Abstract
Over the last decade the zebrafish has emerged as a major genetic model organism. While stimulated originally by the utility of its transparent embryos for the study of vertebrate organogenesis, the success of the zebrafish was consolidated through multiple genetic screens, sequencing of the fish genome by the Sanger Centre, and the advent of extensive genomic resources. In the last few years the potential of the zebrafish for in vivo cell biology, physiology, disease modeling and drug discovery has begun to be realized. This review will highlight work on cardiac electrophysiology, emphasizing the arenas in which the zebrafish complements other in vivo and in vitro models; developmental physiology, large scale screens, high-throughput disease modeling and drug discovery. Much of this work is at an early stage, and so the focus will be on the general principles, the specific advantages of the zebrafish and on future potential.
Introduction
Spontaneous arrhythmias have proven difficult to model in vivo, in part due to our incomplete understanding of the etiology of most clinical rhythm disorders. Multiple elements including genetic predisposition, extrinsic injury, environmental exposures and stochastic processes all contribute to the final arrhythmia, but may vary widely in their contributions to individual arrhythmias or even to distinct arrhythmic events in the same individual(Keating and Sanguinetti 2001; Nerbonne and Kass 2005). Implicit in this multi-step model is the fact that, for most arrhythmias, individuals may exhibit several of the elements of an arrhythmogenic state, but relatively few ever experience a clinical event(Keating and Sanguinetti 2001; Roden et al. 2002). An ideal animal model would recapitulate not just individual components of the causal chain leading to an arrhythmia, but each step along the way. Though there is a wealth of animal models, several areas of in vivo electrophysiology have remained relatively inaccessible including; the developmental patterning of myocardial excitability and coupling, in vivo cell biology and the efficient identification of channel partner proteins. The classic genome-wide screening techniques which have made genetic models such as yeast, nematode or drosophila such powerful tools in the dissection of cell signaling and other phenotypes have not proven feasible for many physiologic traits including cardiac electrophysiology(Warren and Fishman 1998; Jorgensen and Mango 2002; St Johnston 2002). The advent of such models would not only facilitate the rigorous exploration of arrhythmia mechanisms, but also allow systematic approaches to the discovery and testing of novel drugs, or other interventions(Warren and Fishman 1998; Briggs 2002; MacRae and Peterson 2003).
Rationale for the zebrafish
The zebrafish emerged originally as a model for the study of vertebrate development, largely as a result of its transparency, fecundity and diploid genome(Haffter and Nusslein-Volhard 1996). The impetus behind its transformation, in the mid-1990s, into a major genetic model organism came from several large genetic screens that were focused on vertebrate organogenesis(Driever et al. 1996; Haffter et al. 1996). The zebrafish filled a niche as a screenable model organism in the phylogenetic gap between invertebrates such as nematode and Drosophila and the genetically tractable mammals, specifically rodents. The early decision to sequence the zebrafish genome consolidated the organism as a model(Vogel 2000).
The zebrafish is in many ways ideal for phenotype-based screens(Patton and Zon 2001; Grunwald and Eisen 2002; Anderson and Ingham 2003). Its genome and body plan are similar to other vertebrates, but its optical transparency and external development make real time observation of its internal organs straightforward (Fig. 1). The optical clarity of the zebrafish embryo allows the use of fluorescent markers that highlight the locations or activities of specific populations of cells(Gong et al. 2001; Beis and Stainier 2006). Many transgenic zebrafish lines have been created that express fluorescent proteins (Fig. 2) in locations ranging from the presomitic mesoderm(Gajewski et al. 2003) to the pituitary gland(Liu et al. 2003). While the original genetic screens were based on ENU mutagenesis, insertional mutagenesis using retroviruses or transposons also enables efficient genetic screens or enhancer trap approaches to physiologic phenotypes(Amsterdam et al. 1999; Balciunas et al. 2004; Asakawa et al. 2008). The advent of FRET-based and other reporter systems has extended the scope of such transgenic tools from anatomy to physiology, and is facilitating a new generation of genetic screens(Chi et al. 2008). Numerous zebrafish disease models ranging from congenital heart defects to cancers have been identified in screens or developed(Amatruda et al. 2002; Penberthy et al. 2002; Shin and Fishman 2002), and the zebrafish is genetically and pharmacologically similar to humans(Langheinrich 2003; Milan et al. 2003).
Figure 1.
Phenotyping is straightforward as zebrafish larvae are transparent
(a), Phenotype at 48 hpf of an embryo injected with mismatch control oligo compared with a morphant (injected with 25 ng of morpholino dsc2) embryo at the same stage (b). High-resolution pictures of the heart of morphant embryo (c) compared to control-injected embryo (d). Transmission electron microscopy of cardiac desmosomes from a control embryo (e) and morphant embryo (f) at 48 hpf.
Figure 2.

Transgenic fluorescent reporters
In this figure a transgenic construct with a fluorescent reporter downstream of a hypertrophy responsive promoter enable direct in vivo assessment of physiologic pathways. This approach can readily be adapted to automated screening.
Adults reach 3 cm in length, but during the embryonic and larval stages the zebrafish is only about 1–2 mm long. During these stages, developing fish can live for days in the wells of a standard 96 or 384-well plate (Fig. 3), surviving on nutrients stored in their yolk sacs(Barut and Zon 2000). Zebrafish are inexpensive to raise, and a single pair of adults can routinely lay hundreds of fertilized eggs in a single morning. Consequently, even a small zebrafish facility can generate many thousands of embryos per day, making it possible to perform large-scale, phenotype-based screens(Driever, Solnica-Krezel et al. 1996; Haffter and Nusslein-Volhard 1996).
Figure 3.

Zebrafish can be maintained in multiwell plates
The size of the zebrafish enables several embryos to survive for days in the wells of 96 or 384 well plates. Combining this with automated phenotyping allows directed genetic or chemical screens of a scale similar to those undertaken in invertebrate model organisms, yet for vertebrate traits
The ease with which zebrafish phenotypes can be identified has resulted in their use in numerous genetic and chemical screens(Anderson and Ingham 2003; MacRae and Peterson 2003). For the cardiac electrophysiologist, an additional adavntage is the ability of the zebrafish to survive for several days by diffusion alone in the absence of any cardiovascular function, enabling screens for extreme phenotypes (Warren and Fishman 1998). Of note, because screening can be performed in the whole organism, perturbation of potential therapeutic targets by mutations or small molecules reveals the effects of such perturbations on the integrated physiology of the entire organism(MacRae and Peterson 2003; Zon and Peterson 2005).
As zebrafish have become more widely used, additional genetic and genomic technologies have been developed, increasing the utility of the system further. The zebrafish genome project is now nearly complete, and DNA microarrays have been generated for expression profiling studies(Stickney et al. 2002; Ton et al. 2002). Antisense morpholino oligos have proven to be an effective means of “knocking down” gene function(Nasevicius and Ekker 2000). More recently, reverse genetic approaches using zinc finger nucleases have been developed for the zebrafish, enabling researchers to generate mutations in virtually any gene of interest(Wienholds et al. 2002). The zebrafish is rapidly becoming a mature model organism, armed with an impressive collection of genomic and physiologic tools. These tools not only broaden the scope of whole-organism genetic or chemical screens that can be imagined, but also facilitate translation at every level to other model systems, or to human(Shin and Fishman 2002).
Developmental electrophysiology
While the zebrafish may offer advantages over the mouse in the modeling of some adult electrophysiologic phenomena(Salama and London 2007), it is as a developmental organism that the organism is currently enjoying its greatest popularity. The earliest electrophysiologic phenomena in cardiogenesis are accessible in the zebrafish. It is possible to record calcium and voltage transients in the cardiac primordia prior to their midline migration and fusion into the primitive heart tube(Creton et al. 1998). As each physiological event occurs during development it can be visualized in vivo. While similar approaches are feasible in chick and Xenopus, the genetic tractability of the zebrafish allows unique insights into function at stages of cardiogenesis when events are barely accessible in mammals(Moorman and Christoffels 2003; Sedmera et al. 2003).
Several of the morphologic mutants identified in the original morphological screens (for defects in organogenesis) exhibited arrhythmias recognizable with a simple dissection microscope. These included the bradycardic line slo mo, identified in the India strain background prior to mutagenesis, and several mutants with variable degrees of sinoatrial or atrioventricular heart block(Baker et al. 1997; Arnaout et al. 2007). Mutants were also identified with silent chambers, though several of these were rapidly shown, using simple calcium sensitive dyes, to represent defects in excitation-contraction coupling, or contractility rather than failures of electrical impulse propagation(Chen et al. 1996; Stainier et al. 1996). In addition to these recognizable orthologs of human arrhythmias, there were several mutants that exhibited recessive lethal phenotypes which suggested that they might offer insights into cardiac electrophysiology inaccessible in other models. These included mutants such as tremblor (episodic irregular waves of contraction across the entire heart)(Langenbacher et al. 2005), island beat (isolated atrial cell contractions with a silent ventricle)(Rottbauer et al. 2001) and reggae (sinoatrial exit block and rare impulse propagation to the atrium and ventricles)(Hassel et al. 2008). Calcium imaging using dyes such as calcium green established the primary electrical nature of these mutations, but as positional cloning of these novel mutants began, the phenotypic observations resulted in efforts to begin more detailed electrophysiological exploration of the zebrafish heart(Sedmera, Reckova et al. 2003).
Baker and colleagues were able to isolate and patch clamp not only adult cardiomyocytes,(Baker, Warren et al. 1997) but also those from early embryonic stages. Though technically challenging as a result of the cell size (capacitances are in the 1–2 pF range), they were able to identify multiple physiologic currents as early as 3 days post fertilization. The recapitulation of the majority of cardiac conductances combined with the similarities between human and zebrafish heart rates and action potential durations suggested that for many electrophysiological phenomena the zebrafish might prove to be a faithful model(Baker, Warren et al. 1997; Warren and Fishman 1998).
These parallels were subsequently extended to cardiac pharmacology. Several groups have demonstrated substantial homology in the pharmacological responses of the heart to a broad range of pharmacological agents(Langheinrich et al. 2003; Milan, Peterson et al. 2003). Therapeutic and toxic responses for both cardiac and non-cardiac drugs are reproduced, usually by simple immersion of the fish in drug. In many instances, the dose response curves are shifted sharply to the right, but ex vivo analyses have established that this is almost always related to effects of the physicochemical properties of the drug on penetration into the embryo(Milan, Peterson et al. 2003; Milan et al. 2006). These problems are largely predictable based on indices such as the logP, and can be overcome by direct injection(Peterson et al. 2000; Zon and Peterson 2005). In these early studies of cardiac pharmacology, it was possible to exploit automated microscopy to assess simple physiologic endpoints in high throughput in multiwell-plates(Milan, Peterson et al. 2003; Burns et al. 2005). Together these data suggested that not only would drugs identified in the zebrafish potentially be of use in humans, but also that large scale screening for physiological or pharmacological phenotypes would ultimately be feasible(MacRae and Peterson 2003). As the original screen mutants were cloned, so the importance of a developmental electrophysiological model such as the zebrafish has been reinforced. Many of these mutants could not have been identified or studied in higher organisms as the phenotypes would have resulted in death in utero.
One of the earliest mutants to be cloned was island beat, found to be a null allele in the L-type calcium channel(Rottbauer, Baker et al. 2001). The absence of a ventricular action potential during the first 48 hours of development is perhaps not surprising given the emergence of sodium current later in cardiogenesis and the consequent dependence of the primitive heart tube on calcium current. However, the mechanism by which loss of calcium current results in uncoupling of the atrial cardiomyocytes remains obscure. The independent depolarization and contraction of individual atrial cells is quite distinct from atrial fibrillation, but more extensive characterization of the effects of the loss of calcium channel conductance on cell-cell coupling has not yet been performed. The failure of pharmacologic inhibition of the L-type calcium channel to recapitulate this phenomenon may suggest that scaffolding, transcriptional or other functions of the channel may be perturbed in the mutant(Rottbauer, Baker et al. 2001).
The tremblor phenotype was found to result from homozygosity for a null allele in a cardiac sodium-calcium exchanger: NCX1(Ebert et al. 2005; Langenbacher, Dong et al. 2005). This loss of calcium extrusion from the cell appears to not only result in effects on cardiomyocyte differentiation but also leads to cyclical changes in impulse propagation throughout the heart. The presumed calcium overload leads to episodic waves of atrial arrhythmia interspersed with sinus rhythm, suggesting that there are other less efficient mechanisms for calcium extrusion that can intermittently suffice in the atrium(Ebert, Hume et al. 2005). However, the ventricle is virtually silent apparently as a result of calcium overload. There are also profound effects on ventricular cardiomyocyte differentiation and sarcomere organization that are not present in either island beat mutants or in knockdown of SERCA2 function using drug or morpholino(Ebert, Hume et al. 2005).
Two distinct classes of mutant from the original screen have been demonstrated to result from defects in the zebrafish ortholog of ether–a-go-go or KCNH2. Several alleles of breakdance, exhibiting variable atrioventricular block, have been identified and found to result from null or hypomorphic KCNH2 alleles(Langheinrich, Vacun et al. 2003; Arnaout, Ferrer et al. 2007). These phenotypes resemble those observed in severe forms of human long QT where markedly prolonged action potentials lead to intermittent heart block even in the fetus(Presbitero et al. 1989). Conversely, activating mutations in KCNH2 lead in homozygous form to the reggae phenotype with only intermittent exit of impulses from the sinus venosus, and a failure of atrial or ventricular escape(Hassel, Scholz et al. 2008). In this mutant the possibility of atrial fibrillation was again raised but the efficient entrainment of the atrium by external stimuli suggests that this is unlikely to be the case. Heterozygous forms of both these classes of KCNH2 mutant are viable and the development of techniques for the stable recording of adult electrocardiograms ex aqua has confirmed the predicted effects on QT interval(Milan, Jones et al. 2006). Together these data display the remarkably faithful relationship between human cardiac repolarization and that of the zebrafish as early as 48 hours post fertilization. The dependence of the fish heart rate on IKr observed by multiple investigators had led to more extensive efforts to exploit the organism in this arena (vide infra).
These early studies of electrical patterning have energized efforts to explore normal electrophysiological development in the zebrafish. Even in the initial morphologic zebrafish screens investigators had observed a clear transition from an initial peristaltic conduction across the entire heart tube to a sequential pattern of atrioventricular conduction with clear slowing at the AV canal(Stainier, Fouquet et al. 1996; Milan et al. 2006). Using calcium imaging with non-ratiometric dyes it was possible to define the functional transition in electrophysiologic properties in a ring of atrioventricular conduction tissue (only two or three cells across) at 40 hours post fertilization. Analysis of the zebrafish mutant cloche, which fails to develop endothelium, revealed a requirement for endocardial signals in this functional transition(Stainier, Fouquet et al. 1996; Milan, Giokas et al. 2006). With the range of physiologic manipulations feasible in the zebrafish, and its ability to survive for some time without any cardiac function, it was possible to characterize the differentiation of these specialized atrioventricular ring cells. Unlike adjacent endocardial cushions and valves, their specification is not dependent on blood flow or cardiac contraction. Exploiting morpholinos it was possible to demonstrate that neuregulin and notch1b signals are necessary for the development of atrioventricular conduction tissue. The combination of rapid genetic manipulation and high-resolution physiology allowed definition of the endocardial signals required for patterning central ‘slow’ conduction tissue, while revealing the operation of distinct local endocardial–myocardial interactions within the developing heart tube. The efficient nature of in vivo pathway exploration that is possible in the zebrafish with gene knockdown technologies in many ways parallels cell culture, yet in context of all of the native molecular, cellular and physiologic cues(Nasevicius and Ekker 2000).
The accessibility of these early events in the cell biology of cardiomyocytes will enable the study of many phenomena which have previously been relatively obscure; the emergence of excitation-contraction coupling, the serial appearance of distinct membrane currents and associated changes in the organization of the cell. For the first time the electrical coupling changes that accompany cardiomyocyte cell division can be explored in vivo. The ability to study this ontogeny is leading to a proliferation of studies designed to understand the role of electrical phenomena in myocardial patterning, the emergence of specialized conduction tissues and the complex innervation of the heart(Peterson, Link et al. 2000).
In elegant studies using a first generation transgenic gCAMP reporter(Chi, Shaw et al. 2008), it has been possible to follow in vivo some of the early electrical transitions that occur during cardiogenesis and myocardial maturation. These investigators were able to define discrete transitions in impulse propagation during the first few days of development, following the heart as it begins to conduct impulses more rapidly across the ventricular surfaces of growing trabeculae. While these reporters may lack the dynamic range necessary for some key electrophysiologic endpoints, they nevertheless illustrate the tremendous potential of the zebrafish as a model system for the study of physiology in vivo(Tallini et al. 2006). As novel functional reporters of other pathways emerge these can readily be deployed in the fish, and there are ongoing efforts to characterize a host of cellular, organelle and subcellular biology in vivo. Using optical mapping at millisecond resolution with voltage sensitive dyes and calcium imaging with ratiometric dyes such as Fura-2 it is possible to use electrical or calcium transients to follow cell fates within the developing myocardium, bypassing the need for specific transcriptional reporters. In this way detailed maps of the physiologic characteristics over time for each of the 300–350 cells within the organ can be constructed for the early stages of heart development. Ultimately it should prove possible to explore the mechanisms of cross-talk between electrical, mechanical and flow based modulation of cardiac development(Hove et al. 2003; Hove 2004; Hove 2006; Lee et al. 2006). These strategies to understand the sophisticated interactions between function and form cannot easily be matched in other organisms.
High-throughput cardiac biology
For some time investigators have recognized a need for high-throughput model systems capable of rapidly exploring the genetic basis of physiologic phenotypes, defining gene-gene and gene-environment interactions, and ultimately enabling a ‘systems approach’ to the genetics and genomics of integrative physiology(Walhout et al. 2002; Ge et al. 2003). In vitro systems enable high-throughput testing, but fail to recapitulate the complexity of the intact organism(Pepperkok and Ellenberg 2006), while traditional in vivo models are low-throughput and expensive(Kasarskis et al. 1998). In the last decade the zebrafish has emerged as a tool for rapidly assessing gene and small molecule function(Ekker 2000; Peterson, Link et al. 2000; MacRae and Peterson 2003). Investigators have explored the utility of the larval zebrafish for the defining small molecule targets based on phenoclustering, for high-throughput in vivo approaches to the study of drug toxicity and for empirical pathway discovery or the testing of large numbers of variables in genetically faithful disease models(Peterson, Link et al. 2000; Rual et al. 2004; Burns, Milan et al. 2005; Milan, Jones et al. 2006). Within 48 hours of fertilization, the larval fish has an established complex physiology, yet can be sustained in large numbers for days in multi-well plates(Briggs 2002). It has proven possible to develop techniques for measuring physiological variables in an automated or semi-automated fashion at high-throughput(Burns, Milan et al. 2005). In addition, a repertoire of secondary higher resolution assays is emerging including cardiomyocyte number, optical voltage mapping, Ca2+ imaging, and specific transgenic reporters for mitochondrial Ca2+, classic transcriptional hypertrophy markers and or disease pathways (Fig. 4) (Gerull et al. 2004; Peterson et al. 2004; Burns, Milan et al. 2005; Schonberger et al. 2005; Burns and MacRae 2006; Heuser et al. 2006; Lee, Yu et al. 2006). These tools are enabling efficient large-scale pathway exploration of genomic datasets in a completely native context(MacRae and Peterson 2003; Peterson, Shaw et al. 2004).
Figure 4.
Staged screening
By combining different assays of varying throughput and resolution with the genetic tools available for the zebrafish, it is possible to optimize assays for both sensitivity and specificity. In this example, a highly sensitive (but relatively non-specific) first round assay (heart rate and AV block), which can be completely automated, is combined in series with high-resolution second round assays (calcium and voltage imaging), which offer markedly higher specificity. Together with known disease mutants and functional genomics approaches these can be used to develop a screening tool for drug-induced repolarization toxicity or in discovery mode for pharmacogenetic gene identification.
For example, the zebrafish is accelerating investigation of the desmosomal genes implicated in arrhythmogenic right ventricular cardiomyopathy (ARVC)(Heuser, Plovie et al. 2006). It has proven possible to model multiple disease alleles, including desmocollin, desmoplakin and plakophilin 2 mutations, and define synergies between these genes in myocardial biology. The earliest stages of pathogenesis for many human cardiac diseases can be modeled and the electrophysiological effects of these processes can be explored prior to the confounding secondary effects of chronic myocyte dysfunction or damage(Xu et al. 2002; Bassett and Currie 2004).
Genetic and chemical screens
Where it is possible to develop representative models for arrhythmias in the fish, the organism can be used for unbiased phenotype-driven screens focused on the genetic or chemical dissection of normal or abnormal cardiac electrical function. Phenotype-driven screens offer several advantages including: the potential for extreme perturbation of each gene, the possibility of true saturation, and the generation of massive numbers of informative meioses capable of illuminating gene-gene or gene-environment interactions. Where parallel phenotypes are available, human and model organism genetics complement each other directly. Representative and cost-effective in vivo models will be required for the detailed exploration of the effects of multiple linked genes in different tissues necessary to elucidate the effects of each locus identified in human genome wide association studies(Hirschhorn and Daly 2005; McCarthy and Hirschhorn 2008). The exploration of loci of modest effect size and gene-gene interactions may be prohibitively expensive for human studies, but the combination of systematic modeling with subsequent human validation can render these questions tractable(McCarthy and Hirschhorn 2008). For many traits of medical or physiological importance model organism screens may offer the only feasible solution.
The parallels observed between zebrafish and human cardiac repolarization and the importance of drug-induced repolarization toxicity made this an attractive area for early physiological and pharmacological screens. Combining an initial high-throughput screen for abnormal heart rate response to dofetilide with a second high-resolution assay in which confirmed mutants are studied using optical mapping(Burns, Milan et al. 2005; Milan, Giokas et al. 2006) it is possible to devise staged screening strategies with optimized throughput and resolution. This approach identifies mutants based on bimodal distributions of heart rate response to dofetilide. Subsequent testing in the absence of dofetilide allows discrimination between pure drug response phenotypes and intrinsic heart rate defects. Detailed electrophysiologic assessment can then be undertaken only for the screen early round ‘hits’.
Using this strategy in an initial shelf screen of 340 insertional mutants, 15 genes with major effects on repolarization were identified, none of which had previously been implicated in this process. Interestingly, the majority of these genes appear to belong to an integrin associated network modulating channels and their adaptor proteins (Figure 5). Investigators in the KORA and Framingham studies were able to establish that some of these same genes modify human repolarization, confirming the utility of zebrafish screens for gene discovery in physiologic or pharmacologic pathways (Refs pending).
Figure 5.
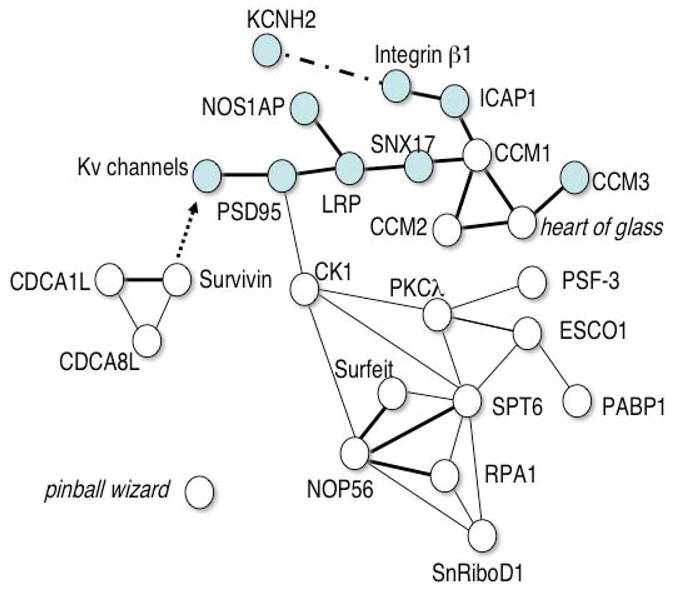
Repolarization network identified by zebrafish screening
Interactions between known repolarization genes (blue symbols) and the genes identified in a phenotype driven screen are depicted. Single lines indicate genetic interactions supported by data from multiple model organisms. Bold lines show direct physical interactions. A dashed line represents a physical interaction that may not be direct. The arrow represents a downstream regulatory effect, the mechanism of which is unknown.
The relatively high percentage of positive results in the zebrafish screen reflects the fact that the panel of insertional mutants used was originally selected on the basis of early recessive morphologic phenotypes identified by light microscopy(Amsterdam, Burgess et al. 1999). Cardiac developmental phenotypes had a high prevalence, presumably because the heart is readily visualized in the developing embryo and cardiac dysfunction leads to an obvious phenotype(Chen, Haffter et al. 1996). Electrophysiological perturbations frequently accompany disorders of cardiac form and function(Martin et al. 1994). Nevertheless, there were many mutants that, despite severe structural and functional cardiac defects, did not display an altered response to dofetilide.
The cross-talk between human and model systems is bidirectional with this type of integrated approach: phenotype-driven studies in the fish not only enable validation of loci implicated by genome-wide association, but also lead to the identification of novel genes regulating repolarization, which can then be confirmed in human populations. The work described above focused on a single complex trait, cardiac repolarization, but the scale of investigation feasible in multiple organ systems in the zebrafish strongly suggests that this integrative strategy will be useful both to aid in the identification of loci with modest effects from human studies and to explore the biologic mechanisms of the underlying genes. This type of scalable comparative physiology will be crucial to understanding the heritable basis of complex traits(Ge, Walhout et al. 2003).
New screens
Other potential screens are readily imagined. For example, robust reporters of repolarization might be exploited as a screen to identify cardiotoxicity early in the drug development process. Indeed, the scale of investigation that is feasible in the zebrafish might allow very detailed assessment of proarrhythmic potential across a broad range of compounds, enabling precise estimates of risk. Thus, the cardiotoxicity risks could be used in the selection of lead compounds, and titrated to the clinical problem in hand(Langheinrich 2003; Lieschke and Currie 2007).
As disease models are generated in the zebrafish, genetic or chemical modifier screens can readily be undertaken in vivo(Zon and Peterson 2005; Lieschke and Currie 2007). The feasibility of this approach has already been demonstrated for some structural phenotypes, where mutants with specific defects were exposed to 5,000 compounds from a diverse small molecule library. Two structurally related compounds were identified that completely restore the mutants to normal without causing additional developmental defects(Peterson, Shaw et al. 2004). The development of innovative automated screens for multiple physiologic parameters, allow this type of systematic chemical screen to be scaled to tens of thousands of compounds(Burns, Milan et al. 2005). Importantly, where identified those compounds that suppress disease phenotypes in zebrafish appear to have similar effects in mammalian systems and ultimately may have direct utility as lead compounds for human therapies. It is also possible that small molecule suppressor screens will reveal novel drug targets and mechanisms by which diseases can be modified. These targets could then be used for conventional drug development.
Regeneration
One area of particular interest in the zebrafish has been the study of cardiac regeneration. Poss and colleagues first demonstrated that distal amputation of the ventricle in the adult zebrafish, while resulting in some initial mortality (~20%), would lead to the formation of a coagulum with subsequent complete regeneration of the apex of the heart(Poss et al. 2002). Subsequent work, from Poss and others, has defined myocardial and epicardial contributions to this regenerative process with apparent dedifferentiation of myocytes, cell division and repopulation of the distal ventricle with differentiated cardiomyocytes(Lepilina et al. 2006). This remarkable process offers a unique experimental setting in which to explore the electrophysiology of cardiomyocyte differentiation. Understanding how new myocytes might successfully couple with existing myocardium will be critical if human myocardial regenerative medicine is ever to become a reality(Wu et al. 2008). In addition, understanding why complete regeneration is feasible in some species, yet postnatal cardiomyocyte cell division is so limited in other species may enable the manipulation of the biology for clinical use. It will also be important to understand how localized damage without tissue amputation will modify the regenerative process. Approaching the cell biology and electrophysiology of injured tissue in vivo, but in an organism where scar formation may not occur, or where it is altered by active cardiomyocyte regeneration, will hopefully offer insights into the mechanisms of ischemic ventricular arrhythmias. Interestingly, preliminary data suggest that after distal amputation the mode of death observed in the minority of fish is sudden and may be tachyarrhythmic.
In the embryonic zebrafish regeneration may be approachable in higher throughput. Several transgenic systems for cell-specific chemical ablation have been developed that enable the destruction of cardiomyocytes or subsets of cardiomyocytes in large numbers of embryos in parallel(Curado et al. 2007; Davison et al. 2007). Given that the key cellular components of the vertebrate heart have been identified in the zebrafish including endocardium, epicardium, chamber-specific myocardium, conduction system and even representation of the second heart field(Schoenebeck and Yelon 2007), it should prove possible to systematically address the effects of cell or tissue ablation rather than gene ablation on the electrophysiology of the heart. Where tissue specific promoters do not exist, lower throughput laser ablation studies can be used to define the effects of destruction of specific cell types on the development of normal physiology(Childs et al. 2002).
Genetic approaches to the logic of cardiomyocyte proliferation and differentiation have proven challenging, though progress is being made(Rossant 2008). Functional and other epigenetic inputs are emerging as substantial contributors to cell fate decisions and to the reinforcement of terminally differentiated cellular phenotypes, but have proven difficult to study systematically(Lammert et al. 2001; Cleaver and Melton 2003; Hove, Koster et al. 2003; Wu, Chien et al. 2008). Traditional analyses of development have focused largely on molecular markers to categorize cell identity, but the relationship between such markers and the cellular physiology is not always straightforward(Cleaver and Melton 2003; Epstein and Parmacek 2005). To overcome some of these limitations it may be possible to design strategies to annotate cardiomyocyte progenitor cell proliferation and differentiation in vivo using physiologic assays, generating a functional fate map of the early heart(Wu, Chien et al. 2008). Understanding such a fate map and its regulation will allow systems biology to begin to access not just the molecular networks but also the cellular networks of cardiogenesis.
Emerging areas of interest
While there has been a rapid proliferation in the use of the zebrafish as a genetic model for cardiac electrophysiology, to date investigators have only begun to scratch the surface of what is possible. Functional fate mapping was outlined above, but as the heterogeneity of myocardial cells is defined, determining the role of developmental physiology in cardiomyocyte differentiation will prove a major hurdle. As we understand the normal transitions between different stages of electrical development in health and disease, it is conceivable that these might be manipulated for therapeutic advantage, not only in hereditary arrhythmia, but also in other forms of congenital and acquired cardiac disease. Characterizing the physiologic changes during cardiomyocyte division will be necessary if we are to use cellular therapies while minimizing the risk of arrhythmia. It will also be possible to exploit the ontogeny of cardiac innervation not only to explore the interactions between the heart and its ingressing neurons, but to understand the impact of innervation on the components of arrhythmogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a cancer model system. Cancer Cell. 2002;1(3):229–31. doi: 10.1016/s1535-6108(02)00052-1. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13(20):2713–24. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV, Ingham PW. The transformation of the model organism: a decade of developmental genetics. Nat Genet. 2003;33(Suppl):285–93. doi: 10.1038/ng1105. [DOI] [PubMed] [Google Scholar]
- Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DY, Tristani-Firouzi M, Chi NC. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci U S A. 2007;104(27):11316–21. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105(4):1255–60. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K, Warren KS, Yellen G, Fishman MC. Defective “pacemaker” current (Ih) in a zebrafish mutant with a slow heart rate. Proc Natl Acad Sci U S A. 1997;94(9):4554–9. doi: 10.1073/pnas.94.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5(1):62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barut BA, Zon LI. Realizing the potential of zebrafish as a model for human disease. Physiol Genomics. 2000;2(2):49–51. doi: 10.1152/physiolgenomics.2000.2.2.49. [DOI] [PubMed] [Google Scholar]
- Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clin Exp Pharmacol Physiol. 2004;31(8):537–40. doi: 10.1111/j.1440-1681.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- Beis D, Stainier DY. In vivo cell biology: following the zebrafish trend. Trends Cell Biol. 2006;16(2):105–12. doi: 10.1016/j.tcb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Briggs JP. The zebrafish: a new model organism for integrative physiology. Am J Physiol Regul Integr Comp Physiol. 2002;282(1):R3–9. doi: 10.1152/ajpregu.00589.2001. [DOI] [PubMed] [Google Scholar]
- Burns CG, MacRae CA. Purification of hearts from zebrafish embryos. Biotechniques. 2006;40(3):274, 276, 278. passim. [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1(5):263–4. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Nusslein-Volhard C. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, Scott I, Beis D, Xiao T, Baier H, Jan LY, Tristani-Firouzi M, Stainier DY. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs S, Chen JN, Garrity DM, Fishman MC. Patterning of angiogenesis in the zebrafish embryo. Development. 2002;129(4):973–82. doi: 10.1242/dev.129.4.973. [DOI] [PubMed] [Google Scholar]
- Cleaver O, Melton DA. Endothelial signaling during development. Nat Med. 2003;9(6):661–8. doi: 10.1038/nm0603-661. [DOI] [PubMed] [Google Scholar]
- Creton R, Speksnijder JE, Jaffe LF. Patterns of free calcium in zebrafish embryos. J Cell Sci. 1998;111(Pt 12):1613–22. doi: 10.1242/jcs.111.12.1613. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304(2):811–24. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A. 2005;102(49):17705–10. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC. Morphants: a new systematic vertebrate functional genomics approach. Yeast. 2000;17(4):302–306. doi: 10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Parmacek MS. Recent advances in cardiac development with therapeutic implications for adult cardiovascular disease. Circulation. 2005;112(4):592–7. doi: 10.1161/CIRCULATIONAHA.104.479857. [DOI] [PubMed] [Google Scholar]
- Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D. Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development. 2003;130(18):4269–4278. doi: 10.1242/dev.00627. [DOI] [PubMed] [Google Scholar]
- Ge H, Walhout AJ, Vidal M. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 2003;19(10):551–60. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. Mutations in the desmosomal protein plakophilin–2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36(11):1162–4. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- Gong Z, Ju B, Wan H. Green fluorescent protein (GFP) transgenic fish and their applications. Genetica. 2001;111(1–3):213–25. doi: 10.1023/a:1013796810782. [DOI] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS. Headwaters of the zebrafish -- emergence of a new model vertebrate. Nat Rev Genet. 2002;3(9):717–24. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Haffter P, Nusslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int J Dev Biol. 1996;40(1):221–7. [PubMed] [Google Scholar]
- Hassel D, Scholz EP, Trano N, Friedrich O, Just S, Meder B, Weiss DL, Zitron E, Marquart S, Vogel B, Karle CA, Seemann G, Fishman MC, Katus HA, Rottbauer W. Deficient zebrafish ether-a-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation. 2008;117(7):866–75. doi: 10.1161/CIRCULATIONAHA.107.752220. [DOI] [PubMed] [Google Scholar]
- Heuser A, Plovie ER, Ellinor PT, Grossmann KS, Shin JT, Wichter T, Basson CT, Lerman BB, Sasse-Klaassen S, Thierfelder L, MacRae CA, Gerull B. Mutant desmocollin-2 causes arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2006;79(6):1081–8. doi: 10.1086/509044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Hove JR. In vivo biofluid dynamic imaging in the developing zebrafish. Birth Defects Res C Embryo Today. 2004;72(3):277–89. doi: 10.1002/bdrc.20019. [DOI] [PubMed] [Google Scholar]
- Hove JR. Quantifying cardiovascular flow dynamics during early development. Pediatr Res. 2006;60(1):6–13. doi: 10.1203/01.pdr.0000219584.22454.92. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–7. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet. 2002;3(5):356–69. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci U S A. 1998;95(13):7485–90. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–80. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294(5542):564–7. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, Philipson KD, Chen JN. Mutation in sodium-calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci U S A. 2005;102(49):17699–704. doi: 10.1073/pnas.0502679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langheinrich U. Zebrafish: a new model on the pharmaceutical catwalk. Bioessays. 2003;25(9):904–12. doi: 10.1002/bies.10326. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Vacun G, Wagner T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol Appl Pharmacol. 2003;193(3):370–82. doi: 10.1016/j.taap.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, Mericko P, Stadtfeld M, Zhou D, Cheng L, Graf T, MacRae CA, Lepore JJ, Lo CW, Kahn ML. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11(6):845–57. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Liu NA, Huang H, Yang Z, Herzog W, Hammerschmidt M, Lin S, Melmed S. Pituitary corticotroph ontogeny and regulation in transgenic zebrafish. Mol Endocrinol. 2003;17(5):959–66. doi: 10.1210/me.2002-0392. [DOI] [PubMed] [Google Scholar]
- MacRae CA, Peterson RT. Zebrafish-based small molecule discovery. Chem Biol. 2003;10(10):901–8. doi: 10.1016/j.chembiol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Martin AB, Garson A, Jr, Perry JC. Prolonged QT interval in hypertrophic and dilated cardiomyopathy in children. Am Heart J. 1994;127(1):64–70. doi: 10.1016/0002-8703(94)90510-x. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–65. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133(6):1125–32. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- Milan DJ, I, Jones L, Ellinor PT, MacRae CA. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am J Physiol Heart Circ Physiol. 2006;291(1):H269–73. doi: 10.1152/ajpheart.00960.2005. [DOI] [PubMed] [Google Scholar]
- Milan DJ, Peterson TA, Ruskin JN, Peterson RT, MacRae CA. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107(10):1355–8. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- Moorman AF, V, Christoffels M. Development of the cardiac conduction system: a matter of chamber development. Novartis Found Symp. 2003;250:25–34. discussion 34–43, 276–9. [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26(2):216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. [see comments.] Nature Genetics. 2000;26(2):216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85(4):1205–53. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2(12):956–66. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Shafizadeh E, Lin S. The zebrafish as a model for human disease. Front Biosci. 2002;7:d1439–53. doi: 10.2741/penber. [DOI] [PubMed] [Google Scholar]
- Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7(9):690–6. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97(24):12965–9. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22(5):595–9. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Presbitero P, Mangiardi L, Antolini R. Congenital long QT syndrome inducing 2:1 atrioventricular block: early detection in fetal life. Int J Cardiol. 1989;24(1):109–12. doi: 10.1016/0167-5273(89)90049-1. [DOI] [PubMed] [Google Scholar]
- Roden DM, Balser JR, George AL, Jr, Anderson ME. Cardiac ion channels. Annu Rev Physiol. 2002;64:431–75. doi: 10.1146/annurev.physiol.64.083101.145105. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132(4):527–31. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1(2):265–75. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14(10B):2162–8. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama G, London B. Mouse models of long QT syndrome. J Physiol. 2007;578(Pt 1):43–53. doi: 10.1113/jphysiol.2006.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Yelon D. Illuminating cardiac development: Advances in imaging add new dimensions to the utility of zebrafish genetics. Semin Cell Dev Biol. 2007;18(1):27–35. doi: 10.1016/j.semcdb.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37(4):418–22. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- Sedmera D, Reckova M, deAlmeida A, Sedmerova M, Biermann M, Volejnik J, Sarre A, Raddatz E, McCarthy RA, Gourdie RG, Thompson RP. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284(4):H1152–60. doi: 10.1152/ajpheart.00870.2002. [DOI] [PubMed] [Google Scholar]
- Shin JT, Fishman MC. From Zebrafish to human: modular medical models. Annu Rev Genomics Hum Genet. 2002;3:311–40. doi: 10.1146/annurev.genom.3.031402.131506. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–88. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–92. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Stickney HL, Schmutz J, Woods IG, Holtzer CC, Dickson MC, Kelly PD, Myers RM, Talbot WS. Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res. 2002;12(12):1929–34. doi: 10.1101/gr.777302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, Kotlikoff MI. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A. 2006;103(12):4753–8. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton C, Stamatiou D, Dzau VJ, Liew CC. Construction of a zebrafish cDNA microarray: gene expression profiling of the zebrafish during development. Biochem Biophys Res Commun. 2002;296(5):1134–42. doi: 10.1016/s0006-291x(02)02010-7. [DOI] [PubMed] [Google Scholar]
- Vogel G. Genomics. Sanger will sequence zebrafish genome. Science. 2000;290(5497):1671. [PubMed] [Google Scholar]
- Walhout AJ, Reboul J, Shtanko O, Bertin N, Vaglio P, Ge H, Lee H, Doucette-Stamm L, Gunsalus KC, Schetter AJ, Morton DG, Kemphues KJ, Reinke V, Kim SK, Piano F, Vidal M. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr Biol. 2002;12(22):1952–8. doi: 10.1016/s0960-9822(02)01279-4. [DOI] [PubMed] [Google Scholar]
- Warren KS, Fishman MC. “Physiological genomics”: mutant screens in zebrafish. Am J Physiol. 1998;275(1 Pt 2):H1–7. doi: 10.1152/ajpheart.1998.275.1.H1. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297(5578):99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132(4):537–43. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30(2):205–9. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]




