Abstract
The H3K9me2 histone methyltransferases G9a and GLP repress Mage-a class cancer germline (CG) antigen gene expression in murine ES cells but the role of these enzymes in CG antigen gene regulation in human cancer cells is unknown. Here we show that while independent or dual knockdown of G9a and GLP in human cancer cells leads to reduced global and CG antigen promoter-associated H3K9me2 levels it does not activate CG antigen gene expression. Moreover, CG antigen gene repression is maintained following pharmacological targeting of G9a or treatment of G9a knockdown cells with the HDAC inhibitor Trichostatin A. However, G9a knockdown cells display increased sensitivity to CG antigen gene activation mediated by the DNA methyltransferase inhibitor decitabine. To account for these findings, we examined DNA methylation at CG antigen gene promoters in both cell types. We found robust DNA hypomethylation in G9a/GLP targeted murine ES cells but a lack of DNA methylation changes in G9a/GLP targeted human cancer cells; intriguingly this distinction also extended to markers of global DNA methylation. These data reveal that G9a/GLP is required for DNA methylation of CG antigen genes and genomic DNA in murine ES cells but not human cancer cells and implicate DNA methylation status as the key epigenetic mechanism involved in CG antigen gene repression.
Keywords: cancer-germline antigens, cancer-testis antigens, DNA methylation, G9a, GLP, EuHMTaseI, histone methylation, epigenetics
Introduction
Cancer/germline (CG) antigens (also known as cancer-testis antigens) are targets of tumor vaccines currently undergoing clinical evaluation world-wide (1). The rationale for vaccine-based targeting of CG antigens in cancer includes both the inherent immunogenicity of these antigens, as well as their restricted expression in normal human tissues and widespread expression in human tumors (1, 2). CG antigen genes appear to be principally regulated at the transcriptional level by epigenetic signals. Evidence for this includes: 1) promoter DNA hypomethylation correlates with CG antigen gene expression in human tumor and normal tissues (3, 4); 2) treatment of non-expressing cancer cells with epigenetic modulatory drugs, including DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors, induces CG antigen gene expression (5-7); and 3) specific histone H3 modification patterns are associated with CG antigen gene expression status in human cancer cells (8-10). More detailed elucidation of the epigenetic mechanisms regulating CG antigen gene expression in human cancer cells and tumors should help facilitate the clinical development of CG antigen-targeted therapeutic approaches (2).
The enzymes that mediate histone H3, lysine 9 (H3K9) methylation have been identified, and these include Suv39-class enzymes, which target heterochromatic loci and catalyze H3K9 trimethylation (H3K9me3), and G9a and GLP (also known as EuHMTaseI), which target euchromatic loci and catalyze H3K9 dimethylation (H3K9me2) (11-14). Similar to DNA methyltransferase I (DNMT1) knockout, G9a or GLP knockout causes embryonic lethality in mice, implicating a crucial role for epigenetic repression in early mammalian development (12, 13, 15). Gene expression profiling of G9a-knockout murine ES cell lines identified Mage-a class CG antigen genes as targets of epigenetic repression by G9a, and subsequent experiments demonstrated a similar role for GLP (12, 13). H3K9me2 levels are reduced both genome-wide and at Mage-a promoters in G9a or GLP knockout ES cells (12, 13). These data suggest that H3K9me2 may play a primary role in mediating CG antigen gene repression in murine ES cells, which appears to conflict with the prevailing view that DNA methylation is the primary mediator of CG antigen gene silencing in human cancer (1, 4, 8).
We previously established the human colon adenocarcinoma cell lines HCT116 and RKO as useful models to study the mechanisms of CG antigen gene repression in human cancer (5, 8). CG antigen genes are transcriptionally silenced in HCT116 and RKO cells by DNA hypermethylation and are activated by pharmacological or genetic targeting of DNMT enzymes in these cells (5, 8). Moreover, CG antigen gene induction in DNMT-deficient HCT116 cells coincides with both DNA hypomethylation and remodeling of histone code patterns at CG antigen gene promoters (8). The fact that CG antigen genes are infrequently expressed in human colorectal cancer also supports the relevance of this experimental model (1).
In the current study we examined the role of G9a and GLP in regulating CG antigen expression in human cancer cells, using HCT116 and RKO colorectal cancer cells as models. We hypothesized that G9a/GLP may repress CG antigen genes in non-expressing human cancer cell lines, as H3K9me2 and DNA methylation signals are often interdependent, and because G9a propagates epigenetic repression at the mammalian replication fork via its interaction with DNMT1 (16). Our data reveal that in human cancer cells, unlike mouse ES cells, G9a and GLP are dispensable for CG antigen gene repression, despite playing a role in the maintenance of H3K9 methylation in both cell types. Most importantly, we find that CG antigen genes are DNA hypomethylated in G9a or GLP null murine ES cells, but their DNA methylation status is unchanged in human cancer cells following G9a and/or GLP targeting. These data reinforce the notion that epigenetic control processes are regulated in distinct ways in ES cells and cells of somatic origin, including cancer cells. Furthermore, they implicate DNA methylation as the primary repression mechanism for CG antigen gene expression.
Results
CG antigen gene expression and histone modifications in G9a-knockdown human cancer cells
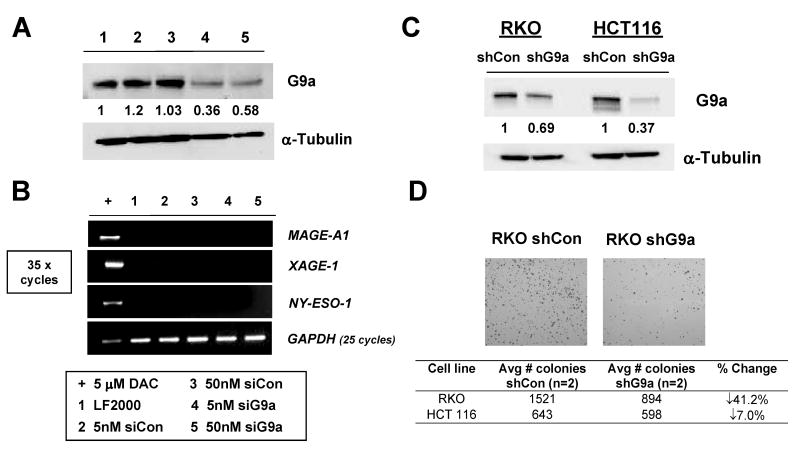
To assess the role of G9a in CG antigen gene repression in human cancer cells, we initially developed an effective transient siRNA G9a targeting strategy in RKO cells (Fig. 1A). Low siRNA transfection efficiency prevented implementation of this strategy in HCT116 cells (data not shown). RT-PCR analysis revealed that efficient G9a knockdown did not induce expression of three representative CG antigen genes MAGE-A1, XAGE-1, and NY-ESO-1 (8) in RKO cells (Fig. 1B). Because sustained knockdown might be required to affect CG antigen gene expression, and to validate these findings in another cell type, we established clonal G9a stable knockdown cell lines in both HCT116 and RKO cells (Fig. 1C). Interestingly, during derivation of the these cell lines, we observed that cancer cells expressing G9a shRNA showed substantially reduced cell clonogenicity, particularly in RKO cells (Fig. 1D). We also observed reduced cell viability following siRNA mediated G9a knockdown in RKO (data not shown). These data suggest that G9a contributes to cell growth and/or survival in colorectal cancer cells, consistent with previous reports using mice and prostate cancer cells (12, 17). The role of G9a in cell survival may explain why it was difficult to achieve complete G9a knockdown in human cancer cells (Fig. 1A and C).
FIGURE 1.
Transient and stable G9a knockdown in human cancer cells. A. Transient siRNA knockdown of G9a in RKO cells. Control and G9a-specific siRNAs were administered at either 5 or 50nM, cells were harvested at day 5, and G9a expression was measured by Western blot. α-tubulin expression was measured as control for protein input, and band densitometry was performed as described in the Materials and Methods. The sample key is shown below panel B. B. End-point RT-PCR analysis of MAGE-A1, NY-ESO-1, and XAGE-1 for the experiment shown in panel A. GAPDH expression was analyzed to control for cDNA input, and amplification of cDNA from RKO cells treated with 5 μM DAC (decitabine) served as a positive control for CG antigen gene expression. CG antigen gene amplification was performed for 35 cycles, while GAPDH was performed for 25 cycles. The sample key is shown below panel B. LF2000= lipofectamine-only transfection control. C. Western blot analysis of G9a protein expression in control shRNA (shCon) and G9a shRNA (shG9a) stable RKO and HCT116 cell lines. α-tubulin expression was measured as control for protein input, and band densitometry was performed as described in the Materials and Methods. D. shG9a stable-expressing cells have reduced clonogenicity. Upper: Representative images of crystal violet-stained colonies of control and G9a knockdown stable RKO cells. Lower: Quantification of colony number in control and G9a knockdown stable RKO and HCT116 cell lines.
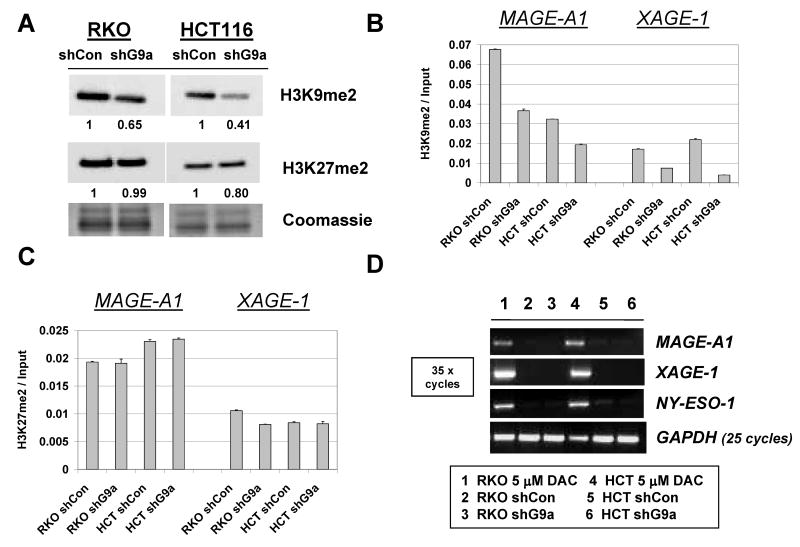
To explore the phenotype of stable G9a knockdown human cancer cells, we used Western blot analysis to measure global H3K9me2 levels and H3K27me2 levels, as these two modifications have been previously linked to G9a function (18, 19). Notably, H3K9me2 levels were reduced in both cell types while H3K27me2 levels were not significantly altered (Fig. 2A). A lack of activity of G9a towards H3K27me2 is consistent with the results of recent studies of G9a function in different model systems (20-23). To assess whether G9a knockdown altered histone modifications at CG antigen gene promoters, we utilized quantitative ChIP-PCR (qChIP) to determine H3K9me2 levels at the MAGE-A1 and XAGE-1 promoters as described previously (8). Similar to the effect on global levels, H3K9me2 levels are reduced at both MAGE-A1 and XAGE-1 in G9a stable knockdown HCT116 and RKO cells (Fig. 2B). In contrast, H3K27me2 levels are unchanged at these promoters (Fig. 2C). While reduced H3K9me2 suggested that gene expression might be altered, CG antigen genes remained repressed in both G9a knockdown cell lines (Fig. 2D). This observation was repeatable as independently derived stable G9a knockdown RKO and HCT116 cell clones also did not express CG-X genes (data not shown). Stable G9a knockdown cells also showed slight increases in the global level of the active histone mark H3K9ac (Fig. 3A). However, consistent with a lack of CG antigen expression in the stable G9a knockdown cells, ChIP analyses revealed that the levels of H3K9ac and H3K4me2 at CG gene promoters, which are associated with CG gene expression (8), were not increased (Fig. 3B-E).
FIGURE 2.
Characterization of stable G9a knockdown human cancer cells. A. Western blot analysis of H3K9me2 and H3K27me2 levels in control shRNA and G9a shRNA expressing stable cell lines. Coomassie staining confirmed equivalent protein input, and band densitometry was performed as described in the Materials and Methods. B-C. qChIP-PCR analysis of H3K9me2 (B) and H3K27me2 (C) levels at the MAGE-A1 and XAGE-1 5′ CpG island regions. Error bars indicate + 1SD. D. RT-PCR analysis of MAGE-A1, XAGE-1, and NY-ESO-1 expression. PCR conditions and controls are the same as described in Figure 1B, and the sample key is shown below panel D.
FIGURE 3.
H3K9ac and H3K4me2 in G9a knockdown human cancer cells. A. Western blot analysis of H3K9ac and H3K4me2 levels in control shRNA and G9a shRNA expressing stable RKO and HCT116 cell lines. Coomassie staining confirmed equivalent protein input, and band densitometry was performed as described in the Materials and Methods. (B-C) qChIP-PCR analysis of H3K9ac levels at the MAGE-A1 and XAGE-1 5′ CpG island regions in G9a targeted RKO (B) and HCT116 (C) cell lines. (D-E) qChIP-PCR analysis of H3K4me2 levels at the MAGE-A1 and XAGE-1 5′ CpG island regions in G9a targeted RKO (D) and HCT116 (E) cell lines. RKO cells treated with decitabine (DAC) served as a positive control for both histone modifications in RKO cells, while DNMT1-/-,3b-/- HCT116 cells (DKO) served as a positive control for both histone modifications in HCT116. Error bars indicate + 1SD.
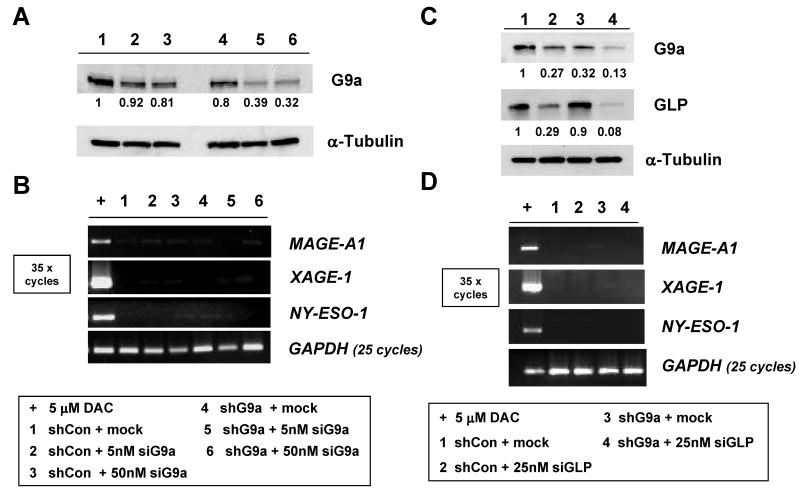
As G9a knockdown was robust but not complete in G9a shRNA knockdown clones, we further targeted G9a in stable G9a knockdown RKO cells using transient siRNA transfection (Fig. 4A). While G9a siRNA transfection resulted in further diminishment of G9a protein, CG-X genes remained repressed (Fig. 4A-B).
FIGURE 4.
CG-X antigen genes are repressed in RKO cells following dual shRNA/siRNA G9a knockdown, or dual G9a/GLP knockdown. A. RKO cells stably expressing control shRNA or G9a shRNA were mock transfected or transfected with a G9a-specific siRNA. Cells were harvested five days post transfection and G9a protein levels were measured by Western blot. α-tubulin expression was measured as control for protein input, and band densitometry was performed as described in the Materials and Methods. The sample key is shown below panel B. B. RT-PCR analysis of CG antigen gene expression. PCR conditions and controls are the same as described in Figure 1B, and the sample key is shown below panel B. C. RKO cells stably expressing control shRNA or G9a shRNA were mock transfected or transfected with a GLP-specific siRNA. Five days post-transfection, G9a and GLP protein levels were measured by Western blot. α-tubulin expression was measured as control for protein input, and band densitometry was performed as described in the Materials and Methods. The sample key is shown below panel D. D. RT-PCR analysis of CG antigen gene expression. PCR conditions and controls are the same as described in Figure 1B, and the sample key is shown below the panel.
Dual targeting of G9a and EuHMTaseI/GLP in human cancer cells
GLP (EuHMTaseI) has similar substrate specificity as G9a, and has a non-redundant and essential role in mammalian development (13). Despite this non-redundancy in mouse development, it remained plausible that GLP could compensate for G9a loss in human cancer cells with regards to CG antigen gene repression. To test this possibility we utilized a siRNA approach. Treatment of RKO cells with GLP siRNA resulted in effective GLP knockdown and, notably, also reduced G9a protein levels (Fig. 4C). This may be explained by the fact that GLP and G9a form an intracellular complex that stabilizes the G9a protein (13). Consistent with this idea, GLP knockdown did not affect G9a mRNA expression (data not shown). Despite effective reduction of both G9a and GLP in RKO cells, CG antigen genes remained repressed (Fig. 4D). Furthermore, we did not observe an additional decrease of H3K9me2 levels at CG antigen gene promoters following GLP knockdown in stable G9a knockdown cells (data not shown). These data suggest that redundancy between G9a and GLP does not account for the repression of CG antigen genes in cancer cells sustaining G9a knockdown.
Pharmacological targeting of G9a in human cancer cells
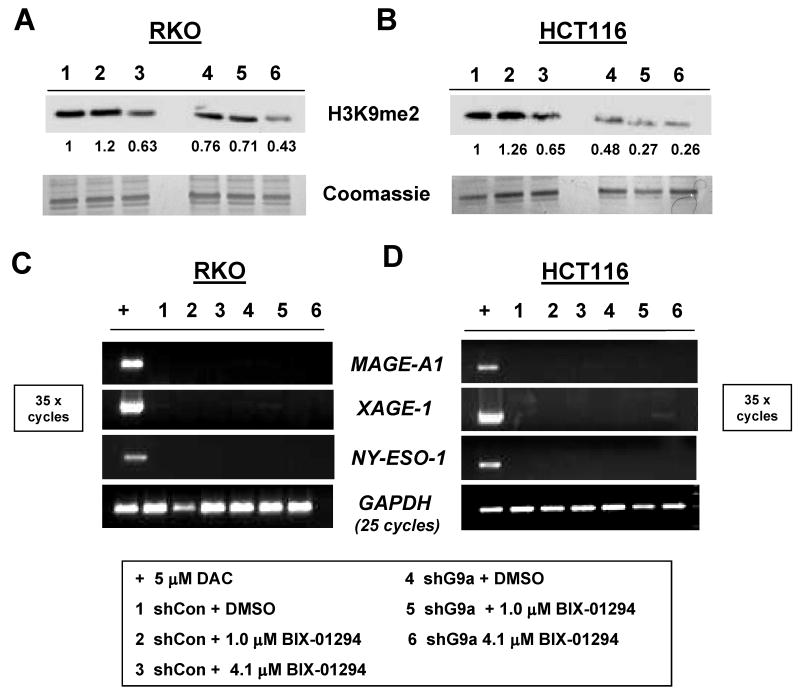
Recently, a novel small molecule inhibitor of G9a was identified by high throughput screening (23). This agent, BIX-01294, was shown to selectively target G9a, and to a lesser extent GLP, and to reduce H3K9me2 levels in mammalian cells at low micromolar concentrations (23). Notably, BIX-01294 treatment was also shown to activate Mage-A2 expression in wildtype mouse ES cells (23). Here, we sought to complement the genetic knockdown approach described above by examining the effect of BIX-01294 treatment on CG antigen gene expression in human cancer cells. BIX-01294 treatment reduced global H3K9me2 levels in both control and G9a stable knockdown HCT116 and RKO cells, validating the activity of this agent in human cancer cells (Fig. 5A-B). However, BIX-01294 treatment failed to induce CG-X gene expression in either cell type (Fig. 5C-D). These data provide additional evidence that G9a is not required for CG antigen gene repression in human cancer cells.
FIGURE 5.
Combined genetic and pharmacological targeting of G9a in human cancer cells. (A-B) RKO (A) and HCT116 (B) cells were treated with DMSO (vehicle) or BIX-01294, cells were harvested 48 hours later, and H3K9me2 levels were determined by Western blot analyses. Coomassie staining confirmed equal protein input, and band densitometry was performed as described in the Materials and Methods. The sample key is shown at the bottom of the figure. (C-D) RT-PCR analysis of CG antigen gene expression in the experiment described in panels A-B. PCR conditions and controls are the same as described in Figure 1B, and the sample key is shown below the figure.
Combined G9a knockdown and HDAC inhibitor treatment effect on CG antigen gene expression in human cancer cells
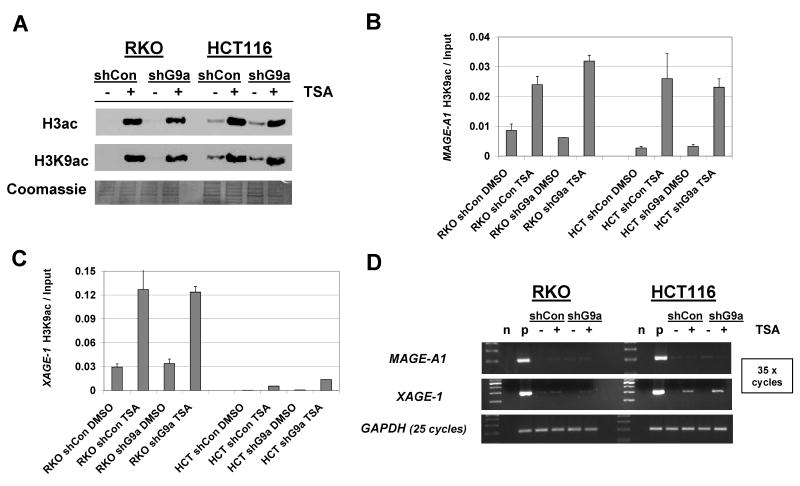
The observation that G9a knockdown results in reduced H3K9me2 at CG antigen promoters without altering H3K9ac raised the question of whether forced induction of H3K9ac in G9a knockdown cells could elicit CG antigen expression. To test this, we utilized the classical HDAC inhibitor Trichostatin A (TSA) to treat control and stable G9a knockdown cells. As anticipated, TSA treatment markedly increased global H3 acetylation and H3K9 acetylation levels in both control and stable G9a knockdown cells (Fig. 6A). Moreover, TSA induced H3K9ac levels at CG antigen promoters (Fig. 6B-C). Despite this effect, MAGE-A1 remained silent in both RKO and HCT116 cells, while XAGE-1 remained silent in RKO (Fig. 6D). For XAGE-1 expression in HCT116, TSA treatment led to a moderate level of induction, suggesting that histone acetylation regulates this gene in certain cellular contexts (Fig. 6D). Taken together though, these data suggest that remodeling the histone code at CG antigen gene promoters is generally insufficient to provoke CG antigen gene expression. A possible explanation for this phenomenon is that CG antigen gene promoters are densely DNA methylated in these cell types (8).
FIGURE 6.
Combined G9a knockdown and HDAC inhibitor treatment effect on CG antigen gene expression. RKO and HCT116 stable control shRNA and stable G9a knockdown cells were treated with the histone deacetylase inhibitor TSA at 600nM concentration for 24 hours prior to harvesting. A. Histone protein extracts were prepared as described in Materials and Methods, and utilized for Western blot analysis of acetylated Histone H3 or acetylated Histone H3K9 levels. Coomassie staining confirmed equivalent protein loading. (B-C) Acetylated H3K9 levels at the MAGE-A1 (B) and XAGE-1 (C) promoters were measured by quantitative ChIP-PCR. D. MAGE-A1 and XAGE-1 mRNA expression were measured by end-point RT-PCR. n and p indicate negative (no template) and positive (decitabine-treated cells) PCR controls, respectively.
G9a knockdown sensitizes human cancer cells to 5-aza-2′-deoxycytidine (decitabine)-mediated CG antigen gene activation
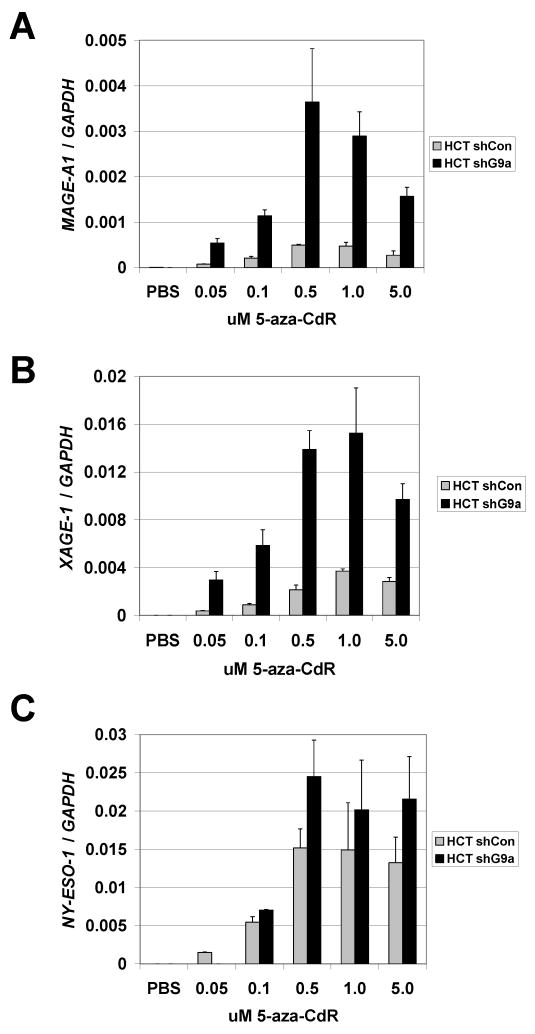
While the results above indicate that inhibition of G9a/GLP is not sufficient to provoke CG antigen gene expression in human cancer cells, either alone or in combination with TSA, it remained plausible that G9a targeting could potentiate CG antigen gene activation by DNMT inhibitors. A similar relationship exists between HDAC inhibitors and DNMT inhibitors in that HDAC inhibition can facilitate the activation of methylation silenced genes by the DNMT inhibitor 5-aza-CdR (24). We thus tested the response of control and G9a knockdown human cancer cells to treatment with decitabine. Notably, stable G9a knockdown HCT116 cells treated with decitabine showed significantly increased MAGE-A1 and XAGE-1 gene expression, and moderately increased NY-ESO-1 expression, as compared to control cells treated with decitabine alone (Fig. 7A-C). In contrast to HCT116, an enhanced CG antigen expression effect was only marginal in decitabine-treated G9a stable knockdown RKO cells (data not shown). These data demonstrate that G9a knockdown can synergize with DNMT inhibition to promote CG antigen gene expression, but that this effect is in part dependent cell context dependent.
FIGURE 7.
Stable G9a knockdown HCT116 cells show increased sensitivity to decitabine-mediated CG antigen gene activation. HCT116 cell lines stably expressing control or G9a-specific shRNAs were treated with the indicated concentrations of decitabine (5-aza-CdR) for 48 hours, RNA extracts were harvested, and (A) MAGE-A1 (B) XAGE-1 and (C) NY-ESO-1 expression was measured using qRT-PCR. Expression data from all three genes were normalized to GAPDH as a cDNA input control. In all panels, mean expression values and + 1SD of triplicate data points are plotted.
CG antigen gene regulation and DNA methylation in G9a and GLP knockout mouse ES cells
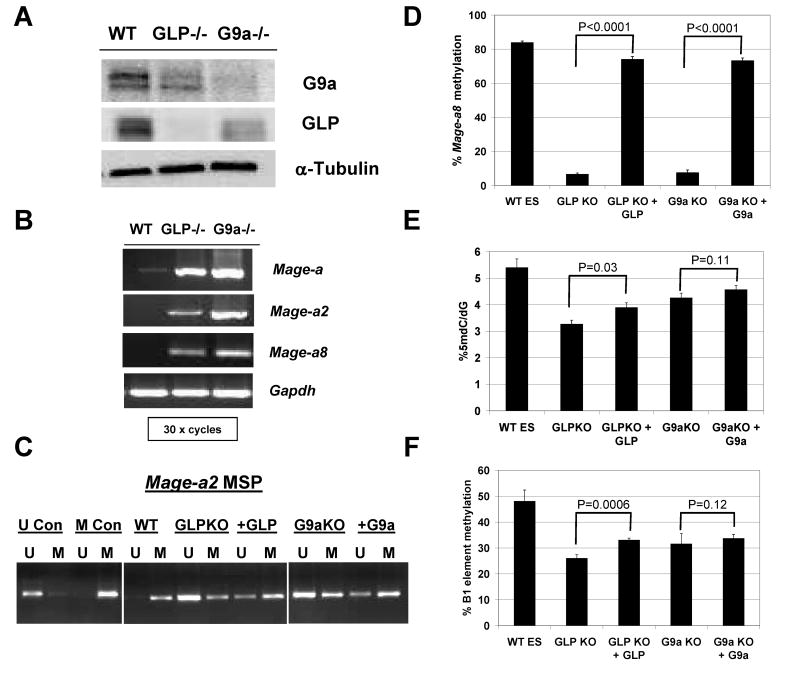
In contrast to our data using human cancer cells, G9a and GLP knockout mouse ES cells show robust expression of Mage-a class CG antigen genes (12, 13). Because loss of H3K9me2 globally and at CG antigen gene promoters is observed in both mouse ES cells and human cancer cells responding to G9a and/or GLP targeting [(12, 13, 17); Figure 2], reduction of this repressive epigenetic histone mark does not appear to explain the differential response. Of note, previous studies have observed altered DNA methylation in G9a knockout mouse ES cells (18, 25, 26). We hypothesized that differences in promoter DNA methylation status at CG antigen genes might explain the distinct response of mouse ES and human cancer cells to G9a/GLP targeting. To test this, we initially utilized G9a and GLP knockout murine ES cell lines (Fig. 8A). Using pan-Mage-a primers that amplify murine Mage-a2, -a5, -a6, and -a8, we observe that G9a and GLP knockout ES cells express high levels of Mage-a genes (Fig. 8B). Of the murine Mage-a gene family members, only Mage-a2 and –a8 contain classical 5′ CpG islands (27) and thus we focused further attention on these two genes. Both Mage-a2 and Mage-a8 were induced in G9a and GLP knockout ES cells (Fig. 8B). We next utilized methylation specific PCR (MSP) to determine the methylation status of the 5′ CpG region of Mage-a2 in wild-type, G9a knockout, and GLP knockout ES cells. As shown in Fig. 8C, Mage-a2 was hypermethylated in wildtype ES cells and becomes dramatically hypomethylated in GLP and G9a knockout cells. Moreover, reintroduction of either GLP or G9a into the corresponding knockout cell line led to partial re-methylation of Mage-a2 (Fig. 8C). This re-methylation correlated with repression of Mage-a2 (12, 13). Similar to the Mage-a2 result, bisulfite pyrosequencing analysis indicated that Mage-a8 was dramatically hypomethylated in G9a and GLP null ES cells and re-methylated following reintroduction of the relevant protein (Fig. 8D). To determine if the effect on DNA methylation in G9a and GLP knockout ES cells extended to markers of global DNA hypomethylation, we utilized two different measures of global DNA methylation status: 1) 5-methyl-2′-deoxycytidine (5mdC) levels, and 2) bisulfite pyrosequencing of the murine repetitive element B1 (28). Both knockout cell types display sizable reduction of 5mdC levels, which was partially restored following reintroduction of the corresponding proteins (Fig. 8E). A similar phenotype was observed for methylation of the repetitive B1 element (Fig. 8F). These data reveal that G9a and GLP are required for DNA methylation of CG antigen genes as well as other regions of the genome in murine ES cells, and provide a plausible explanation for the activation of CG antigen genes in this cell type.
FIGURE 8.
CG antigen gene expression and DNA methylation in G9a and GLP knockout mouse ES cells. A. Western blot analysis of G9a and GLP protein expression. α-tubulin expression was measured as a control for protein input. B. RT-PCR analysis of Mage-a, Mage-a2, and Mage-a8 expression. Gapdh was amplified as a control for cDNA input. All reactions utilized 30 PCR cycles. Specific amplification of Mage-a2 and Mage-a8 was confirmed by DNA sequencing of the PCR amplicons. C. MSP analysis of Mage-a2 5′ region methylation. Results are shown for control DNAs, wildtype ES cells, and G9a and GLP knockout ES cells before and after re-introduction of the knocked-out protein. U and M lanes correspond to PCR amplifications specific for unmethylated and methylated DNA, respectively. U and M control DNAs are described in the Materials and Methods. D. Bisulfite pyrosequencing analysis of Mage-a8 5′ region methylation. Samples are same as described in panel C. E. LC-MS analysis of total genomic 5-methyldeoxycytidine (5mdC). Samples are same as described in panel C. F. Bisulfite pyrosequencing analysis of B1 repetitive element methylation. Samples are same as described in panel C. Statistical analysis shown in Panels D-F utilized one-tailed T-Test (GraphPad Prism) and the relevant P-values are indicated on the figure.
CG antigen gene and global genomic DNA methylation in G9a knockdown human cancer cells
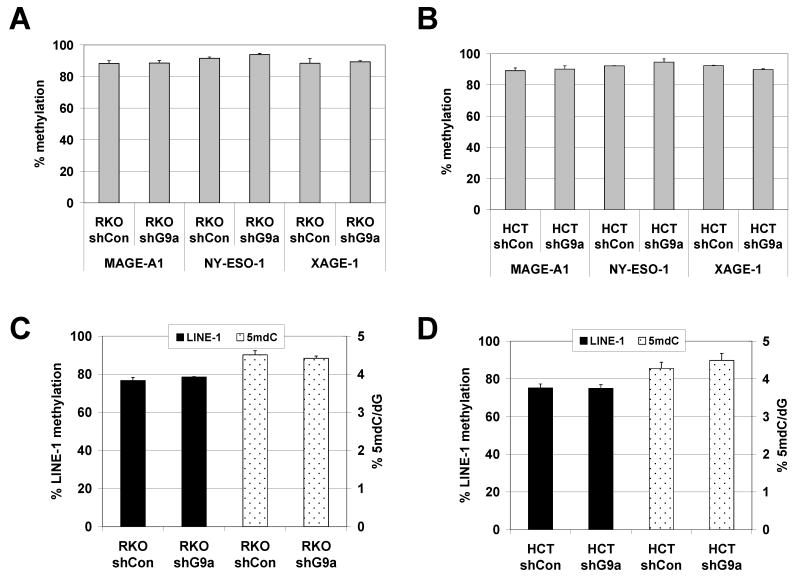
We next examined DNA methylation in stable G9a and GLP knockdown human cancer cells. We utilized quantitative bisulfite pyrosequencing to determine the methylation level of the 5′ CpG island promoters of MAGE-A1, NY-ESO-1, and XAGE-1. As expected, in both the RKO and HCT116 cell lines, stable G9a knockdown did not alter CG antigen promoter methylation levels (Fig. 9A-B). Standard sodium bisulfite sequencing and MSP analyses confirmed the pyrosequencing data (data not shown). In addition, stable G9a knockdown in HCT116 and RKO cells did not lead to altered global DNA methylation, as assessed by either 5mdC levels or by pyrosequencing of the human LINE-1 repetitive DNA element (Fig. 9C-D). Consistent with the findings using stable G9a knockdown cells, transient knockdown of G9a or GLP in the stable G9a knockdown RKO cell line, or combined stable G9a knockdown and TSA treatment in either HCT116 or RKO, also did not alter CG antigen or global DNA methylation (data not shown).
FIGURE 9.
DNA methylation in stable G9a knockdown human cancer cells. (A-B) Bisulfite pyrosequencing analysis of MAGE-A1, NY-ESO-1, and XAGE-1 5′ promoter CpG island region methylation in (A) RKO and (B) HCT116 control and stable G9a knockdown cell lines. (C-D) Global DNA methylation analyses of (C) RKO and (D) HCT116 control and stable G9a knockdown cell lines. Two measurements of global DNA methylation were performed: bisulfite pyrosequencing analysis of LINE-1 repetitive element methylation, and LC-MS determination of total 5mdC levels.
Discussion
We previously reported that disruption of DNMT enzymes in human cancer cells results in reduced H3K9me2 levels at CG antigen gene promoters, correlating with increased gene expression (8). In addition, it was previously shown that murine ES cells deficient for G9a or GLP express Mage-a class CG antigen genes, coincident with reduced H3K9me2 levels at their promoters (12, 13). These data suggested that H3K9me2 and/or G9a/GLP may play a primary role in CG antigen gene repression in human cancer cells. However, our current data argue against this model. First, either transient or stable G9a knockdown in human cancer cell lines does not induce CG antigen gene expression, despite loss of H3K9me2 both globally throughout the genome and specifically at CG antigen promoter regions. Second, simultaneous knockdown of G9a and GLP, and combined genetic and pharmacological inhibition of G9a also failed to induce CG gene expression. Third, combined G9a knockdown and HDAC inhibitor treatment generally failed to induce CG gene expression. Moreover, we previously found that unlike double DNMT1/3b knockout cells, single DNMT1-/- or DNMT3b-/- HCT116 cells show reduced H3K9me2 at CG-X gene promoters in the absence of DNA methylation changes or expression of CG genes (8). Collectively, these data suggest that G9a/GLP knockdown and/or altered histone modification states are not sufficient alone to reactivate the expression of CG antigen genes in human cancer cells. Moreover, our data argue that promoter DNA hypomethylation is the chief epigenetic signal leading to CG antigen expression in human cancer cells. In agreement with our findings for CG antigen genes, it was recently reported that G9a knockdown in human prostate cancer cells leads to the up-regulation of very few genes in a genome-wide microarray screen and does not cause DNA hypomethylation of methylation-silenced tumor suppressor genes (17).
Based on our data from human cancer cells, we re-examined the mechanism of CG antigen gene activation in G9a or GLP deficient murine ES cells (12, 13). We hypothesized that G9a or GLP loss could lead to CG gene promoter DNA hypomethylation in murine ES cells, but not in human cancer cells, thus explaining the differential outcomes in the two systems. Our data validated this hypothesis. In both G9a and GLP null ES cells, the Mage-a2 and Mage-a8 promoters were severely hypomethylated coincident with gene activation. That this effect was specific was validated by experiments that showed that reintroduction of wild-type G9a or GLP led to re-methylation and silencing of these genes. Interestingly, G9a and GLP null ES cells also displayed both a global loss of methylation at the non-LTR short interspersed repetitive element (SINE) B1 and a reduction of total genomic 5mdC levels. In stark contrast, in human cancer cells in which G9a and/or GLP were down-regulated by RNAi, both CG promoter and global DNA methylation levels (either of the LINE-1 element or 5mdC levels) were unaltered. This was true despite significant reduction of both global and promoter-specific H3K9me2 levels in these cells. These data emphasize a primary regulatory role for DNA methylation in CG antigen gene regulation. The apparently tight dependence of CG gene silencing on DNA methylation may be accounted for by direct inhibition of Ets transcription factor binding, the recruitment of methylated DNA binding proteins at CG antigen gene promoter regions, or other as yet undefined mechanisms (5, 29, 30).
Our finding of DNA hypomethylation in G9a/GLP null murine ES cells is consistent with other reports (18, 25, 26). DNA hypomethylation in G9a-null ES cells was initially described for the Prader-Willi imprinting center, and it was later shown that methylation of the Oct3/4 gene during differentiation is blocked in G9a-null ES cells (25, 26). In addition, a recent study reported that 32/1300 (∼2.5%) genomic loci are hypomethylated in G9a -/- ES cells, implying a more widespread effect (18). Here we show that G9a or GLP null ES cells display significant reductions in 5mdC levels and hypomethylation of the murine B1 SINE repetitive DNA element. Similarly, Dong et al. recently reported that murine LTR and non-LTR LINE elements are DNA hypomethylated in G9a null ES cells (31). Together, these data suggest that G9a or GLP loss disrupts maintenance and/or de novo DNA methylation. Loss of DNA methylation in G9a and GLP knockout ES cells does not appear to result from decreased expression of Dnmt genes or proteins (31, 32). Instead, the mechanistic basis for the DNA hypomethylation effect may reflect either reduced recruitment of Dnmt1 to target sites during DNA replication, or reduced Dnmt3a and/or Dnmt3b targeting to non-replicating chromatin (16, 31, 32). Intriguingly, recent data suggest that DNA hypomethylation in G9a or GLP null murine ES cells may result from a loss of a function unrelated to their histone methyltransferase activity (31, 32).
It is important to note that although H3K9me2 may not be sufficient for CG antigen gene repression in human cancer cells, it does appear to play a secondary role. Notably, we found that certain G9a knockdown human colorectal cancer cells show an increased sensitivity to decitabine-mediated CG antigen gene induction. Additionally, it was recently shown that G9a and DNMT1 knockdown cooperatively facilitate induction of the DNA methylation-silenced tumor suppressor gene MASPIN in breast cancer cells (33). In ES cells, unlike human cancer cells (or other somatic cells), it appears that either H3K9 methylation or DNA methylation is sufficient for transcriptional repression (17, 32). This may reflect a “dual silencing” mechanism to maintain silent states upon cellular differentiation, which is uniquely important in ES cells (17, 32).
We demonstrate here that murine ES cells are more susceptible to the loss of DNA methylation at CG antigen genes following G9a or GLP loss than are human cancer cells. One possible explanation for the differential effect in the two cell types is based on the structure of CG antigen genes in the two species. In humans, CG antigen genes have promoter regions of relatively high CpG density while murine Mage-a class CG genes generally have CpG poor promoter regions [(8) and data not shown]. To mitigate this issue, our study focused on the two murine Mage-a genes (Mage-a2 and Mage–a8) that have CpG dense promoter regions. Therefore it is unlikely that CpG density of the studied CG gene promoters accounts for the different results observed in the two cell types. Of note, the orthologous human genes MAGE-A2 and MAGE-A8 were not activated in human cancer cells sustaining G9a knockdown (data not shown). Moreover, the effects of G9a or GLP loss on global DNA methylation (e.g. 5mdC levels) in the two cell types are also distinct, despite widespread conservation of the mouse and human genomes. A second possible explanation for the differential effect relates to the two cell types under study. We speculate that the dramatic loss of DNA methylation in G9a or GLP null ES cells reflects a greater level of “epigenetic plasticity” in this cell type than is seen in somatic cells (including cancer cells). ES cells may require higher epigenetic plasticity since the ultimate fate of these cells is epigenetic reprogramming along specific cell lineages (34). One example of the epigenetic plasticity of ES cells is the bivalent chromatin domains consisting of H3K4me3 and H3K27me3 often found at the promoters of developmentally regulated genes in ES cells (35). Also consistent with this explanation, G9a knockout embryos (somatic cells) do not display DNA hypomethylation at target loci, despite loss of H3K9 methylation at these regions (32). Finally, studies of the impact of DNA methylation on chromatin structure and histone modifications suggest that there are important distinctions between epigenetic regulation in ES cells and somatic cells (36).
In summary, our data emphasize a primary regulatory role for DNA methylation in CG antigen gene regulation in human cancer. There has been increasing interest in the utilization of epigenetic modulatory drugs to promote CG antigen gene expression, in order to augment CG antigen vaccine efficacy in human cancer patients (2, 37). Our data suggest that these approaches may need to include direct inhibition of DNMT enzymes as a required step to promote CG antigen gene expression. However, it remains plausible that agents that modify histone modification status, including HDAC or H3K9 methyltransferase inhibitors, may prove useful to enhance the effect of DNMT inhibitors in this context.
Materials and Methods
Cell culture and drug treatments
Colon adenocarcinoma cell lines RKO and HCT116 (ATCC, Rockville, MD, USA) were cultured as described previously (8). Mouse embryonic stem (ES) cells (wild type (TT2), G9a-/- (22-10), GLP-/- (CD-10)) and G9a and GLP reintroduction cell lines were described previously (12, 13). Control or stable G9a knock-down HCT116 or RKO cell lines (described below) were treated with 5-aza-2′-deoxycytidine (DAC; decitabine) (Sigma, St Louis, MO) dissolved in phosphate-buffered saline (PBS), and were harvested 48 hours post-treatment. In other experiments, cells were treated for 24 hours with 600 nM Trichostatin A (TSA) (Sigma) solubilized in DMSO. The small molecule G9a inhibitor BIX-01294 (23) was solubilized in DMSO and used to treat control or stable G9a knock-down HCT116 or RKO cell lines for 48 hours.
Transient and stable RNAi knockdown experiments
For transient knockdown of G9a or GLP (EuHMTaseI), RKO cells were transfected with 5-50 nM siRNA (Ambion, Austin, TX) using Lipofectamine 2000 (Invitrogen). As a negative control, RKO cells were transfected with 5-50 nM of siControl Non-Targeting siRNA #2 (Dharmacon, Lafayette, CO.). siRNA transfections occurred on day zero and day two, and cells were harvested on day five. For stable knockdown, a shRNA specific for human G9a was designed using the shRNA insert design tool (Ambion). The shRNA duplex was ligated into the pSilencer 3.1 H1 neo vector (Ambion). After sequence verification, the G9a shRNA plasmid, or a control shRNA vector (Ambion) were transfected into RKO and HCT116 cells using Lipofectamine 2000 (Invitrogen). Antibiotic selection was performed using 1.5 mg/ml (RKO) or 400 μg/ml (HCT116) G418 (Mediatech, Herndon, VA) and was continued until all mock-transfected control cells were killed. G9a shRNA and control shRNA expressing cell clones were picked and expanded while maintaining in normal media supplemented with G418 at one half of the antibiotic kill concentration. In combination shRNA/siRNA experiments, control or stable G9a knockdown RKO cells were transfected with various siRNAs using Lipofectamine 2000 on day 0 and day 2, and cells were harvested on day 5. siRNA and shRNA sequences are shown in Supplemental Table 1.
Colony formation assays
Equivalent numbers of RKO and HCT116 cells were transfected with control or G9a shRNA plasmids, and antibiotic selection was performed on cells grown in 15 cm tissue culture plates. For colony counts, cells were washed in PBS, fixed in 10% formaldehyde, and stained with 0.1% crystal violet in methanol. Colonies were counted manually.
Reverse transcriptase PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA), and cDNA was generated using the First Strand cDNA synthesis kit (Fermentas, Hanover, MD). RT-PCR and qRT-PCR amplifications of human MAGE-A1, NY-ESO-1, XAGE-1, and GAPDH were performed as described previously (8). Primer sequences for murine Mage-a, Mage-a2, Mage-a8, and Gapdh were designed using Primer 3 (38) and are shown in Supplemental Table 1. Murine pan Mage-a primers were adapted from a previous study (39), and amplify mouse Mage-a2, -a5, -a6, and –a8. Primers were obtained from IDT (Coralville, IA).
Western blot analyses of G9a and GLP
Western blots of whole cell protein extracts were performed as described previously (8). G9a was detected using the D141-3 primary antibody (MBL International Corp. Woburn, MA) at a 1:100 dilution followed by Protein A HRP (Amersham Biosciences, Piscataway, NJ) at a 1:1000 dilution. Alternatively, G9a was detected using a polyclonal antibody (kind gift from Dr. Sriharsa Pradhan, New England Biolabs) at a 1:1000 dilution followed by anti-Rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:10,000 dilution. GLP (EuHMTaseI) was detected using primary antibody D220-3 (MBL International, Woburn, MA) at 1 μg/ml, in combination with Protein A HRP at a 1:1000 dilution. α-Tubulin was detected as described previously (5). Proteins were detected using Western Lightning Chemiluminescence Reagent (Perkin Elmer Life Sciences, Boston, MA) and the Versadoc imaging system (BioRad, Hercules, CA).
Measurement of Histone Modifications
Global levels of histone H3 lysine 9 dimethylation (H3K9me2), lysine 27 dimethylation (H3K27me2), and lysine 9 acetylation (H3K9ac) were measured by Western blotting of acid-soluble nuclear protein extracts. Extracts were prepared according to the Upstate Acid Extraction protocol (Upstate Biotechnology) and dialysis was accomplished using extra strength 10,000 MWCO Slide-A-Lyzer Dialysis Cassettes (Pierce, Rockford, IL). Extracts were fractionated by SDS-PAGE and Coomassie staining was used to confirm equivalent histone protein loading. Primary antibodies for Western blot and ChIP analysis specific for H3K9me2 (Cat #07-441), H3K27me2 (Cat #07-452), H3K9ac (Cat#07-352), and H3K4me2 (Cat #07-030) were obtained from Upstate Biotechnology (Waltham, MA), and secondary antibodies were obtained from Amersham Biosciences. Western blot detection was performed as described above. Quantitative chromatin immunoprecipitation assays (qChIP) were used to detect histone code modifications at the MAGE-A1 and XAGE-1 promoters as described previously (8).
Densitometry analysis of Western blots
Protein densitometry was used to quantify the extent of siRNA and shRNA knockdowns and histone modification levels on Western blots. Chemiluminescent imaging of Western blots utilized VersaDoc (Biorad) and signal quantification was performed with Quantity One software (Biorad), using background signal correction. G9a and GLP levels in total protein extracts were normalized by the level of alpha-tubulin in sequentially probed blots. Histone modification levels in acid soluble protein extracts were not normalized; however, Coomassie staining was used to visually confirm equivalent protein input. All Western blots were loaded with equivalent amounts of protein per lane.
DNA methylation analysis of CG antigen genes
DNA methylation levels of human MAGE-A1, NY-ESO-1, and XAGE-1 were determined using quantitative sodium bisulfite pyrosequencing (40). NY-ESO-1 pyrosequencing primers were described previously (41). Pyrosequencing assays specific for the 5′ CpG island or CpG-rich regions of human MAGE-A1 and XAGE-1 were designed using the Pyrosequencing Assay Design Software (Biotage) and the primers are shown in Supplemental Table 1. All primers were obtained from IDT. Genomic DNA samples were isolated using the Puregene kit (Gentra Systems, Minneapolis, MN) and sodium bisulfite conversion and quantitative pyrosequencing was accomplished as described previously (10, 41). Of the murine Mage-a genes, only Mage-a2 (Region: -179 to +54, 234 bp, 51% GC, Obs/Exp = 0.6) and Mage-a8 (Region: -127 to +92, 220 bp, 52% GC, Obs/Exp = 0.6) contain 5′ CpG islands meeting the classical definition (27) (data not shown), and thus were selected for further analysis. Mage-a2 methylation was determined using methylation specific PCR (MSP) (42), using the primers shown in Supplemental Table 1. Mage-a8 methylation was determined using bisulfite pyrosequencing as described above, using the primers shown in Supplemental Table 1. Controls included DNA methylated in vitro with SssI (New England Biolabs), and unmethylated DNA generated as described previously (43).
Global genomic DNA methylation analyses
Global genomic DNA methylation was measured by determination of 5-methyl-deoxycytidine (5mdC) levels using a quantitative liquid chromatography mass spectrometry (LC-MS) method (44), or by sodium bisulfite pyrosequencing of the murine SINE B1 or human LINE-1 repetitive elements as described previously (28, 41). Primers were obtained from IDT.
Supplementary Material
Acknowledgments
We thank Dr. E. Michael August of Boehringer Ingelheim for kindly providing BIX-01294. This work was supported by the NIH (RO1CA11674), The Ralph Wilson Biomedical Research Foundation, and Phi Beta Psi (to A.R.K), and by NCI Center Grant CA16056 (to RPCI).
References
- 1.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 2.Karpf AR. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics. 2006;1:116–20. doi: 10.4161/epi.1.3.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Smet C, Loriot A, Boon T. Promoter-Dependent Mechanism Leading to Selective Hypomethylation within the 5′ Region of Gene MAGE-A1 in Tumor Cells. Molecular and cellular biology. 2004;24:4781–90. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Molecular and cellular biology. 1999;19:7327–35. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol. 2004;65:18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 6.Weber J, Salgaller M, Samid D, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54:1766–71. [PubMed] [Google Scholar]
- 7.Weiser TS, Guo ZS, Ohnmacht GA, et al. Sequential 5-Aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother. 2001;24:151–61. doi: 10.1097/00002371-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 8.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25:6975–85. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 9.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–49. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 10.Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- 11.Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–24. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 12.Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–26. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteve PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S. Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic acids research. 2005;33:3211–23. doi: 10.1093/nar/gki635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 16.Esteve PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo Y, Shen L, Ahmed S, et al. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikegami K, Iwatani M, Suzuki M, et al. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. The Journal of biological chemistry. 2001;276:25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 20.Chin HG, Pradhan M, Esteve PO, Patnaik D, Evans TC, Jr, Pradhan S. Sequence specificity and role of proximal amino acids of the histone H3 tail on catalysis of murine G9A lysine 9 histone H3 methyltransferase. Biochemistry. 2005;44:12998–3006. doi: 10.1021/bi0509907. [DOI] [PubMed] [Google Scholar]
- 21.Collins RE, Tachibana M, Tamaru H, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. The Journal of biological chemistry. 2005;280:5563–70. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nature immunology. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 23.Kubicek S, O'Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 25.Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–94. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 26.Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. The Journal of biological chemistry. 2003;278:14996–5000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 27.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. Journal of molecular biology. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 28.Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res. 2008;6:1365–74. doi: 10.1158/1541-7786.MCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7149–53. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wischnewski F, Friese O, Pantel K, Schwarzenbach H. Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters. Mol Cancer Res. 2007;5:749–59. doi: 10.1158/1541-7786.MCR-06-0364. [DOI] [PubMed] [Google Scholar]
- 31.Dong KB, Maksakova IA, Mohn F, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. The EMBO journal. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. The EMBO journal. 2008;27:2681–90. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 34.Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell stem cell. 2008;2:123–34. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 36.Takebayashi S, Tamura T, Matsuoka C, Okano M. Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Molecular and cellular biology. 2007;27:8243–58. doi: 10.1128/MCB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigalotti L, Coral S, Fratta E, et al. Epigenetic modulation of solid tumors as a novel approach for cancer immunotherapy. Seminars in oncology. 2005;32:473–8. doi: 10.1053/j.seminoncol.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Rozen S, Skaletsky H. Methods in molecular biology. Vol. 132. Clifton, NJ: 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–86. [DOI] [PubMed] [Google Scholar]
- 39.De Plaen E, De Backer O, Arnaud D, et al. A new family of mouse genes homologous to the human MAGE genes. Genomics. 1999;55:176–84. doi: 10.1006/geno.1998.5638. [DOI] [PubMed] [Google Scholar]
- 40.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 41.Woloszynska-Read A, Mhawech-Fauceglia P, Yu J, Odunsi K, Karpf AR. Intertumor and Intratumor NY-ESO-1 Expression Heterogeneity Is Associated with Promoter-Specific and Global DNA Methylation Status in Ovarian Cancer. Clin Cancer Res. 2008;14:3283–90. doi: 10.1158/1078-0432.CCR-07-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochemical and biophysical research communications. 2005;329:219–23. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- 44.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–10. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.