Abstract
Sertoli cells provide nutritional and physical support to germ cells during spermatogenesis. Sox8 encodes a high mobility group transcription factor closely related to Sox9 and Sox10. Sertoli cells produceSOX8 protein, and its elimination results in an age-dependent deregulation of spermatogenesis resulting in male infertility. This suggests that Sox8 is a critical regulator of Sertoli cell function for the maintenance of male fertility beyond the first wave of spermatogenesis. To better understand the roles of Sox8 in testicular development and maintenance of male fertility, we have performed a detailed analysis to explore its downstream genes. We have used mRNA expression profiling to identify affected genes in Sertoli cells in the mutant testes of 2-month-old mice. Expression profiling of the heterozygous and homozygous Sox8 mutant testes indicates alterations in several important spermatogenesis and blood-testis barrier genes, providing insight into the molecular basis of the defects in Sox8–/– testes beyond the first wave of spermatogenesis.
Key Words: Blood-testis barrier, Male fertility, Sertoli cell, Sry-box, Spermatogenesis
Introduction
Sox8 isa member of the SRY-box containing gene family and encodes a high mobility group (HMG) transcription factor that possesses the ability to bind and bend DNA, thus allowing the trans-activation of target genes (Giese et al., 1994; Pontiggia et al., 1994). The Sox family has been divided into 10 subgroups, A–J. Sox8 along with Sox9 and Sox10 compose the SoxE subgroup (Bowles et al., 2000). Sox8 is a product of Sertoli cells and is critical for the maintenance of adult male fertility (O’Bryan et al., 2008). Apart from their obvious supportive role for the seminiferous epithelium, the Sertoli cells harbor clones of differentiating germ cells in cytoplasmic crypts and provide the germ cells with nutrients and growth factors throughout their development (Petersen and Soder, 2006). Sertoli cells regulate the flow of nutrients, growth factors, and mitogens by tight junctions between adjacent cells, creating a structure called blood-testis barrier (Lui and Cheng, 2007). Disruption of Sertoli cell function is suggested to be the cause of pathogenesis in the testicular dysgenesis syndrome (TDS) (Skakkebaek et al., 2001).
Based on its expression pattern in developing testes and ability to induce Amh expression in vitro, Sox8 was initially believed to be a regulator of male sex determination, differentiation, or germ cell development (Schepers et al., 2002, 2003). However, genetic ablation of Sox8 in mice does not result in abnormal sexual development. The lack of defects in sex determination in the mutant mice could be the result of a functional compensation by other SoxE family members (Chaboissier et al., 2004; Koopman, 2005). Detailed analysis revealed that the Sox8–/– mice showed decreased adiposity (Sock et al., 2001) and premature osteoblast differentiation, which resulted in poor tarsal development and low bone density (Sock et al., 2001; Schmidt et al., 2005). We independently generated a Sox8 mutant mouse strain and found that Sox8–/– males rarely produced litters, while Sox8+/– males and Sox8–/– females appeared to be reproductively normal (O’Bryan et al., 2008). In embryonic stages, no overt morphological disruption of cord formation was noted in Sox8–/–embryos (O’Bryan et al., 2008). Very small numbers of young Sox8–/– mice can sire pups with reduced litter size suggesting that at least some sperm resulting from the first few waves of spermatogenesis can be fertile and then they progressively become sterile. Testes weight was normal up to 35 days after birth, but by 2 months of age, the testes of Sox8–/– mice were significantly smaller than those of control animals. Sox8–/– adult mice (35 days) displayed an obvious infertility phenotype after the first wave of spermatogenesis (O’Bryan et al., 2008). A failure of spermiation was evident in stage IX tubules in 1-month-old mice and was seen more frequently in 2- and 5-month-old mutant animals, and a complete loss of the cycle of the seminiferous epithelium was evident by 9 months. In the gonad, Sox8 is expressed in Sertoli cells and not in germ cells (Kennedy et al., 2007). SOX8 localization varied within Sertoli cells based on the stage of the spermatogenic cycle. Specifically, within stage I–IX tubules SOX8 protein is localized in nucleo- and cytoplasm, whereas in stage X–XII tubules SOX8 protein was seen only in Sertoli cell cytoplasm (O’Bryan et al., 2008). Sertoli cells usually create 4 to 5 different and ever changing microenvironments concurrently around germ cells. Taken together this suggests that Sox8 expression in Sertoli cells regulates a set of genes that are essential for male germ cell differentiation and spermatogenesis. Sertoli cells in Sox8–/– mice lose this ability in an age-dependent manner, which results in infertility due to the inability to establish full spermatogenesis and shows substantial deregulation of spermatogenesis at 2 months of age.
To gain insight into the molecular basis of this phenotype found in Sox8–/– males, we have performed a detailed expression analysis using testes of 2-month-oldhomozygousand heterozygous mutant mice to identify the downstream genes of Sox8 that would affect male fertility. By microarray analysis we have determined that loss of Sox8 affects the expression levels of mRNAs that are important for the functions of Sertoli cells, germ cells, and the blood-testis barrier. The abnormal expression of these genes suggests an involvement of Sox8,in part, in the regulation of testicular genes required for male fertility.
Materials and Methods
Mice
The generation of Sox8 mutantmice has been previously described (O’Bryan et al., 2008). In brief, a 2.6-kb locus of Sox8 including exons 1 and 2 was replaced with the lacZ expression cassette and the floxed Pgk-neo cassette. The Pgk-neo cassette was removed after germline transmission of the mutation by breeding with Mox2-Cre mice. All animal studies were approved by the National Institute of Environmental Health Sciences Animal Care and Use Committee. All experimental data were collected from a minimum of 3 animals of each genotype and stage.
Genotyping
Genotypes of pups were determined by PCR analysis of genomic DNA with primers S8a: 5′-GAG GAC AAA GAT TGG GTC CTG C-3′ and S8b: 5′-GAA GCG TTC GTC TGC TGC C-3′ to detect the wild-type allele (299 bp); and primers S8a: 5′-GAG GAC AAA GAT TGG GTC CTG C-3′ and S8c: 5′-GAT GAA ACG CCG AGT TAA ACG C-3′ to detect the mutant allele (553 bp).
Microarray Analysis
Total RNA was isolated from 2-month-old heterozygous and homozygous mutant mice using TRIzol (Invitrogen). Gene expression analysis was conducted using Agilent Whole Mouse Genome 4×44 multiplex format oligo arrays (014868) (Agilent Technologies) following the Agilent 1-color microarray-based gene expression analysis protocol. Starting with 500 ng of total RNA, Cy3 labeled cRNA was produced according to the manufacturer's protocol. For each sample, 1.65 mg of Cy3 labeled cRNA was fragmented and hybridized for 17 h in a rotating hybridization oven. Slides were washed and then scanned with an Agilent Scanner. Data were obtained using the Agilent Feature Extraction software (v9.5), using the 1-color defaults for all parameters.
The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. The resulting data were processed using the Rosetta Resolver® system (version 7.0) (Rosetta Biosoftware, Kirkland, WA). In order to identify differentially expressed genes, analysis of variance (ANOVA) was used to determine if there was a statistical difference between the mean heterozygous and homozygous Sox8 mutants. Specifically, an error-weighted ANOVA with multiple test corrections was performed using Rosetta Resolver. The Benjamini Hochberg False Discovery Rate multiple test correction was used to reduce the number of false positives. Probes with p <0.01 were considered to be differentially expressed.
Quantitative PCR
Heterozygous and homozygous mutant mice were euthanized on postnatal day 15 (P15), P20, P25, P35, and at 2, 3, and 5 months of age. RNA was isolated from testes using TRIzol (Invitrogen), and cDNA was synthesized using the SuperScript First-Strand Synthesis System (Invitrogen) with random hexamer primers. Quantitative real-time reverse transcriptase PCR (Q-RT-PCR) measurements of individual cDNAs were performed with the ABI Prism 7700 sequence detection system. Gene-specific primers for Pde4D (Mm01304777_m1), Cd164 (Mm01189607_m1), Cep2 (Mm01329789_m1), Cldn3 (Mm00515499_s1), and Cldn23 (Mm00510971_s1) were purchased as pre-designed TaqMan gene-specific probe and primer mixtures (PE Applied Biosystems). The TaqMan rodent glyceraldehyde-3-phosphate dehydrogenase (Gapdh) control reagent (PE Applied Biosystems) was used as an internal control. All measurements were performedin triplicate. Values were normalized to Gapdh using the 2–ΔΔCt method and expressed as means ± SEM.
Results
Expression Profiling and Validation
Sox8 is critical for the maintenance of male fertility (O’Bryan et al., 2008). Homozygous mutant mice were infertile except for a few that produced small litters of pups while young and then subsequently became infertile (O’Bryan et al., 2008). Weights of testes from Sox8–/– mice were normal at 20 and 35 days after birth, suggesting that alteration of gene expression may not be apparent at this age. The obvious infertility phenotypes were apparent in homozygous mutant mice at 2 months of age; testes were significantly smaller, the seminiferous epithelium showed histological abnormalities, and the number and mobility of sperm were significantly reduced (O’Bryan et al., 2008). Based on these previous observations, we chose 2-month-old testes for gene expression profiling to identify the gene alteration in Sertoli cells.
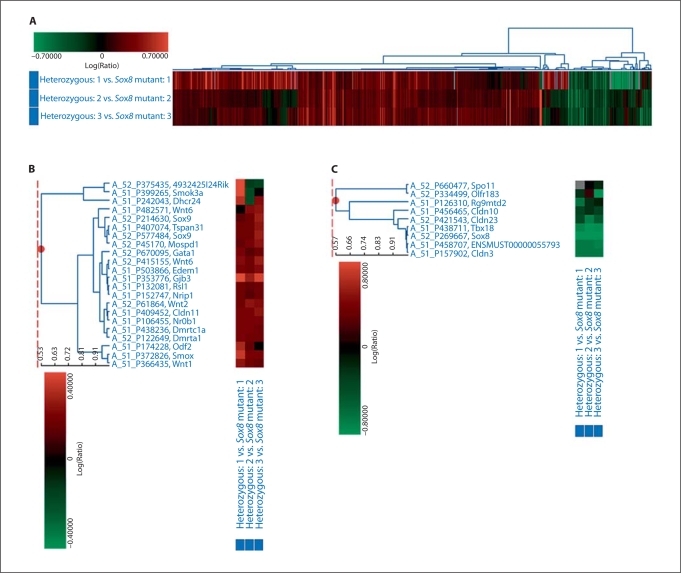
We performed three independent microarray comparisons of mRNA from testes of heterozygous and homozygous mice at 2 months of age. Statistical analysis identified 1,601 probes as differentially expressed between the homozygous Sox8 mutant testes and the heterozygous testes (fig. 1A). Of these, 1,314 probes were upregulated, and 108 thereof showed a 2-fold induction or more in the mutant testes (fig. 1B, table 1). 286 probes were downregulated, and 46 of these showed a 2-fold reduction or more in mutant testes at 2 months of age (fig. 1C, table 2). The targeted disruption of the Sox8 locus resulted in a complete absence of Sox8 mRNA. Neither the targeted exons (exon 1 and 2) nor the residual coding exon (exon 3) were expressed in the mutant animals (O’Bryan et al., 2008). As expected, Sox8 was listed as the most reduced gene in the mutant testes by the microarray comparison (26-fold reduction at 2 month of age).
Fig. 1.
Microarray comparison of the levels of gene expression in heterozygous and homozygous mutant testes. A Induced genes are indicated in shades of red, and repressed genes are indicated in shades of green for the 3 comparisons shown. B Representative graph of the upregulated genes in testes of Sox8–/–mice. C Representative graph of the downregulated genes in testes of Sox8–/–mice. Genes listed in B and C that are known to be involved in testicular development were selected from the results shown in A.
Table 1.
Genes with at least 2-fold increased expression in Sox8−/− testes at 2 months of age
| Sequence code | Gene symbol | Sequence description | Fold change | ANOVA p value (p <0.01) |
|---|---|---|---|---|
| A_51_P370363 | 1700030L22Rik | RIKEN cDNA 1700030L22 gene | 77.4 | 4.2E-14 |
| A_51_P433789 | Agr3 | Anterior gradient homolog 3 (Xenopus laevis) | 26.3 | 4.2E-14 |
| A_51_P433796 | Agr3 | Anterior gradient homolog 3 (Xenopus laevis) | 14.0 | 4.2E-14 |
| A_52_P731333 | 5730526G10Rik | 3 days neonate thymus cDNA, RIKEN full-length enriched library, clone: | 10.3 | 4.2E-14 |
| A630054F14 product: unclassifiable, full insert sequence | ||||
| A_52_P499523 | Scfd1 | Sec1 family domain containing 1 | 9.3 | 4.2E-14 |
| A_52_P739568 | Npas3 | 0 day neonate cerebellum cDNA, RIKEN full-length enriched library, clone: | 7.0 | 7.0E-06 |
| C230053P15, product: unclassifiable, full insert sequence | ||||
| A_52_P333567 | Adam33 | A disintegrin and metallopeptidase domain 33 | 6.3 | 2.0E-10 |
| A_51_P335555 | Snap25 | Synaptosomal-associated protein 25 | 5.3 | 7.7E-04 |
| A_51_P464420 | 4921508M14Rik | RIKEN cDNA 4921508M14 gene | 5.0 | 5.1E-04 |
| A_51_P458638 | Spink8 | Serine peptidase inhibitor, Kazal type 8 | 4.8 | 4.2E-14 |
| A_51_P180423 | Camp | Cathelicidin antimicrobial peptide | 4.2 | 4.3E-09 |
| A_51_P514177 | Lrrc2 | Leucine rich repeat containing 2 | 4.0 | 8.1E-07 |
| A_52_P420608 | Gm1631 | Gene model 1631NCBI | 3.9 | 4.0E-05 |
| A_52_P188295 | A930021C24Rik | RIKEN cDNA A930021C24 gene | 3.9 | 5.9E-10 |
| A_51_P131408 | Tnfrsf12a | Tumor necrosis factor receptor superfamily, member 12a | 3.8 | 1.0E-05 |
| A_51_P452533 | D1Ertd471e | DNA segment, Chr 1, ERATO Doi 471, expressed | 3.8 | 9.0E-05 |
| A_51_P340456 | Ela3 | Elastase 3, pancreatic | 3.7 | 4.2E-14 |
| A_51_P324633 | Elovl3 | Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 | 3.6 | 2.0E-05 |
| A_52_P339959 | Rab3b | RAB3B, member RAS oncogene family | 3.4 | 4.2E-14 |
| A_52_P298002 | Gch1 | GTP cyclohydrolase 1 | 3.4 | 6.0E-05 |
| A_51_P465211 | Wfdc2 | WAP four-disulfide core domain 2 | 3.3 | 4.2E-14 |
| A_52_P420357 | Slc15a1 | Solute carrier family 15 (oligopeptide transporter), member 1 | 3.3 | 3.3E-03 |
| A_51_P180091 | Cyp2s1 | Cytochrome P450, family 2, subfamily s, polypeptide 1 | 3.3 | 4.2E-14 |
| A_51_P302942 | Rasl10a | RAS-like, family 10, member A | 3.3 | 6.2E-11 |
| A_52_P261184 | Il1rl2 | Interleukin 1 receptor-like 2 | 3.2 | 1.0E-05 |
| A_52_P520788 | Scfd1 | Sec1 family domain containing 1 | 3.2 | 3.9E-09 |
| A_51_P340699 | Rasl11a | RAS-like, family 11, member A | 3.2 | 4.8E-09 |
| A_51_P304478 | Tmem28 | Transmembrane protein 28 | 3.2 | 5.0E-05 |
| A_51_P153042 | Psg16 | Pregnancy specific glycoprotein 16 | 3.2 | 1.5E-11 |
| A_52_P159365 | Sall3 | Sal-like 3 (Drosophila) | 3.2 | 5.8E-06 |
| A_52_P21550 | Gcnt1 | Glucosaminyl (N-acetyl) transferase 1, core 2 | 3.1 | 9.9E-06 |
| A_51_P385030 | Svs6 | Seminal vesicle secretory protein 6 | 3.1 | 4.2E-14 |
| A_52_P452256 | NAP055974-1 | Unknown | 3.1 | 1.1E-03 |
| A_51_P464387 | Hspb8 | Heat shock protein 8 | 3.0 | 4.2E-12 |
| A_52_P418952 | Krt79 | Keratin 79 | 3.0 | 4.2E-14 |
| A_52_P214347 | Cst11 | Cystatin 11 | 3.0 | 1.6E-10 |
| A_51_P138939 | Robo4 | Roundabout homolog 4 (Drosophila) | 2.9 | 1.6E-06 |
| A_51_P239203 | Mapk13 | Mitogen activated protein kinase 13 | 2.9 | 4.6E-09 |
| A_51_P308048 | Cmtm8 | CKLF-like MARVEL transmembrane domain containing 8 | 2.8 | 4.2E-14 |
| A_51_P496905 | Cfi | Complement component factor i | 2.8 | 1.4E-08 |
| A_52_P180933 | Defb8 | Defensin beta 8 | 2.7 | 5.0E-05 |
| A_52_P10622 | Emb | Embigin | 2.7 | 4.2E-14 |
| A_52_P211418 | 6030408C04Rik | RIKEN cDNA 6030408C04 gene | 2.7 | 1.0E-03 |
| A_51_P465449 | Mybpc3 | Myosin binding protein C, cardiac | 2.7 | 5.0E-07 |
| A_51_P382849 | Emb | Embigin | 2.6 | 1.6E-12 |
| A_51_P510466 | Pldn | Pallidin | 2.6 | 4.2E-14 |
| A_52_P964651 | 2310033K02Rik | RIKEN cDNA 2310033K02 gene | 2.6 | 1.0E-02 |
| A_51_P157255 | Sdc2 | Syndecan 2 | 2.4 | 4.2E-14 |
| A_51_P362638 | Trf | Transferrin | 2.4 | 4.2E-14 |
| A_52_P244682 | 5430435G22Rik | RIKEN cDNA 5430435G22 gene | 2.4 | 2.8E-06 |
| A_52_P644297 | Pafah1b2 | Adult male spinal cord cDNA, RIKEN full-length enriched library, clone: | 2.4 | 4.4E-03 |
| A330108B13, product: unclassifiable, full insert sequence | ||||
| A_52_P65237 | Zbtb7c | Zinc finger and BTB domain containing 7C | 2.4 | 4.2E-14 |
| A_51_P440365 | Frrs1 | Ferric-chelate reductase 1 | 2.4 | 2.0E-05 |
| A_51_P100327 | Tap1 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 2.4 | 3.0E-05 |
| A_52_P407049 | Hoxd10 | Homeo box D10 | 2.4 | 1.8E-07 |
| A_51_P267754 | Icam2 | Intercellular adhesion molecule 2 | 2.4 | 1.3E-07 |
| A_52_P217796 | 1500035H01Rik | RIKEN cDNA 1500035H01 gene | 2.3 | 3.0E-05 |
Table 2.
Genes with at least 2-fold decreased expression in Sox8−/− testes at 2 months of age
| Sequence code | Gene symbol | Sequence description | Fold change | ANOVA p value (p <0.01) |
|---|---|---|---|---|
| A_52_P269667 | Sox8 | SRY-box containing gene 8 | −25.65 | 6.1E-09 |
| A_52_P462657 | TC1060275 | Unknown | −14.25 | 2.3E-03 |
| A_51_P259848 | Ccdc32 | Coiled-coil domain containing 32 | −11.68 | 1.0E-03 |
| A_52_P421413 | Cd164 | Mus musculus adult male thymus cDNA, RIKEN full-length enriched library, clone: 5830453P09, product: CD164 antigen, full insert sequence | −10.20 | 4.2E-14 |
| A_51_P245324 | 4931402G19Rik | RIKEN cDNA 4931402G19 gene | −7.57 | 4.2E-14 |
| A_52_P269672 | Sox8 | SRY-box containing gene 8 | −5.31 | 4.2E-14 |
| A_51_P438711 | Tbx18 | T-box18 | −4.99 | 4.3E-08 |
| A_51_P120990 | Pde4d | Phosphodiesterase 4D, cAMP specific | −4.51 | 5.3E-03 |
| A_51_P458707 | ENSMUST00000055793 | OLFACTORY RECEPTOR GA_X6K02T2NHDJ-9643949-9642957 | −4.07 | 7.7E-03 |
| [Source: SPTREMBL; Acc: Q7TS05] [ENSMUST00000055793] | ||||
| A_51_P157902 | Cldn3 | Claudin 3 | −3.88 | 4.2E-14 |
| A_52_P392216 | Dab1 | Disabled homolog 1 (Drosophila) | −3.87 | 4.2E-14 |
| A_52_P529790 | TC1034687 | AK122264 mKIAA0377 protein (Mus musculus), partial (7%) [TC1034687] | −3.74 | 4.2E-14 |
| A_51_P115178 | Scara3 | Scavenger receptor class A, member 3 | −3.70 | 4.2E-14 |
| A_51_P131494 | Foxk2 | RIKEN cDNA 6230415M23 gene | −3.64 | 4.0E-05 |
| A_51_P326994 | 2810048G17Rik | RIKEN cDNA 2810048G17 gene | −3.57 | 6.2E-11 |
| A_51_P497295 | Qpct | Glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | −3.45 | 3.6E-13 |
| A_51_P176811 | Tspan10 | Tetraspanin 10 | −3.33 | 3.5E-09 |
| A_52_P1202828 | 6620401J10Rik | RIKEN cDNA 6620401J10 gene | −3.32 | 9.9E-04 |
| A_52_P1027837 | 9330175M20Rik | RIKEN cDNA 9330175M20 gene | −3.15 | 5.0E-05 |
| A_52_P246831 | Cep27 | Centrosomal protein 27 | −3.10 | 4.2E-14 |
| A_52_P192085 | Kcng2 | Potassium voltage-gated channel, subfamily G, member 2 | −2.94 | 4.2E-14 |
| A_51_P271200 | Slco1a5 | Solute carrier organic anion transporter family, member 1a5 | −2.81 | 6.8E-10 |
| A_51_P302831 | Dbc1 | Deleted in bladder cancer 1 (human) | −2.77 | 8.0E-03 |
| A_52_P377838 | AY172399 | Cell-line M2-86 immunoglobulin heavy chain variable region | −2.68 | 8.5E-03 |
| A_52_P110604 | Hmg20a | High mobility group 20A | −2.67 | 3.3E-03 |
| A_52_P421543 | Cldn23 | Claudin 23 | −2.56 | 2.5E-12 |
| A_52_P253044 | Syt13 | Synaptotagmin XIII | −2.47 | 4.2E-14 |
| A_51_P114094 | Clstn3 | Calsyntenin 3 | −2.44 | 4.2E-14 |
| A_51_P128807 | 4930485G23Rik | MAX gene associated | −2.42 | 2.6E-12 |
| A_52_P541095 | Unc5a | Unc-5 homolog A (C. elegans) | −2.39 | 4.2E-14 |
| A_52_P116102 | Tpi1 | Triosephosphate isomerase 1 | −2.39 | 5.0E-09 |
| A_51_P496253 | Slc6a4 | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | −2.34 | 1.2E-04 |
| A_51_P155152 | Ank | Progressive ankylosis | −2.34 | 1.1E-03 |
| A_52_P322096 | Patl2 | RIKEN cDNA 4930424G05 gene | −2.31 | 3.8E-04 |
| A_51_P258894 | Chst2 | Carbohydrate sulfotransferase 2 | −2.27 | 4.7E-04 |
| A_52_P1149183 | AI841723 | Transcribed locus | −2.22 | 6.0E-03 |
| A_51_P249335 | Sds | Serine dehydratase | −2.20 | 1.9E-07 |
| A_52_P185705 | Acn9 | Mus musculus adult male corpora quadrigemina cDNA, RIKEN full-length enriched library, clone: B230384K14, product: hypothetical ACN9 containing protein, full insert sequence | −2.15 | 8.6E-03 |
| A_51_P424739 | Nmnat3 | Nicotinamide nucleotide adenylyltransferase 3 | −2.15 | 3.6E-03 |
| A_52_P304105 | Ppp1r16b | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | −2.13 | 4.8E-10 |
| A_52_P723436 | D130062J10Rik | RIKEN cDNA D130062J10 gene | −2.08 | 1.3E-03 |
| A_52_P674189 | 4930512B01Rik | RIKEN cDNA 4930512B01 gene | −2.08 | 9.9E-03 |
| A_52_P67270 | 4930515G01Rik | Mus musculus RIKEN cDNA 4930515G01 gene (4930515G01Rik), mRNA | −2.06 | 6.3E-03 |
| A_52_P168575 | EG434197 | Predicted gene, EG434197 | −2.06 | 2.0E-05 |
| A_52_P599317 | Hs6st2 | Heparan sulfate 6-O-sulfotransferase 2 | −2.01 | 4.2E-14 |
| A_51_P386069 | Rab9b | RAB9B, member RAS oncogene family | −2.00 | 4.2E-14 |
Among the genes identified, some are known regulators of testicular development. These include Sex comb on midleg-like 4 (Scml4, 2.1-fold induction); SRY-box containing gene 9 (Sox9, 1.4-fold induction); cytochrome P450, family 2, subfamily s, polypeptide 1 (Cyp2s1, 3.3-fold induction); DMRT-like family C1a (Dmrtc1a, 1.3-fold induction); regulator of sex limited protein 1 (Rsl1, 1.3-fold induction); and Anti-Mullerian hormone type 2 receptor (Amhr2, 1.5-fold induction). Some of the chromatin remodeling factors known for gene repression were also upregulated in the testes of homozygous mice. These include histone deacetylases 6 (Hdac6, 1.3-fold induction) and chromodomain helicase DNA binding protein 6 (Chd6, 1.7-fold induction). In addition, the expression of spermatogenesis-associated genes was altered in the mutant testes, including phosphodiesterase 4D (Pde4D, 4.5-fold reduction), centrosomal protein 27 (Cep27, 3.1-fold reduction), sperm motility kinase 3A (Smok3a, 1.2-fold induction), and spermatogenesis associated, serine-rich 1 (Spats1, 1.2-fold reduction). Furthermore, there were changes in the expression of genes for proteins perhaps associated with the blood-testis barrier, including occludin (Ocln, 1.4-fold induction), claudin 11 (Cldn11, 1.4-fold induction), claudin 12 (Cldn12, 1.4-fold induction), claudin 10 (Cldn10, 1.5-fold reduction), claudin 3 (Cldn3, 3.9-fold reduction), and claudin 23 (Cldn23, 2.6-fold reduction) (table 1 and 2).
Expression Levels of the Downstream Genes for Sox8 in the Mutant Testes at 2 Months of Age
To validate the microarray results we further analyzed mRNA expression levels of genes by quantitative real-time reverse transcriptase PCR (Q-RT-PCR) in 2-month-old testes. Cldn23, Cep27, Cldn3, and Pde4D were chosen because their expression levels were downregulated more than 2-fold in the mutant, and they were known for their involvement in spermatogenesis and formation of the blood-testis barrier. We also included Cd164 because it encodes a cell surface antigen expressed higher in developing testes than in developing ovaries (McClive et al., 2003) and found it to be highly downregulated (10.2-fold reduction) in the 2-month-old mutant testes (table 2).
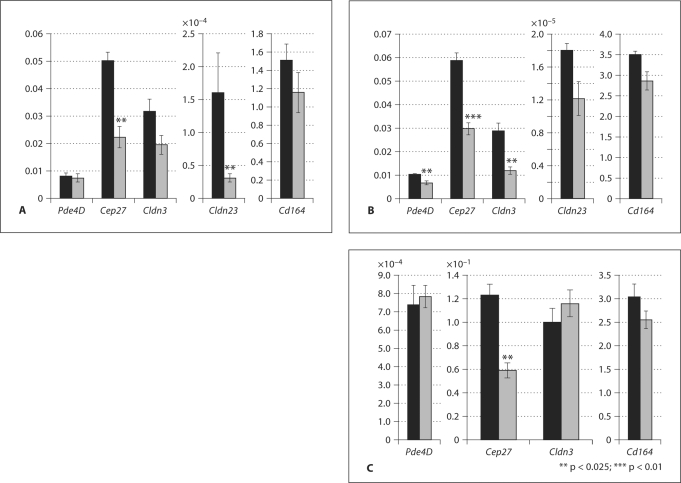
Studies of human sperm centrioles showed a greater incidence of centriolar abnormalities in nonprogressively motile spermatozoa when compared with normally motile sperm (Sathananthan, 1994). Males homozygous for a Sox8 mutation displayed an age-dependent decrease in the percentage of progressively motile sperm (O’Bryan et al., 2008). Together, these data suggest that downregulation of Cep27 in the mutant testes could be the cause of immotile sperm. Claudins are known as the major component of tight junctions in several tissues (Tsukita and Furuse, 2000; Van Itallie and Anderson, 2004). A number of claudins have been shown to be expressed in testes (Lui et al., 2003). Cldn3 was concentrated in the tight junctions near the basal lamina by P20 (Meng et al., 2005). Pde4D is expressed in pachytene spermatocytes and during the spermatid elongation phase (Salanova et al., 1999). Based on the localization and function of these repressed genes, we assayed mRNA expression by Q-RT-PCR. As expected, expression levels of all 5 genes were reduced in the mutant testes at 2 months of age (fig. 2A). The expression level of Cldn23 was reduced most in mutant testes compared to the other 4 genes. Cep27,Cldn3, and Cd164 also showed reduced expression levels in the homozygous mutant testes compared to the heterozygous testes. The change in expression level of Pde4D was relatively minor in the homozygous mutant testes at 2 months of age compared to the heterozygous testes.
Fig. 2.
Temporal pattern of expression of putative downstream genes of Sox8 in adult testes. Quantitative RT-PCR determined the relative abundance of Pde4D, Cep27, Cldn3, Cldn23, and Cd164 transcripts compared to Gapdh mRNA in heterozygous (black bars) and homozygous (gray bars) mutant testes for Sox8 at 2 months (A), 3 months (B) and 5 months (C) after birth. The data are expressed as mean ± SEM.
Age-Dependent Expression of the Downstream Target Genes for Sox8 in Mutant Testes
Previously, we have shown that spermiation failure was evident in 1-month-old homozygous mutant mice but becomes more apparent in 2- and 5-month-old homozygous mutant mice, resulting in complete sterility (O’Bryan et al., 2008). This led us to investigate the expression of these selected genes at 3 and 5 months of age. At 3 months, the expression of all 5 genes was reduced in homozygous mutant testes compared to heterozygous testes. The expression level of Cldn3 in mutant testes was greatly reduced at 3 months of age relative to the 2-month-old animals (fig. 2B). The level of Cldn23 expression was higher at 3 months relative to the level in testes of 2-month-old mutants but reduced in comparison to the level in testes of heterozygous animals. The Cep27 mRNA expression level was reduced at 3 months and 5 months of age in homozygous testes compared to heterozygous testes. In contrast, the expression levels of Pde4D, Cldn3, and Cd164 at 5 months of age were unchanged or showed only small changes in homozygous compared to heterozygous testes (fig. 2C), suggesting that these genes are required for male fertility in a particular time frame during spermatogenesis. Expression levels of Cldn23 in 5-month-old testes were too low to detect.
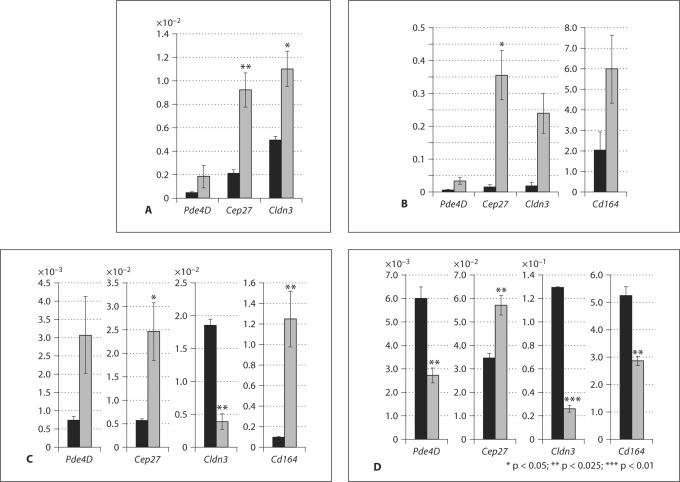
Sertoli cells proliferate during fetal life until puberty and then cease to divide. In wild type mice, Sertoli cells cease proliferation by about P15 and undergo maturation (Sharpe et al., 2003; Walker, 2003). At the start of puberty (P25) the tight junctions of the barrier are formed (Skinner and Griswold, 1980). Following the establishment of full spermatogenesis at P35 in the mouse (Kramer and Erickson, 1981), each Sertoli cell concurrently supports 4 or 5 different types of germ cells. Our previous study demonstrated that 80% of Sox8 mutant males were sterile, and ∼20% were able to produce small litters at young age (O’Bryan et al., 2008). Where specific expression changes were seen in microarray expression analysis at 2 months of age, we investigated expression at earlier time points, prior to the time that changes in spermatogenesis are observed in Sox8 mutant testes. To investigate the expression of downstream genes of Sox8, RNA wasextracted from P15, P20, P25, and P35 heterozygous and homozygous mutant testes.
In contrast to the findings from adult mice, the expression levels of Pde4D, Cep27, and Cldn3 were elevated at P15 in the Sox8 mutant testes before puberty and blood-testis-barrier formation (fig. 3A). Similar results were obtained from P20 samples for Pde4D, Cep27, Cldn3, and Cd164 (fig. 3B). The expression levels of Pde4D, Cep27, and Cd164 were increased in Sox8–/– mice at P25, at the time of blood-testis-barrier formation (fig. 3C). At this stage, Cldn3 expression was decreased in Sox8 mutant testes (fig. 3C). At the establishment of full spermatogenesis (P35) expression levels of Pde4D,Cldn3, and Cd164, but not Cep27 were decreased in the homozygous mutant testes (fig. 3D). These results suggested that the loss of Sox8 in Sertoli cells might be the cause of differential changes in expression of these genes in an age-dependent manner and in different stages of testicular maturation.
Fig. 3.
Effect of Sox8mutation on expression of selected downregulated genes before puberty (A and B, P10 and P20, respectively) at puberty (C, P25) and during the establishment of full spermatogenesis (D, P35). The relative abundance of Pde4D, Cd164, Cep27, Cldn3, and Cldn23 transcripts in heterozygous (black bars) and homozygous (gray bars) mutant testes compared to Gapdh mRNA as an external control is shown (mean ± SEM).
Sequence analysis 2.0 kb upstream of the Pde4d and Cldn23 promoter revealed the presence of a putative SOX binding site (cacacac) between −748 and −742 bp (Pde4d) as well as a SOX4 binding site (tctttctctc) between −2095 and −2086 bp and a SOX2 binding site (cacacacaca) between −940 and −931 bp (Cldn23) from the initiation codon. Quantitative mRNA expression analysis of the downregulated genes in Sox8 heterozygous and homozygous mutant testes revealed age-dependent expression changes in the homozygous mutant testes. These data strongly suggest that ablation of Sox8 results in an age-dependent and within a definite time frame progressive decrease in the expression level of the selected downregulated genes.
Discussion
In the present study we have investigated transcriptional changes in the Sox8 mutant testis during the stages immediately preceding extensive germ cell loss in the Sox8–/–mice. Expression profiling identified 108 mRNAs whose expression is significantly (>2-fold) elevated, and 46 mRNAs whose expression is significantly reduced (>2-fold) at 2 months of age, providing a suite of candidate effectors of Sox8 function for further investigation. We reported earlier that Sox8 mutant mice displayed an obvious mutant phenotype after the first wave of spermatogenesis in adult mice (35 days), and this became progressively more pronounced in 2-month- and 5-month-old Sox8 mutant animals (O’Bryan et al., 2008). Some of the genes shown in table 1 and 2 might become apparent due to the block in spermiogenesis or germ cell loss and their effect on whole tissue microarray expression data. Therefore, we chose genes for further analyses based on their expression patterns, i.e., genes specifically expressed in Sertoli cells and repressed more than 2-fold.
Among the genes altered in the Sox8–/–testes there are several proteins important for testicular development, spermatogenesis, and formation of tight junctions.
Pde4D is expressed during spermatogenesis in round spermatids and pachytene spermatocytes (Salanova et al., 1999) in addition to Sertoli cells (Levallet et al., 2007). As we reported, in Sox8 mutant testes spermatogenesis is arrested at stage IX and the formation of round spermatids to elongated spermatids failed (O’Bryan et al., 2008). In Sox8 mutant testes, expression of Pde4D measured by quantitative RT-PCR decreased from 2 to 3 months of age, but then remained unchanged at 5 months of age. This finding suggested that Pde4D might be required for spermatogenesis at a specific age. At the same time, it appears that Sox8 is required for specific timing of Pde4D expression. The decrease of the expression of Pde4D in the mutant testes may be, at least in part, explained by a reduction of elongated spermatids where Pde4D is expressed. However, presence of a SOX binding site strongly suggests that SOX8 might directly regulate the expression of Pde4D in Sertoli cells. Further investigation is required to define molecular mechanisms on how SOX8 regulates expression of Pde4D.
We have found a number of genes up- and downregulated in Sox8 mutant testes that are important for tight junctions and the blood-testis barrier, for instance Cldn3 and Cldn23 which have a reduced expression between puberty and adulthood. Formation of the blood-testis barrier starts around P11. CLDN3 is first detected at P15 in a diffusive manner and then in the blood-testis barrier at P20 in newly formed tight junctions that regulate the permeability of the barrier as germ cells move from the basal to the adluminal compartment (Meng et al., 2005). Tight junctions in other tissues are shown to regulate the selective transport of ions and nutrients from the blood stream and interstitial fluid (Goodenough, 1999). We also found that Cldn11 and Cldn12, which are crucial for testis tight junction formation (Gow et al., 1999; Bronstein et al., 2000), were upregulated 1.4-fold in the microarray analyses. Lower expression of Cldn3 and Cldn23 in the Sox8 mutant testes suggested that this might be the cause of an altered function of the blood-testis barrier and contributes to male infertility. However, we previously found no overt changes in the expression of Espin and Vinculin (markers for the blood-testis barrier) in Sox8 mutant testes relative to control testes (O’Bryan et al., 2008). It is possible that the blood-testis barrier structurally forms in the Sox8 mutant testes, but with an altered function. Thus, further investigation of the blood-testis barrier function in Sox8 mutant testes is needed.
Spermatogenesis in seminiferous tubules proceeds if an appropriate environment is provided by Sertoli cells (Clermont, 1972). This notion is supported by the study reported here, which demonstrates that the loss of Sox8 in Sertoli cells downregulates the expression of a group of genes resulting in male infertility. By using microarray analysis and quantitative RT-PCR in a complementary strategy, we have identified expression changes associated with the loss of Sox8 and consequent loss of spermatogenesis in the mutant mice. Some of these genes are expressed in Sertoli cells and may be direct or indirect targets of SOX8. Further investigation into the function of these genes will help to explain their specific roles in testicular development, male fertility, and its maintenance in an adult age.
Acknowledgements
The authors would like to dedicate this manuscript to Dr. Shun Harada, a great colleague and a wonderful friend, who made significant contributions to the field of SOX biology. We gratefully thank the NIEHS Microarrays Core Facility led by Dr. Rich Paules. We thank Dr. Leping Li for sequence analyses, Drs. Mitch Eddy, Manas Ray, and Greg Scott for helpful discussion and critical reading of the manuscript. We also thank Mss. Ijeoma Nwosu, Gloria MacDonald, and Kelly McCann for their technical support, Ms. Tonya Miller for her excellent service of maintenance of the mouse colonies. This work was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES071003-11), by the Indo-US Collaboration on Environment and Occupational Health Grant, and by a gift from RIKEN Brain Science Institute (833–3944) to Y.M.
References
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Bronstein JM, Chen K, Tiwari-Woodruff S, Kornblum HI. Developmental expression of OSP/claudin-11. J Neurosci Res. 2000;60:284–290. doi: 10.1002/(SICI)1097-4547(20000501)60:3<284::AID-JNR2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52:198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R. Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc Natl Acad Sci USA. 1994;91:3368–3372. doi: 10.1073/pnas.91.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough DA. Plugging the leaks. Proc Natl Acad Sci USA. 1999;96:319–321. doi: 10.1073/pnas.96.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, et al. CNS myelin and Sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell. 1999;99:649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- Kennedy CL, Koopman P, Mishina Y, O'Bryan MK. Sox8 and Sertoli-cell function. Ann N Y Acad Sci. 2007;1120:104–113. doi: 10.1196/annals.1411.007. [DOI] [PubMed] [Google Scholar]
- Koopman P. Sex determination: a tale of two Sox genes. Trends Genet. 2005;21:367–370. doi: 10.1016/j.tig.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Erickson RP. Developmental program of PGK-1 and PGK-2 isozymes in spermatogenic cells of the mouse: specific activities and rates of synthesis. Dev Biol. 1981;87:37–45. doi: 10.1016/0012-1606(81)90058-0. [DOI] [PubMed] [Google Scholar]
- Levallet G, Levallet J, Bouraima-Lelong H, Bonnamy PJ. Expression of the cAMP-phosphodiesterase PDE4D isoforms and age-related changes in follicle-stimulating hormone-stimulated PDE4 activities in immature rat Sertoli cells. Biol Reprod. 2007;76:794–803. doi: 10.1095/biolreprod.106.055343. [DOI] [PubMed] [Google Scholar]
- Lui WY, Cheng CY. Regulation of cell junction dynamics by cytokines in the testis: a molecular and biochemical perspective. Cytokine Growth Factor Rev. 2007;18:299–311. doi: 10.1016/j.cytogfr.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WY, Mruk DD, Lee WM, Cheng CY. Adherens junction dynamics in the testis and spermatogenesis. J Androl. 2003;24:1–14. [PubMed] [Google Scholar]
- McClive PJ, Hurley TM, Sarraj MA, van den Bergen JA, Sinclair AH. Subtractive hybridisation screen identifies sexually dimorphic gene expression in the embryonic mouse gonad. Genesis. 2003;37:84–90. doi: 10.1002/gene.10231. [DOI] [PubMed] [Google Scholar]
- Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier. Proc Natl Acad Sci USA. 2005;102:16696–16700. doi: 10.1073/pnas.0506084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryan MK, Takada S, Kennedy CL, Scott G, Harada S, et al. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol. 2008;316:359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Soder O. The Sertoli cell – a hormonal target and ‘super’ nurse for germ cells that determines testicular size. Horm Res. 2006;66:153–161. doi: 10.1159/000094142. [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME. Sex-reversing mutations affect the architecture of SRY-DNA complexes. Embo J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanova M, Chun SY, Iona S, Puri C, Stefanini M, Conti M. Type 4 cyclic adenosine monophosphate-specific phosphodiesterases are expressed in discrete subcellular compartments during rat spermiogenesis. Endocrinology. 1999;140:2297–2306. doi: 10.1210/endo.140.5.6686. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH. Functional competence of abnormal spermatozoa. Baillieres Clin Obstet Gynaecol. 1994;8:141–156. doi: 10.1016/s0950-3552(05)80029-x. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- Schepers G, Wilson M, Wilhelm D, Koopman P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem. 2003;278:28101–28108. doi: 10.1074/jbc.M304067200. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Schinke T, Haberland M, Priemel M, Schilling AF, et al. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005;168:899–910. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125:769–784. doi: 10.1530/rep.0.1250769. [DOI] [PubMed] [Google Scholar]
- Skakkebaek NE, Rajpert-De Meyts E, Main KM. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod. 2001;16:972–978. doi: 10.1093/humrep/16.5.972. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Griswold MD. Sertoli cells synthesize and secrete transferrin-like protein. J Biol Chem. 1980;255:9523–9525. [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bosl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Furuse M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann N Y Acad Sci. 2000;915:129–135. doi: 10.1111/j.1749-6632.2000.tb05235.x. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. The role of claudins in determining paracellular charge selectivity. Proc Am Thorac Soc. 2004;1:38–41. doi: 10.1513/pats.2306013. [DOI] [PubMed] [Google Scholar]
- Walker WH. Molecular mechanisms controlling Sertoli cell proliferation and differentiation. Endocrinology. 2003;144:3719–3721. doi: 10.1210/en.2003-0765. [DOI] [PubMed] [Google Scholar]