by fluorescence spectroscopy. (a) Kinetic data using FPP as a substrate. (b) Kinetic data using amide 3 as a substrate. Fluorescence was monitored at 30°C by excitation at 340 nm and emission at 505 nm. (B) .Inhibition of ScPFTase-catalyzed farnesylation of N-dansyl-GCVIA by amide 3. Reactions contained 2.0 μM FPP, 2.0 μM N-dansyl-GCVIA and 3 at varying concentrations and were monitored using a continuous spectrofluorometric assay. Each point is the average of 2-3 determinations with the error bars indicating the standard error for each measurement.
Scheme 2.
Structures of ScPFTase modified N-dansyl-GCVIA peptide products. 11: peptide product from reaction with FPP (1); 12: peptide product from reaction with allylic ester 2; 13: peptide product from reaction with amide 3; 14: peptide product from reaction with alkyl ester 4.
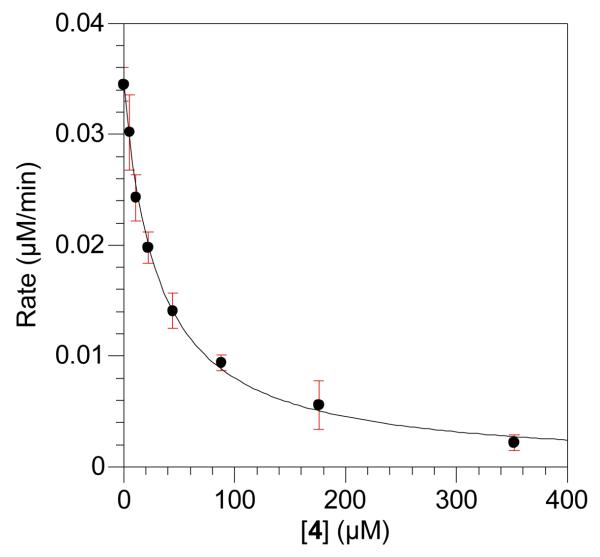
Since DATFP-amide 3 appeared to be a significantly slower substrate than FPP for ScPFTase, the analogue was evaluated as an inhibitor of the reaction between FPP and the peptide substrate N-dansyl-GCVIA. An IC50 value of 50 μM (see Figure 4B) was obtained for the inhibition of ScPFTase by 3 which is significantly higher than the value of 0.72 μM measured for the corresponding ester-linked compound 2.(5) This higher value suggests lower affinity for the amide-containing analogue and may reflect the more rigid nature of the amide- versus ester-linked analogue.
Photolysis Experiments with DATFP-Amide 3
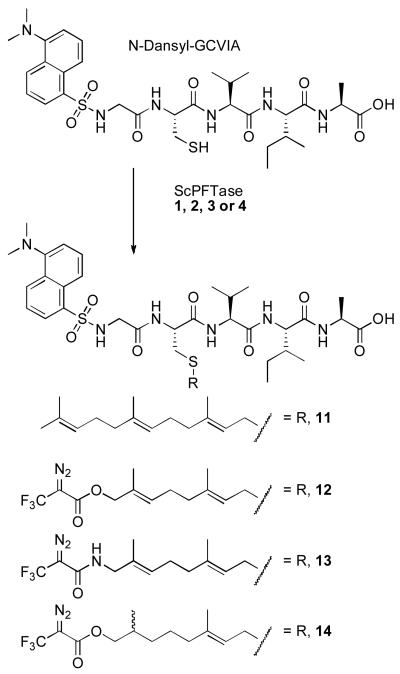
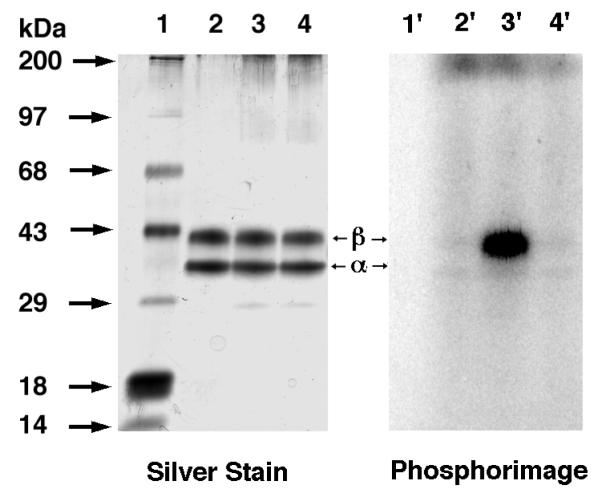
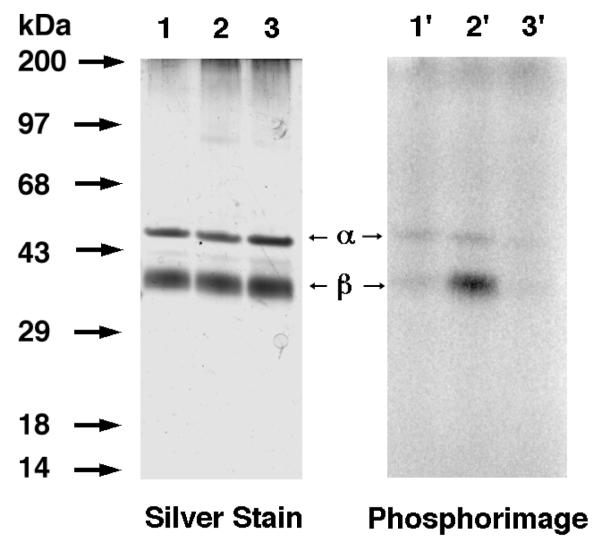
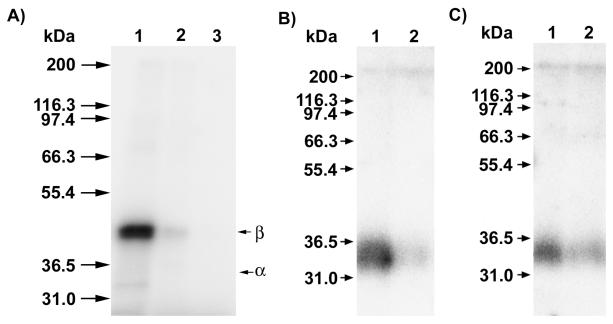
Amide 3 was first evaluated for its ability to label ScPFTase. UV irradiation of ScPFTase in the presence of 15 μM [32P]3 at 254 nm for 1 min resulted in preferential labeling of the 43 kDa β subunit (Figure 5, Lane 3′); inclusion of 30 μM FPP in the reaction mixture gave substantial protection from labeling (Lane 4′, 3% of the crosslinking observed in Lane 3′). This selective labeling of the β-subunit is consistent with previous results with DATFP-esters and suggests that the isoprenoid moiety interacts predominantly with the β-subunit when it binds to the enzymatically active α,β-heterodimer. Similar results were obtained with Homo sapiens protein geranylgeranyltransferase type I (HsPGGTase I) where UV irradiation of purified enzyme in the presence of 15 μM [32P]DATFP-GPP amide at 254 nm for 1 min resulted in preferential labeling of the 43 kDa β subunit (Figure 6, Lane 2′); inclusion of 30 μM GGPP in the reaction mixture resulted in substantial protection from labeling (Lane 3′, 5% of the crosslinking observed in Lane 2′). It is interesting to note that a small amount of labeling of the α-subunit was also observed with this enzyme. This may reflect an alternative binding mode for the probe; crystallographic studies suggest that a second isoprenoid binding site exists in PGGTase that is involved in interactions with the prenylated product.(45) Finally, we explored the photolysis of Rattus norvegicus protein geranylgeranyltransferase type II (RnPGGTase II) in the presence of 3 since there have been no reported studies of photoaffinity labeling of the type II enzyme with DATFP-based probes. UV irradiation of RnPGGTase II in the presence of 10 μM [32P]3 resulted in preferential labeling of the 38 kDa β subunit (Figure 7, Lane 2′). As was observed in the cases noted above, including 30 μM GGPP in the reaction mixtures gave substantial protection from labeling (Lane 3′, 9% of the crosslinking observed in Lane 2′). These results give direct evidence for the involvement of the β subunit of RnGGPTase II in isoprenoid substrate binding which is consistent with crystallographic data on this system.(46, 47) Also, since these experiments were done in the absence of the escort protein, REP-1, the results indicate that this protein is not necessary for isoprenoid substrate binding.
Figure 5.
Analysis of photolabeling of ScPFTase with DATFP-amide ([32P]3) by SDS-PAGE. Lanes 1 and 1′: molecular weight standards. Lanes 2 and 2′: sample containing PFTase and [32P]3, no UV irradiation. Lanes 3 and 3′: PFTase irradiated in the presence of [32P]3. Lanes 4 and 4′: PFTase irradiated in the presence of [32P]3 and FPP (substrate). Lanes 1, 2, 3, and 4 show the silver-stained proteins. Lanes 1′, 2′, 3′, and 4′ show the radiolabeled proteins.
Figure 6.
Analysis of photolabeling of HsPGGTase I with DATFP-amide ([32P]3) by SDS-PAGE. Lanes 1 and 1′ contain samples of PGGTase I and [32P]3 which were not irradiated. Lanes 2 and 2′ contain samples of HsPGGTase I irradiated at 254 nm in the presence of [32P]3. Lanes 3 and 3′ contain samples of PGGTase I irradiated in the presence of [[32P]3 and FPP (substrate). Lanes 1, 2, and 3 show the silver-stained proteins. Lanes 1′, 2′, and 3′ show the radiolabeled proteins.
Figure 7.
Analysis of photolabeling of RnPGGTase II with DATFP-amide ([32P]3) by SDS-PAGE. Lanes 1 and 1′: PGGTase II and [32P]3, no UV irradiation. Lanes 2 and 2′: RnGGPTase II and [32P]3, 1 min UV irradiation. Lanes 3 and 3′: RnPGGTase II, [32P]3, and GGPP, 1 min UV irradiation. Lanes 1, 2, and 3 show the silver-stained proteins. Lanes 1′, 2′, and 3′ show the radiolabeled proteins.
Synthesis of Dihydroester 4
While the amide linkage present in 3 did contribute to its enhanced stability, the rearrangement of the diazoamide to the triazolone shown in Figure 3 limited the utility of this compound. Thus, we undertook the synthesis of dihydroester-linked analogue 4 which was prepared using the route shown in Scheme 3. THP protection of geraniol followed by allylic oxidation and selective reduction of the α,β-unsaturated aldehyde afforded 15 as previously described.(26) That alcohol was acylated with DATFP-Cl (7), deprotected and phosphorylated using the method of Cramer.(38) For photolabeling experiments, radiolabeled 4 was prepared via phosphorylation of alcohol 17 with [32P]H3PO4 using a procedure previously described for related analogues.(31) It should be noted that 4 contains a stereogeneic center (C-7) and is hence chiral. No attempt was made to resolve the enantiomers and thus all experiments were performed with racemic material.
Scheme 3.
Synthesis of dihydroester-linked photoaffinity analog 4.
Stability studies of alkyl and allylic DATFP Esters
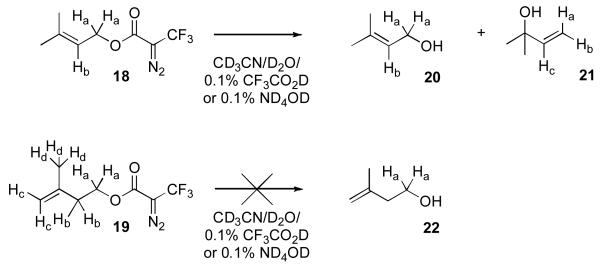
To investigate the stability of the dihydro isoprene unit in 4 compared with its allylic counterpart present in 2, five carbon DATFP-containing model compounds were synthesized. Compound 18 (Scheme 4) contains a DATFP moiety linked to an allylic alcohol to model compound 2 while 19 lacks the allylic alcohol and is hence designed to mimic 4. Model compounds 18 and 19 were synthesized by reaction of DATFP-Cl (7) with dimethylallyl alcohol or isopentenyl alcohol, respectively.
Scheme 4.
DATFP-containing allylic- and alkyl-ester model compounds and their putitive hydrolysis products.
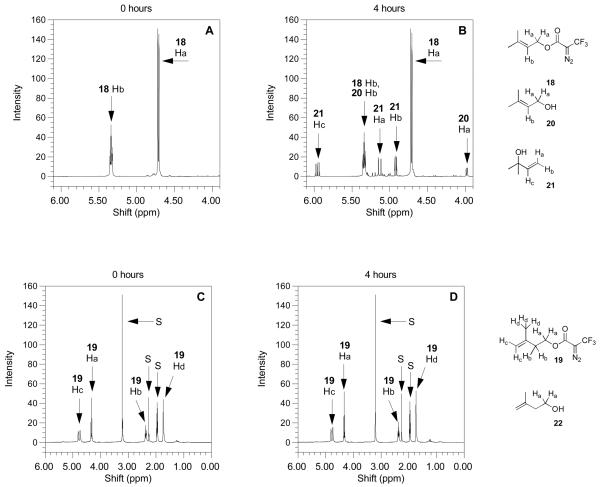
As a test of stability, compounds 18 and 19 were treated with a solvent mixture consisting of CD3CN/D2O (9/1, v/v) containing 0.1% TFA-d. That mixture was used here since it is representative of what is typically employed for peptide extraction from gels and subsequent chromatographic fractionation. Deuterated solvents were used to allow direct analysis of the reaction mixtures to be performed by 1H-NMR. Portions of the spectral data from 18 and 19 prior to reaction are shown Figure 8 (panels A and C, respectively). Treatment of allylic ester 18 in the solvent mixture described above at 100°C for 4 h resulted in substantial hydrolysis to produce primarily tertiary alcohol 21 together with traces of several other products including allylic alcohol 20 as evidenced from the 1H NMR spectrum (Figure 8B) of the reaction mixture; in addition to unreacted starting material, the observation of three doublet of doublets at 4.92, 5.13 and 5.96 ppm confirms the presence of 21. In contrast, incubation of the homoallylic ester 19 under identical conditions resulted in no changes in the 1H NMR spectrum (Figure 8D) of the corresponding reaction mixture (compare Figure 8C with Figure 8D); no evidence for the formation of 22 was observed. These results indicate that 19 is substantially more stable than 18 under these mildly acidic conditions and suggest that crosslinked products derived from reactions with probe 4 (homoallylic ester) should be significantly more resistant to hydrolytic degradation than products obtained with allylic ester 2.
Figure 8.
1H-NMR spectra of DATFP-containing model esters obtained under mild acidolytic conditions. Spectra on the left are the compounds before heating and spectra on the right are the compounds after heating at 100°C for 4 h. (A) Compound 18 treated under acidic conditions (CD3CN/D20 10%/TFA-d 0.1%) before heating. (B) Compound 18 treated under acidic conditions (CD3CN/D20 10%/TFA-d 0.1%) after heating to 100°C for 4 h. The appearance of three new doublets of doublets after heating indicates the presence of 21. (C) Compound 19 treated under acidic conditions (CD3CN/D20 10%/TFA-d 0.1%) before heating. (D) Compound 18 treated under acidic conditions (CD3CN/D20 10%/TFA-d 0.1%) after heating to 100°C for 4 h. Compound 19 shows no visible change in the 1H-NMR spectrum after 4 h. In panels (C) and (D), peaks labeled “S” are from D2O, p-xylene (internal standard for integration) and CD3CN.
Enzymatic Studies with PFTase using Dihydroester 4
In order to evaluate dihydroester 4 as a substrate, we initially utilized the continuous fluorescence assay employed to study amide 3. Those experiments gave similar results as were observed with 3, suggesting that compound 4 was a slow alternative substrate for ScPFTase. However, unambiguous interpretation of the data was made difficult due to slow, time-dependent perturbations of the fluorescence of the putative prenylated peptide product (14, see Scheme 2). It has been previously noted that the fluorescence assay is sensitive to small changes in the reaction conditions (32, 33) and that some isoprenoid analogues that are alternative substrates modulate the fluorescence of the starting peptide and product.(48, 49) Hence we elected to monitor the enzymatic reactions by a method that is less prone to such artifacts. Accordingly, the incorporation of 4 into a peptide substrate was monitored by reversed-phase HPLC using fluorescence detection. When the natural substrate FPP (10 μM) was incubated with N-dansyl-GCVIA (2.4 μM) and ScPFTase (24 nM), near complete conversion (93%) to the prenylated peptide product (11) was observed within 5 min (Figure 9A). The reaction of 4 (10 μM) with N-dansyl-GCVIA (2.4 μM) and ScPFTase (24 nM) to yield 14, however, was still not complete after 23 h (Figure 9B). Based on peak integrations, prenylation of the dansylated peptide with the unnatural substrate 4 was 77% complete after 23 hr, making it approximately 300 times slower than the same reaction with FPP. The lower rate observed with 4 is comparable to that observed with 2 and 3 (see above). However, it should be noted that if extensive conversion is desired, the lower efficiency of incorporation manifested by these compounds can generally be compensated for by the employment of higher enzyme concentrations and longer reaction times. Moreover, methods have been developed for the purification of both prenylated peptides (conventional reversed-phase HPLC) and proteins (cyclodextrin affinity chromatography).(50) Thus, this lower efficiency of incorporation does not represent a serious limitation for applications with these analogues even if the goal is to produce proteins or peptides that incorporate these photoactive isoprenoids.
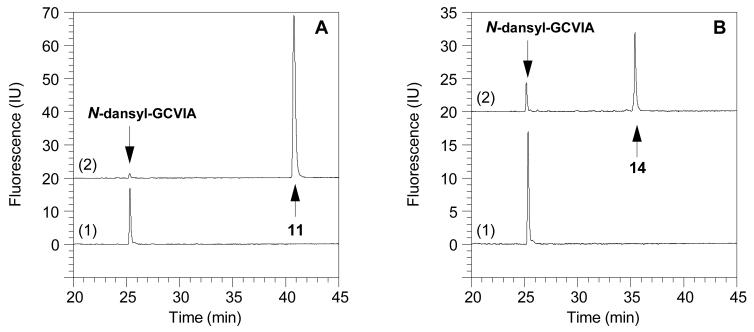
Figure 9.
Reversed-phase HPLC analysis with fluorescence detection of reactions between N-dansyl-GCVIA and FPP (1) or dihydroester 4 catalyzed by ScPFTase. (A) N-dansyl-GCVIA (2.4 μM) incubated with FPP (1, 10 μM) and ScPFTase (24 nM). Chromatogram 1 (bottom): Reaction before the addition of the enzyme with the peptide eluting at tR=25.7 min. Chromatogram 2 (top): Reaction after 5 min with the prenylated peptide (11) appearing at tR=41.5 min. (B) N-dansyl-GCVIA (2.4 μM) incubated with dihydroester (4, 10 μM) and ScPFTase (24 nM). Chromatogram 1 (bottom): Reaction before the addition of the enzyme with the peptide substrate at tR=25.7 min. Chromatogram 2 (top): Reaction after 23 h with the prenylated peptide (14) appearing at tR=36.0 min and some starting material still present.
Since dihydroester 4 appeared to be a significantly slower substrate than FPP for ScPFTase, the analogue was evaluated as an inhibitor of the reaction between FPP and the peptide substrate N-dansyl-GCVIA. An IC50 value of 30 μM (see Figure 10) was obtained for the inhibition of ScPFTase by 4. That value is 1.7-fold lower than that for 3 but still significantly higher than that observed for 2 (0.7 μM);(5) these data show how sensitive the enzyme is to even small perturbations in the isoprenoid structure. This result is perhaps not surprising since both the first and second isoprenoid units from FPP undergo conformational changes as the enzyme proceeds from the ternary complex to the product complex.(51)
Figure 10.
Inhibition of ScPFTase-catalyzed farnesylation of N-dansyl-GCVIA by dihydroester 4. Reactions contained 2.0 μM FPP, 2.0 μM N-dansyl-GCVIA and 4 at varying concentrations and were monitored using a continuous spectrofluorometric assay. Each point is the average of 3 determinations with the error bars indicating the standard error for each measurement.
Photolysis Experiments with Dihydroester 4
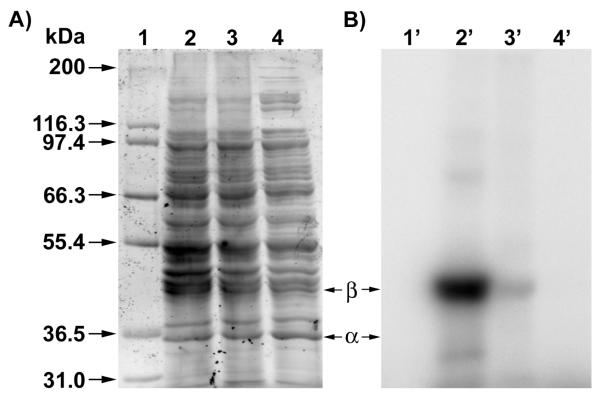
To compare the properties of dihydroester 4 and amide 3, compound 4 was evaluated for its ability to label ScPFTase. UV irradiation of ScPFTase in the presence of 15 μM [32P]4 at 254 nm for 1 min resulted in preferential labeling of the 43 kDa β subunit (see Figure 11A, Lane 1). Inclusion of 30 μM FPP in the reaction mixture gave substantial protection from labeling, reducing the amount of crosslinking to 20% of that obtained in the absence of FPP (Lane 2); no labeling was observed in unphotolyzed reaction mixtures (Lane 3). This selective labeling of the β-subunit is similar to what is observed in experiments with 2 and 3. We next wanted to explore the ability of 4 to label other FPP-utilizing enzymes to gain insight into the scope of this probe. Thus, E. coli farnesyldiphosphate synthase (EcFPPSase) was irradiated in the presence of [32P]4 and the reaction analyzed in a manner analogous to that employed above with ScPFTase. The results of this experiment are shown in Figure 11B; Lane 1 shows significant labeling of the protein while Lane 2 shows a decrease in labeling (29% of the labeling observed in Lane 1) when the photolysis was performed in the presence of both [32P]4 and FPP as a competitor. Similar results were obtained in labeling experiments with a germacrene-producing, sesquiterpene synthase from a cyanobacterial source, Nostoc sp. strain PCC7120 (NoSTSase). Lane 1 (Figure 11C) illustrates the labeling of the enzyme by [32P]4 while Lane 2 shows the results when the photolysis was performed in the presence of both [32P]4 and FPP; a significant (although not as great as for the other enzymes studied) decrease in labeling (58% of the labeling observed in Lane 1) was again observed when the reaction was carried out in the presence of the competitor (FPP). Finally, to evaluate the specificity of [32P]4, photolabeling was performed on a complex mixture of proteins containing ScPFTase present at low concentration. We routinely purify ScPFTase by expressing the genes in E. coli; based on activity assays, we estimate that ScPFTase is present in crude soluble extract as 1% of the total protein. Hence, we considered this protein mixture to be a useful benchmark for examining the specificity of these probes. Accordingly, crude E. coli extract containing ScPFTase was photolyzed in the presence of 5.0 μM [32P]4; results of those experiments are shown in Figure 12. Lane 2 shows the large number of different proteins present in E. coli crude extract while lane 2′ reveals only one major photocrosslinked product corresponding to the ScPFTase β-subunit and a few additional faint bands. As was noted above with photolysis reactions employing purified ScPFTase, inclusion of FPP in the photolysis mixture results in almost complete elimination of photolabeling (Lane 3′). Thus it appears that [32P]4 can selectively label ScPFTase selectively even in the presence of a large number of other proteins.
Figure 11.
Analysis of photolabeling of purified FPP-utilizing enzymes with dihydroester [32P]4. For all samples, photolysis reactions were fractionated by SDS PAGE followed by phosphorimaging analysis to allow visualization of the radiolabeled proteins. In each case, Lane 1 contains enzyme irradiated in the presence of [32P]4 while Lane 2 contains enzyme irradiated in the presence of [32P]4 and FPP (substrate). Where shown, Lane 3 contains [32P]4 incubated in the presence of protein without irradiation. Panel A: Analysis of photolabeling of purified PFTase. Panel B: Analysis of photolabeling of purified EcFPPSase. Panel C: Analysis of photolabeling of purified NoSTSase.
Figure 12.
Selective photolabeling of ScyPFTase in crude E. coli extract using dihydroester 4. Analysis of photolabeling of ScPFTase with [32P]4 by SDS-PAGE. Lanes 1 and 1′: Molecular weight standards. Lanes 2 and 2′: Crude ScPFTase irradiated in the presence of [32P]4. Lanes 3 and 3′: Crude ScPFTase irradiated in the presence of [32P]4 and FPP (substrate). Lanes 4 and 4′: Sample containing crude yPFTase and [32P]4, no UV irradiation. Panel A: Sypro Orange-stained proteins. Panel B: Radiolabeled proteins visualized via phosphorimaging analysis.
Docking Analysis of Dihydroester 4 with RnPFTase
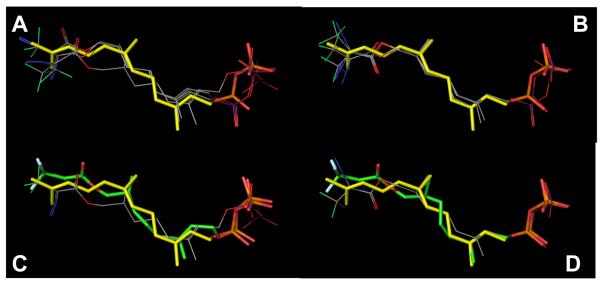
As noted above, excellent selectivity of labeling was obtained with dihydroester 4. However, it is clear that 4 binds to ScPFTase with significantly less affinity than does FPP; a rough estimation of this difference can be obtained from a comparison of the IC50 of 4 reported here for ScPFTase (IC50 = 30 μM) versus the KD of FPP for PFTase (KD = 75 nM).(52) To gain insight into why 4 might bind to ScPFTase with attenuated affinity, docking experiments were performed in which the two enantiomers of dihydroester 4 were docked into the structure of RnPFTase. For each enantiomer, an ensemble of poses was obtained; inspection of those structures indicated that the diazoester moiety was twisted significantly from a planar conformation. Consequently, the top five poses for each enantiomer were further optimized by QM-MM calculations. Interestingly, the rank order (based on energy) of the structures obtained following QM-MM energy minimization was different than the order obtained directly from the docking (based on docking scores). In all subsequent discussion, the term “docked” poses refers to docked poses following QM-MM energy minimization. In general, there was greater variation in the docked R-poses (Figure 13A) compared to the docked S-poses (Figure 13B) with the greatest variation occurring in the position of the DATFP group. The best structures for each of the two enantiomers preserved the 180° dihedral angle between the DATFP-carbonyl and azide groups. Significant deviations in the positioning of the diphosphate groups were observed in the docked R-poses compared to the docked S-poses. Collectively, these docking experiments predict that dihydroester 4binds to RnPFTase in the FPP binding site in a fashion similar to FPP although the significant variations observed in the docked structures make a specific conformation difficult to assign.
Figure 13.
Results of docking experiments with dihydroester 4 and RnPFTase. Top Left (A): Top 3 poses for the R enantiomer of 4 (line representations) docked into RnPFTase. The position of FPP (stick representation) bound to RnPFTase determined via crystallography (pdb code: 1JCR) is shown for comparison. Top Right (B): Top 3 poses for the S enantiomer of 4 docked into RnPFTase. Bottom Left (C): Comparison of highest-scoring docked pose for the R enantiomer of 4 (line representation) with the structure of the protein-bound R enantiomer determined via crystallography (stick representation, green carbons) and FPP (stick representation, yellow carbons). Bottom Right (D): Comparison of highest-scoring docked pose for the S enantiomer of 4 (line representation) with the structure of the protein-bound S enantiomer determined via crystallography (stick representation) and FPP (stick representation, yellow carbons). Colors: C (line representations: grey, stick representations: yellow [FPP], green [crystallographic 4), F (line representations: green, stick representations: white), N (blue), O (red), P (line representations: purple, stick representations: orange). As noted in the text, the term “docked” pose used here refers to the docked poses that have been subjected to energy minimization.
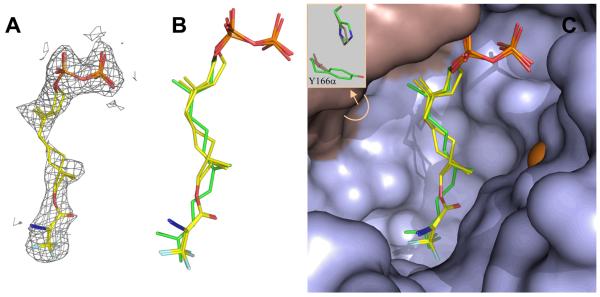
Crystallographic Analysis of Dihydroester 4 bound to RnPFTase
Since the docking experiments described above suggested that compound 4 would bind in the active site of PFTase in a similar manner as FPP, we sought to confirm this by obtaining a structure of the analogue bound to RnPFTase; hence, 4 was soaked into crystals of RnPFTase and the structure determined by X-ray crystallography. Comparison of the protein in the structure of the RnPFTase:4 complex and the RnPFTase:FPP:SCH66336 ternary complex shows a 0.2 Å r.m.s. deviation for 667 of the 716 C-alpha protein atoms, indicating that no significant domain movements have occurred.(36, 53) The most significant side chain movement close to 4 is Tyr166α, which rotates 75° degrees about chi1 resulting in a 5.5Å movement of the tyrosine hydroxyl (Figure 14C). The electron density resulting from the presence of 4 is shown in Figure 14A; since it was not possible to unambiguously determine whether one or both enantiomers were bound, models for both were fit into the density. A superposition of these models with the previously determined structure for bound FPP is presented in Figure 14B along with a view of the enzyme active site showing both the analogue, FPP and the protein surface (Right). In general, the analogue (4) mimics FPP well. The diphosphate group and first isoprenoid units of 4 closely mirror the corresponding positions in FPP although C-1 in the analogue is displaced 0.2 Å for the S-enantiomer and 0.9 Å for the R-enantiomer. Additionally, the DATFP group of 4 is positioned similarly to the third isoprenoid unit of FPP although it is also displaced approximately 1.0 Å to one side. The most significant difference lies in the position of the second isoprenoid unit within the analogue. Introduction of the sp3 hybridized centers at C-6 and C-7 along with the ester linkage present in the analogue result in a significant rotation of the second isoprenoid unit; however, one note of caution that should be acknowledged concerning the structure of the middle isoprenoid element is that the electron density in that region is not well defined (see Figure 14A) making definitive structural assignment difficult. It is interesting to compare the conformation of 4 obtained from crystallography with the best poses obtained from docking. A comparison for the R-enantiomer of 4 is shown in Figure 13C and for the S-enantiomer in Figure 13D. The best docked pose of S-4 is closer to the conformation of crystallographically determined FPP than to crystallographically determined S-4; in contrast, the best docked pose of R-4 differs significantly from the crystallographically determined conformations of both FPP and R-4. These results suggest that while the docking experiments do correctly predict the overall orientation of the bound analogue, they are less accurate in predicting the precise conformation of the bound compound. This may reflect that fact that no specific hydrogen bonds or polar interactions occur between the protein and the ligand (except for the diphosphate moiety). Hence the energetics are dominated by nondirectional Van der Waals interactions.
Figure 14.
Structure of dihydroester 4 bound to RnPFTase determined by X-ray crystallography. (A) Electron density for dihydroester 4 (mesh) contoured at 1.0 σ. The structures of the two enantiomers of 4 (shown in stick representations) are fit within the electron density. (B) Structures of the two enantiomers of 4 superimposed with the structure of FPP (green) bound to RnPFTase. (C) Active site of RnPFTase showing structures of the two enantiomers of 4 superimposed with the structure of FPP (green). The insert shows the difference in conformation for Y166α and H210α between the structures of 4 and FPP. Colors: C (yellow), N (blue), O (red), α-subunit (grey), β-subunit (light blue), Zn (orange).
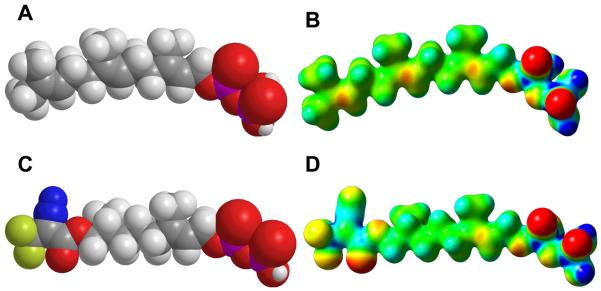
Comparison of Electrostatic Potential Calculations of FPP and Dihydroester 4
While the docking calculations described above in conjunction with the crystallographic experiments demonstrate that compound 4 can be accommodated into the active site of PFTase, they do not explain why the affinity of 4 for the enzyme is so much weaker than that of FPP. To gain insight into that question, DFT calculations were employed to calculate the molecular electrostatic potential maps for FPP and 4 presented in Figure 15 (Panels B and D); standard CPK models are also provided in Figure 15 (Panels A and C) and serve to illustrate that the overall size of the DATFP photophore (compare Figure 15A and 15C) is similar to that of the third isoprene unit in FPP. Clearly, based on geometric considerations alone, 4 and FPP are quite similar. However, inspection of the molecular electrostatic potential maps shows how much these molecules differ (compare Figures 15B and 15D). Note the regions of significant negative potential (red and yellow) in the surface of 4 due to the carbonyl oxygen and fluorine atoms. Similarly, significant positive potential is observed due to the presence of the diazo group and the highly polarized CF3-carbon. Overall, 4 exhibits a substantial increase in total polar surface area of 63 Å2. Not surprisingly, this increase in polarity is accompanied by a decrease in the calculated LogP from 5.0 for FPP to 3.9 for 4. Thus, these calculations suggest that while FPP and 4 are similar in size, the greater polarity of 4 renders its binding within the hydrophobic isoprenoid binding site in PFTase less favorable compared with FPP. However, it should be noted that these calculations, which examine variations in electrostatic potential, may not provide a complete explanation for the behavior of 4. More subtle factors including the greater flexibility of 4 and the concomitant effect this might have on the entropy of binding may also be operating.
Figure 15.
CPK models and molecular electrostatic potential (MEP) maps calculated for FPP and dihydroester 4. Above: CPK model (A) and MEP (B) for FPP. Below: CPK model (C) and MEP (D) for 4. For MEP maps, potentials were plotted on a surface of constant density (density = 0.02). Electronegative areas are shown in red and electropositive areas are shown in blue.
CONCLUSIONS AND FUTURE DIRECTIONS
In an effort to create photoactive analogues of FPP with enhanced stability, two new compounds have been prepared. Probe 3 contains an amide-linked DATFP moiety, is an alternative substrate for ScPFTase and covalently modifies a number of prenyltransferases upon photolysis. However, 3 rearranges to a photochemically inactive triazolone limiting its potential utility. Probe 4 employs an alkyl ester in lieu of the allylic ester found in 2; that substitution renders the linkage between the DATFP group and the isoprenoid significantly more stable to acidic and basic conditions. Compound 4 is an alternative substrate for ScPFTase. Computer assisted docking and crystallographic analysis indicate that 4 binds to RnPFTase in a manner similar (although not identical) to the natural substrate FPP; those suggest that 4 is a reasonable surrogate for a farnesyl group. Photolysis of 4 in the presence of a several different types of FPP-utilizing enzymes results in crosslinking; of particular note, photolysis of 4 in crude E. coli extract expressing ScPFTase results in selective labeling of that enzyme even though it is present as approximately 1% of the total protein. We are currently employing 4 in experiments designed to identify proteins involved in latex and sesquiterpene biosynthesis. Finally, the ability to introduce 4 into peptides, established here, suggests that it should be possible to prepare peptides and proteins that incorporate photoactive isoprenoids that can be used to study the processing of prenylated proteins.(12, 18, 54-56) Given our recent development of cell penetrating prenylated peptides, it should be possible to perform these experiments in living cells.(57) Efforts to accomplish this are currently under way.
Figure 1.
Farnesyl diphosphate and related DATFP-containing photoprobes.
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health Grants GM58442 (M.D.D.) and GM080299 (C.S.-D.) and by a Fellowship from the Beckman Scholars Program (M. L. H.). This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute, instrumentation from the University of Minnesota Center for Mass Spectrometry and Proteomics, and facilities of the Masonic Cancer Center.
Abbreviations
- BSA
bovine serum albumin
- DATFP
diazotrifluoropropanoyl
- DEAD
diethyl azodicarboxylate
- DTT
dithiothreitol
- EcFPPSase
E. coli farnesyl diphosphate synthase
- EDTA
ethylenediaminetetraacetic acid
- ESI-MS
electrospray ionization mass spectrometry
- FAB-MS
fast atom bombardment mass spectrometry
- FPP
farnesyl diphosphate
- GGPP
geranylgeranyl diphosphate
- HPLC
high performance liquid chromatography
- HsPGGTase I
H. sapiens protein geranylgeranyltransferase type 1
- NMR
nuclear magnetic resonance
- NoSTSase
Nostoc sp. strain PCC7120 sesquiterpene synthase PFTase, protein farnesyl transferase
- PPTS
pyridinium p-toluenesulfonate
- QM-MM
quantum mechanics molecular mechanics
- RnPGGTase II
R. norvegicus protein geranylgeranyltransferase type 2
- ScPFTase
S. cerevisiae protein farnesyltransferase
- SDS PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
REFERENCES
- 1.Cane DE. Sesquiterpene biosynthesis: Cyclization mechanisms. Comp Nat Prod Chem. 1999;2:155–200. [Google Scholar]
- 2.Davis EM, Croteau R. Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Topics Curr Chem. 2000;209:53–95. [Google Scholar]
- 3.Loomis WD, Croteau R. Biochemistry of terpenoids. Biochem Plants. 1980;4:363–418. [Google Scholar]
- 4.Zhang FL, Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Ann Rev Biochem. 1996;65:241–69. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 5.Edelstein RL, Distefano MD. Photoaffinity labeling of yeast farnesyl protein transferase and enzymic synthesis of a Ras protein incorporating a photoactive isoprenoid. Biochem Biophys Res Comm. 1997;235:377–82. doi: 10.1006/bbrc.1997.6792. [DOI] [PubMed] [Google Scholar]
- 6.Allen CM, Baba T. Photolabile Analogs of the Allylic Pyrophosphate Substrate of Prenyltransferases. Meth Enzymol. 1985;110:117–24. doi: 10.1016/s0076-6879(85)10066-2. [DOI] [PubMed] [Google Scholar]
- 7.Baba T, Allen CM. Inactivation of Undecaprenylpyrophosphate Synthetase with a Photolabile Analogue of Farnesyl Pyrophosphate. Biochemistry. 1984;23:1312–22. [Google Scholar]
- 8.Baba T, Muth J, Allen CM. Photoaffinity Labeling of Undecaprenyl Pyrophosphate Synthetase with a Farnesyl Pyrophosphate Analogue. J Biol Chem. 1985;260:10467–73. [PubMed] [Google Scholar]
- 9.Omer CA, Kral AM, Diehl RE, Prendergast GC, Powers S, Allen CM, et al. Characterization of Recombinant Human Farnesyl-Protein Transferase: Cloning, Expression, Farnesyl Diphosphate Binding, and Functional Homology with Yeast Prenyl-Protein Transferases. Biochemistry. 1993;32:5167–76. doi: 10.1021/bi00070a028. [DOI] [PubMed] [Google Scholar]
- 10.Das NP, Allen CM. Inhibition of Farnesyl Transferases From Malignant and Non-Malignant Cultured Human Lyphocytes By Prenyl Substrate Analogues. Biochem Biophys Res Comm. 1991;181:729–35. doi: 10.1016/0006-291x(91)91251-7. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama K, McGeady P, Gelb MH. Mammalian Protein Geranylgeranyltransferase-I: Substrate Specificity, Kinetic Mechanism, Metal Requirements, and Affinity Labeling. Biochemistry. 1995;34:1344–54. doi: 10.1021/bi00004a029. [DOI] [PubMed] [Google Scholar]
- 12.Kale TA, Distefano MD. Diazotrifluoropropionamido-Containing Prenylcysteines: Syntheses and Applications for Studying Isoprenoid-Protein Interactions. Org Lett. 2003;5:609–12. doi: 10.1021/ol026752a. [DOI] [PubMed] [Google Scholar]
- 13.Marecak DM, Horiuchi Y, Arai H, Shimonaga M, Maki Y, Koyama T, et al. Benzylphenoxy Analogs of Isoprenoid Diphosphates as Photoactivatable Substrates for Bacterial Prenyltransferases. Bioorg Med Chem Lett. 1997;7:1973–8. [Google Scholar]
- 14.Zhang Y-W, Koyama T, Marecak DM, Prestwich GD, Maki Y, Ogura K. Two Subunits of Heptaprenyl Diphosphate Synthase of Bacillus subtilis Form a Catalytically Active Complex. Biochemistry. 1998;37:13411–20. doi: 10.1021/bi972926y. [DOI] [PubMed] [Google Scholar]
- 15.Tian R, Li L, Tang W, Liu H, Ye M, Zhao ZK, et al. Chemical proteomic study of isoprenoid chain interactome with a synthetic photoaffinity probe. Proteomics. 2008;8:3094–104. doi: 10.1002/pmic.200800021. [DOI] [PubMed] [Google Scholar]
- 16.DeGraw AJ, Zhao Z, Hsieh J, Jefferies M, Distefano MD, Strickland CL, et al. A photoactive isoprenoid diphosphate analogue containing a stable phosphonate linkage: synthesis and structural biochemical studies with prenyltransferases. J Org Chem. 2007;72:4587–95. doi: 10.1021/jo0623033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turek TC, Gaon I, Distefano MD, Strickland CL. Synthesis of Farnesyl Diphosphate Analogues Containing Ether-Linked Photoactive Benzophenones and Their Application in Studies of Protein Prenyltransferases. J Org Chem. 2001;66:3253–64. doi: 10.1021/jo991130x. [DOI] [PubMed] [Google Scholar]
- 18.Kale TA, Raab C, Yu N, Dean DC, Distefano MD. A Photoactivatable Prenylated Cysteine Designed to Study Isoprenoid Recognition. J Am Chem Soc. 2001;123:4373–81. doi: 10.1021/ja0012016. [DOI] [PubMed] [Google Scholar]
- 19.Turek TC, Gaon I, Gamache D, Distefano MD. Synthesis and evaluation of benzophenone-based photoaffinity labeling analogs of prenyl pyrophosphates containing stable amide linkages. Bioorg Med Chem Lett. 1997;7:2125–30. [Google Scholar]
- 20.Turek TC, Gaon I, Distefano MD. Analogs of farnesyl pyrophosphate incorporating internal benzoylbenzoate esters: Synthesis, inhibition kinetics and photoinactivation of yeast protein farnesyltransferase. Tet Lett. 1996;37:4845–8. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- 21.Gaon I, Turek TC, Weller VA, Edelstein RL, Singh SK, Distefano MD. Photoactive Analogs of Farnesyl Pyrophosphate Containing Benzoylbenzoate Esters: Synthesis and Application to Photoaffinity Labeling of Yeast Farnesyltransferase. J Org Chem. 1996;61:7738–45. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- 22.Gaon I, Turek TC, Distefano MD. Farnesyl and geranylgeranyl pyrophosphate analogs incorporating benzoylbenzyl ethers: synthesis and inhibition of yeast protein farnesyltransferase. Tet Lett. 1996;37:8833–6. [Google Scholar]
- 23.Rilling HC. Photoaffinity Substrate Analogs for Eukaryotic Prenyltransferase. Meth Enzmol. 1985;110:125–31. doi: 10.1016/s0076-6879(85)10067-4. [DOI] [PubMed] [Google Scholar]
- 24.Chehade KAH, Kiegiel K, Isaacs RJ, Pickett JS, Bowers KE, Fierke CA, et al. Photoaffinity Analogues of Farnesyl Pyrophosphate Transferable by Protein Farnesyl Transferase. J Am Chem Soc. 2002;124:8206–19. doi: 10.1021/ja0124717. [DOI] [PubMed] [Google Scholar]
- 25.Bikhtiyarov YE, Omer CA, Allen CM. Photoreactive Analogues of Prenyl Diphosphates as Inhibitors and Probes of Human Protein Farnesyltransferase and Geranylgeranyltransferase Type I. J Biol Chem. 1995;270:19035–40. doi: 10.1074/jbc.270.32.19035. [DOI] [PubMed] [Google Scholar]
- 26.Duckworth BP, Xu J, Taton TA, Guo A, Distefano MD. Site-Specific, Covalent Attachment of Proteins to a Solid Surface. Bioconj Chem. 2006;17:967–74. doi: 10.1021/bc060125e. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, DeGraw AJ, Duckworth BP, Lenevich S, Tann C-M, Jenson EC, et al. Synthesis and Reactivity of 6,7-Dihydrogeranylazides: Reagents for Primary Azide Incorporation into Peptides and Subsequent Staudinger Ligation. Chem Biol Drug Des. 2006;68:85–96. doi: 10.1111/j.1747-0285.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee PC, Petri R, Mijts BN, Watts KT, Schmidt-Dannert C. Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab Eng. 2005;7:18–26. doi: 10.1016/j.ymben.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J Bacteriol. 2008;190:6084–96. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhry V, Vaughan R, Westheimer FH. 2-Diazo-3,3,3-trifluoropropionyl chloride: Reagent for photoaffinity labeling. Proc Nat Acad Sci USA. 1976;73:1406–8. doi: 10.1073/pnas.73.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turek TC, Gaon I, Distefano MD. Synthesis and Rapid Purification of 32P-Labeled Photoactive Analogs of Farnesyl Pyrophosphate. J Lab Cmpds Radiopharm. 1997;39:140–6. [Google Scholar]
- 32.Pompliano DL, Gomez RP, Anthony NJ. Intramolecular Fluorescence Enhancement: A Continuous Assay of Ras Farnesyl:Protein Transferase. J Am Chem Soc. 1992;114:7945–6. [Google Scholar]
- 33.Bond PD, Dolence JM, Poulter CD. A Continuous Fluorescence Assay for Protein:Prenyl Transferases. Meth Enzymol. 1995;250:30–43. doi: 10.1016/0076-6879(95)50060-x. [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Long SB, Hancock PJ, Kral AM, Hellinga HW, Beese LS. The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics. Proc Nat Acad Sci USA. 2001;98:12948–53. doi: 10.1073/pnas.241407898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strickland CL, Weber PC, Windsor WT, Wu Z, Le HV, Albanese MM, et al. Tricyclic farnesyl protein transferase inhibitors: crystallographic and calorimetric studies of structure-activity relationships. J Med Chem. 1999;42:2125–35. doi: 10.1021/jm990030g. [DOI] [PubMed] [Google Scholar]
- 37.Cramer F, Rittersdorf W, Boehm W. Chemistry of high-energy phosphates. XVIII. Synthesis of phosphoric acid esters and pyrophosphoric acid esters of the terpene alcohols. Ann. 1962;654:180–8. [Google Scholar]
- 38.Cramer F, Weimann G. Imido esters. VII. Trichloroacetonitrile, a reagent for the selective esterification of phosphoric acid. Chem Ber. 1961;94:996–1007. [Google Scholar]
- 39.Umbreit MA, Sharpless KB. Allylic Oxidation of Olefins by Catalytic and Stoichiometric Selenium Dioxide with tert-Butyl Hydroperoxide. J Am Chem Soc. 1977;99:5526–8. [Google Scholar]
- 40.Mitsunobu O, Wada M, Sano T. Stereospecific and stereoselective reactions. I. Preparation of amines from alcohols. J Amer Chem Soc. 1972;94:697–80. [Google Scholar]
- 41.Mitsunobu O. The use of diethyl azodicarboxylate and triphenylphosphine in synthesis and transformation of natural products. Synthesis. 1981;1:1–28. [Google Scholar]
- 42.Turek TC, Gaon I, Distefano MD. Synthesis and rapid purification of 32P-labeled photoactive analogs of farnesyl pyrophosphate. J Labelled Compds Radiopharm. 1997;39:139–46. [Google Scholar]
- 43.Hahn KM, Hastie SB, Sundberg RJ. Synthesis and evaluation of 2-diazo-3,3,3-trifluopropanoyl derivatives of colchicine and podophyllotoxin as photoaffinity labels: reactivity, photochemistry, and tubulin binding. Photochem Photobiol. 1992;55:17–27. doi: 10.1111/j.1751-1097.1992.tb04204.x. [DOI] [PubMed] [Google Scholar]
- 44.Theodore LJ, Nelson WL, Dave B, Giacomini JC. Studies on Ca2+ channel antagonists. A 2-diazo-3,3,3-trifluoropropionamide derivative related to verapamil as a potential photoaffinity probe. J Med Chem. 1990;33:873–7. doi: 10.1021/jm00164a064. [DOI] [PubMed] [Google Scholar]
- 45.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO Journal. 2003;22:5963–74. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Seabra MC, Deisenhofer J. Crystal Structure of Rab geranylgeranyltransferase at 2.0 A resolution. Structure. 2000;8:241–51. doi: 10.1016/s0969-2126(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 47.Guo Z, Wu Y-W, Das D, Delon C, Cramer J, Yu S, et al. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 2008;27:2444–56. doi: 10.1038/emboj.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzysiak AJ, Rawat DS, Scott SA, Pais JE, Handley M, Harrison ML, et al. Combinatorial Modulation of Protein Prenylation. ACS Chem Biol. 2007;2:385–9. doi: 10.1021/cb700062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosokawa A, Wollack JW, Zhang Z, Chen L, Barany G, Distefano MD. Evaluation of an alkyne-containing analogue of farnesyl diphosphate as a dual substrate for protein-prenyltransferases. Int J Peptide Res. 2007;13:345–54. [Google Scholar]
- 50.Chung JA, Wollack JW, Okesli A, Hovlid ML, Chen Y, Mueller JD, et al. Purification of prenylated proteins by affinity chromatography on cyclodextrin-modified agarose. Anal Biochem. 2009;386:1–8. doi: 10.1016/j.ab.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long SB, Casey PJ, Beese LS. Reaction path of protein farnesyltransferase at atomic resolution. Nature. 2002;419:645–50. doi: 10.1038/nature00986. [DOI] [PubMed] [Google Scholar]
- 52.Dolence JM, Cassidy PB, Mathis JR, Poulter CD. Yeast Protein Farnesyltransferase: Steady-State Kinetic Studies of Substrate Binding. Biochemistry. 1995;34:16687–94. doi: 10.1021/bi00051a017. [DOI] [PubMed] [Google Scholar]
- 53.Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, et al. Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry. 1998;37:16601–11. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- 54.Voelkert M, Uwai K, Tebbe A, Popkirova B, Wagner M, Kuhlmann J, et al. Synthesis and Biological Activity of Photoactivatable N-Ras Peptides and Proteins. J Am Chem Soc. 2003;125:12749–58. doi: 10.1021/ja036178d. [DOI] [PubMed] [Google Scholar]
- 55.Alexander M, Gerauer M, Pechlivanis M, Popkirova B, Dvorsky R, Brunsveld L, et al. Mapping the isoprenoid binding pocket of PDEdelta by a semisynthetic, photoactivatable N-Ras lipoprotein. ChemBioChem. 2009;10:98–108. doi: 10.1002/cbic.200800275. [DOI] [PubMed] [Google Scholar]
- 56.Quellhorst GJ, Jr., Allen CM, Wessling-Resnick M. Modification of Rab5 with a photoactivatable analog of geranylgeranyl diphosphate. J Biol Chem. 2001;276:40727–33. doi: 10.1074/jbc.M104398200. [DOI] [PubMed] [Google Scholar]
- 57.Wollack JW, Zeliadt NA, Mullen DG, Amundson G, Geier S, Falkum S, et al. Multifunctional prenylated peptides for in vivo analysis. J Am Chem Soc. 2009;131:7293–303. doi: 10.1021/ja805174z. [DOI] [PMC free article] [PubMed] [Google Scholar]